Molecular Identification and Genotyping of Cryptosporidium spp. and Blastocystis sp. in Cattle in Representative Areas of Shanxi Province, North China
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Size
- n = the required sample size;
- Pexp = expected prevalence;
- d = desired absolute precision.
2.2. Fecal Sample Collection and Preparation
2.3. DNA Extraction and PCR Amplification
2.4. Sequencing and Phylogenetic Analysis
2.5. Statistical Analysis
3. Results
3.1. The Prevalence of Cryptosporidium spp. and Blastocystis sp. in Cattle in Shanxi Province
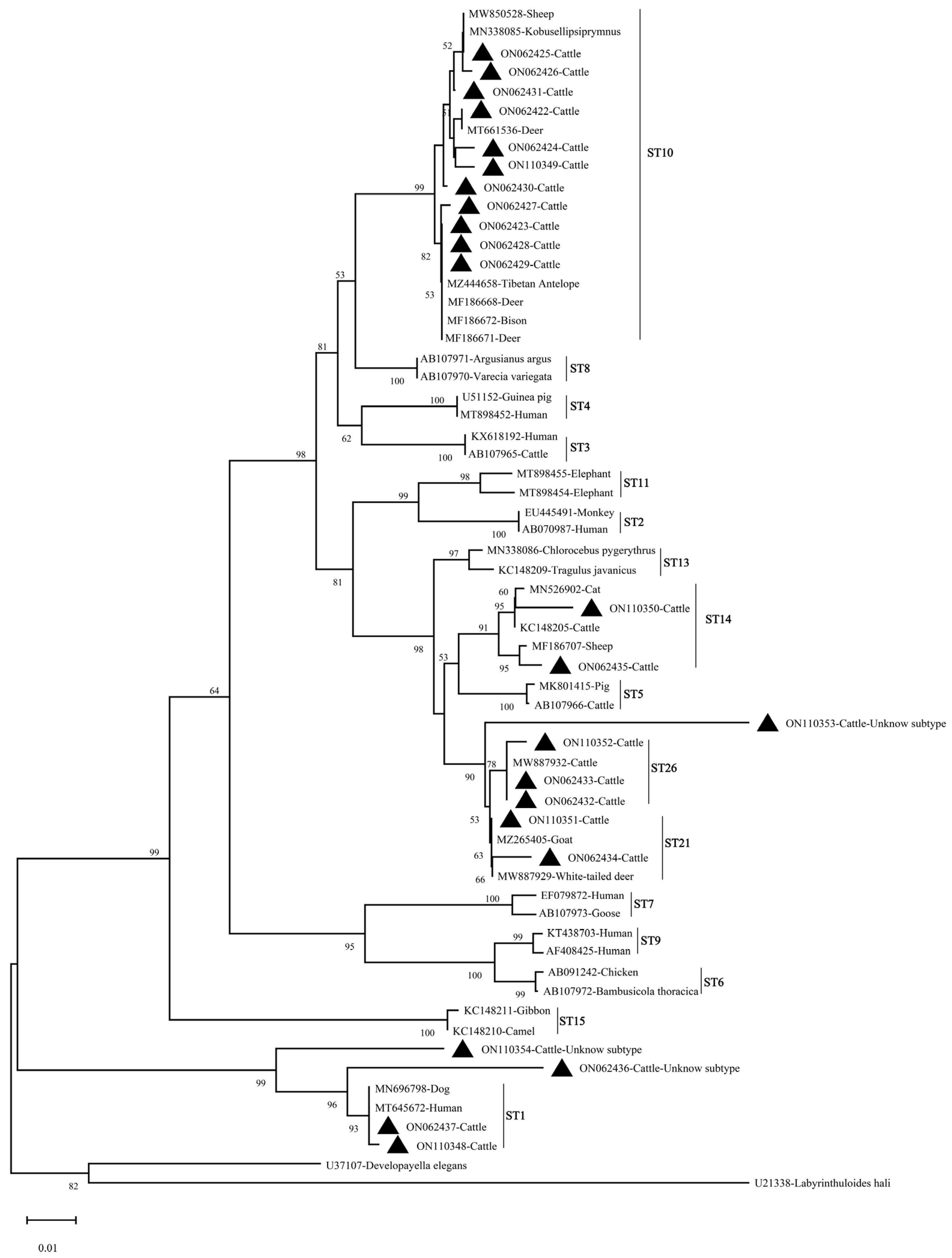
3.2. Distribution of Genetic Diversity and Phylogenetic Analysis of Cryptosporidium spp. and Blastocystis sp. in Cattle in Shanxi Province
4. Discussion
| Area | No. Positive/Total | Prevalence (%) | Gene Locus | Years | References |
|---|---|---|---|---|---|
| Anhui Province | 23/955 | 2.4 | SSU rRNA | 2018 | [6] |
| Beijing city | 21/822 | 2.6 | SSU rRNA, Gp60 | 2014–2015 | [28] |
| Gansu Province | 59/1414 | 4.2 | SSU rRNA, Gp60 | 2015 | [37] |
| Shanghai city | 303/818 | 37.0 | SSU rRNA | 2015–2016 | [38] |
| Guangdong Province | 63/1440 | 4.4 | SSU rRNA | 2016 | [39] |
| Heilongjiang Province | 27/423 | 6.4 | SSU rRNA, Gp60 | 2019 | [40] |
| Jiangxi Province | 71/556 | 12.8 | SSU rRNA | 2019 | [41] |
| Sichuan Province | 40/278 | 14.4 | SSU rRNA, Gp60 | 2016–2017 | [42] |
| Taiwan | 60/226 | 26.5 | SSU rRNA | 2017–2018 | [43] |
| Yunnan Province | 65/442 | 14.7 | SSU rRNA, Gp60 | 2019–2020 | [44] |
| Tibet | 3/442 | 0.7 | SSU rRNA | 2016 | [30] |
| Hebei Provinces and Tianjin city | 10/1040 | 1.0 | SSU rRNA, Gp60 | 2016 | [45] |
| Xinjiang Uyghur Autonomous Region | 70/1827 | 3.8 | SSU rRNA | 2013 | [46] |
| Area | No. Positive/Total | Prevalence (%) | Gene Locus | Years | References |
|---|---|---|---|---|---|
| Qinghai Province | 278/1027 | 27.1 | SSU rRNA | 2016–2017 | [47] |
| Jiangxi Province | 305/556 | 54.9 | SSU rRNA | 2019 | [33] |
| Heilongjiang Province | 14/147 | 9.5 | SSU rRNA | 2010–2016 | [48] |
| Yunnan Province | 119/987 | 12.1 | SSU rRNA | 2017–2018 | a |
| Shaanxi Province | 92/371 | 24.8 | SSU rRNA | 2018 | a |
| Anhui Province | 0/955 | 0 | SSU rRNA | 2018 | a |
| Guangdong Province | 9/479 | 1.9 | SSU rRNA | 2016 | a |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Innes, E.A.; Chalmers, R.M.; Wells, B.; Pawlowic, M.C. A one health approach to tackle cryptosporidiosis. Trends Parasitol. 2020, 36, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Relat, R.M.B.; O’connor, R.M. Cryptosporidium: Host and parasite transcriptome in infection. Curr. Opin. Microbiol. 2020, 58, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.N. Cryptosporidiosis. JAMA 2020, 323, 288. [Google Scholar] [CrossRef]
- Thompson, R.C.; Olson, M.E.; Zhu, G.; Enomoto, S.; Abrahamsen, M.S.; Hijjawi, N.S. Cryptosporidium and cryptosporidiosis. Adv. Parasitol. 2005, 59, 77–158. [Google Scholar] [PubMed]
- Ryan, U.; Zahedi, A.; Feng, Y.; Xiao, L. An update on zoonotic Cryptosporidium species and genotypes in humans. Animals 2021, 11, 3307. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tang, L.; Li, W.; Li, C.; Gu, Y. Prevalence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi from large-scale cattle farms in Anhui Province, China. J. Vet. Med. Sci. 2022, 84, 40–47. [Google Scholar] [CrossRef]
- Caccio, S.M.; Chalmers, R.M. Human cryptosporidiosis in Europe. Clin. Microbiol. Infect. 2016, 22, 471–480. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Tan, K.S.W.; Clark, C.G. Blastocystis. Trends Parasitol. 2020, 36, 315–316. [Google Scholar] [CrossRef]
- Shams, M.; Shamsi, L.; Sadrebazzaz, A.; Asghari, A.; Badali, R.; Omidian, M.; Hassanipour, S. A systematic review and meta-analysis on the global prevalence and subtypes distribution of Blastocystis sp. infection in cattle: A zoonotic concern. Comp. Immunol. Microbiol. Infect. Dis. 2021, 76, 101650. [Google Scholar] [CrossRef]
- Rauff-Adedotun, A.A.; Mohd Zain, S.N.; Farah Haziqah, M.T. Current status of Blastocystis sp. in animals from Southeast Asia: A review. Parasitol. Res. 2020, 119, 3559–3570. [Google Scholar] [CrossRef]
- Yang, X.; Li, Y.; Wang, Y.; Wang, J.; Lai, P.; Li, Y.; Song, J.; Qi, M.; Zhao, G. Molecular characterization of Blastocystis sp. in Camelus bactrianus in Northwestern China. Animals 2021, 11, 3016. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, P.A.; Jaimes, J.E.; Ramírez, J.D. A summary of Blastocystis subtypes in North and South America. Parasites Vectors 2019, 12, 376. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Qiao, H.; Wang, H.; Li, S.; Zhai, P.; Huang, J.; Guo, Y. Molecular prevalence and subtypes of Blastocystis sp. in primates in Northern China. Transbound. Emerg. Dis. 2020, 67, 2789–2796. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Yao, J.; Chen, S.; He, T.; Chai, Y.; Zhou, Z.; Shi, X.; Liu, H.; Zhong, Z.; Fu, H.; et al. First identification and molecular subtyping of Blastocystis sp. in zoo animals in Southwestern China. Parasites Vectors 2021, 14, 11. [Google Scholar] [CrossRef]
- Scanlan, P.D.; Stensvold, C.R. Blastocystis: Getting to grips with our guileful guest. Trends Parasitol. 2013, 29, 523–529. [Google Scholar] [CrossRef]
- Bahrami, F.; Babaei, E.; Badirzadeh, A.; Riabi, T.R.; Abdoli, A. Blastocystis, urticaria, and skin disorders: Review of the current evidences. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1027–1042. [Google Scholar] [CrossRef]
- Thrusfield, M. Veterinary Epidemiology; Blackwell Science Ltd.: New York, NY, USA, 1995; pp. 180–186. [Google Scholar]
- Gong, C.; Cao, X.F.; Deng, L.; Li, W.; Huang, X.M.; Lan, J.C.; Xiao, Q.C.; Zhong, Z.J.; Feng, F.; Zhang, Y.; et al. Epidemiology of Cryptosporidium infection in cattle in China: A review. Parasite 2017, 24, 1. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, B.; Li, J.; Yu, S.; Zhang, N.; Liu, S.; Zhang, Y.; Li, J.; Ma, N.; Cai, Y.; et al. Development of a quantitative real-time PCR assay for detection of Cryptosporidium spp. infection and threatening caused by Cryptosporidium parvum subtype IIdA19G1 in diarrhea calves from Northeastern China. Vector Borne Zoonotic Dis. 2021, 21, 179–190. [Google Scholar] [CrossRef]
- Ma, Y.T.; Liu, Q.; Xie, S.C.; Li, X.D.; Ma, Y.Y.; Li, T.S.; Gao, W.W.; Zhu, X.Q. Prevalence and subtypes of Blastocystis in alpacas, vicugna pacos in Shanxi Province, China. Korean J. Parasitol. 2020, 58, 181–184. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Liang, X.X.; Zou, Y.; Li, T.S.; Chen, H.; Wang, S.S.; Cao, F.Q.; Yang, J.F.; Sun, X.L.; Zhu, X.Q.; Zou, F.C. First report of the prevalence and genetic characterization of Giardia duodenalis and Cryptosporidium spp. in yunling cattle in Yunnan Province, southwestern China. Microb. Pathog. 2021, 158, 105025. [Google Scholar] [CrossRef] [PubMed]
- El-Alfy, E.S.; Nishikawa, Y. Cryptosporidium species and cryptosporidiosis in Japan: A literature review and insights into the role played by animals in its transmission. J. Vet. Med. Sci. 2020, 82, 1051–1067. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ryan, U.; Feng, Y.; Xiao, L. Emergence of zoonotic Cryptosporidium parvum in China. Trends Parasitol. 2022, 38, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Popruk, S.; Adao, D.E.V.; Rivera, W.L. Epidemiology and subtype distribution of Blastocystis in humans: A review. Infect. Genet. Evol. 2021, 95, 105085. [Google Scholar] [CrossRef]
- Rauff-Adedotun, A.A.; Meor Termizi, F.H.; Shaari, N.; Lee, I.L. The coexistence of Blastocystis spp. in humans, animals and environmental sources from 2010–2021 in Asia. Biology 2021, 10, 990. [Google Scholar] [CrossRef]
- Attah, A.O.; Sanggari, A.; Li, L.I.; Nik Him, N.; Ismail, A.H.; Meor Termizi, F.H. Blastocystis occurrence in water sources worldwide from 2005 to 2022: A review. Parasitol. Res. 2023, 122, 1–10. [Google Scholar] [CrossRef]
- Li, F.; Wang, H.; Zhang, Z.; Li, J.; Wang, C.; Zhao, J.; Hu, S.; Wang, R.; Zhang, L.; Wang, M. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Beijing, China. Vet. Parasitol. 2016, 219, 61–65. [Google Scholar] [CrossRef]
- Zahedi, A.; Ryan, U. Cryptosporidium—An update with an emphasis on foodborne and waterborne transmission. Res. Vet. Sci. 2020, 132, 500–512. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, Y.; Chang, Y.; Zhang, X.; Li, D.; Wang, L.; Zheng, S.; Wang, R.; Zhang, S.; Li, J.; et al. Genotyping and identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi from free-range Tibetan yellow cattle and cattle-yak in Tibet, China. Acta Trop. 2020, 212, 105671. [Google Scholar] [CrossRef]
- Murakoshi, F.; Xiao, L.; Matsubara, R.; Sato, R.; Kato, Y.; Sasaki, T.; Fukuda, Y.; Tada, C.; Nakai, Y. Molecular characterization of Cryptosporidium spp. in grazing beef cattle in Japan. Vet. Parasitol. 2012, 187, 123–128. [Google Scholar] [CrossRef]
- Feng, Y.; Xiao, L. Molecular epidemiology of cryptosporidiosis in China. Front. Microbiol. 2017, 8, 1701. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, P.; Zhu, X.Q.; Zou, Y.; Chen, X.Q. Prevalence and genotypes/subtypes of Enterocytozoon bieneusi and Blastocystis sp. in different breeds of cattle in Jiangxi Province, southeastern China. Infect. Genet. Evol. 2022, 98, 105216. [Google Scholar] [CrossRef]
- Deng, L.; Wojciech, L.; Gascoigne, N.R.J.; Peng, G.; Tan, K.S.W. New insights into the interactions between Blastocystis, the gut microbiota, and host immunity. PLoS Pathog. 2021, 17, e1009253. [Google Scholar] [CrossRef]
- Zhao, W.; Zhou, H.; Huang, Y.; Xu, L.; Rao, L.; Wang, S.; Wang, W.; Yi, Y.; Zhou, X.; Wu, Y.; et al. Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: Molecular detection, species/genotype identification and implications for public health. Int. J. Parasitol. Parasites Wildl. 2019, 9, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Livia, K.; Martin-Alonso, A.; Foronda, P. Diversity of Cryptosporidium spp. in wild rodents from the Canary Islands, Spain. Parasites Vectors 2020, 13, 445. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, J.; Chang, Y.; Yu, F.; Zhang, S.; Wang, R.; Zhang, L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Gansu, northwest China. Parasite 2020, 27, 62. [Google Scholar] [CrossRef]
- Cai, M.; Guo, Y.; Pan, B.; Li, N.; Wang, X.; Tang, C.; Feng, Y.; Xiao, L. Longitudinal monitoring of Cryptosporidium species in pre-weaned dairy calves on five farms in Shanghai, China. Vet. Parasitol. 2017, 241, 14–19. [Google Scholar] [CrossRef]
- Liang, N.; Wu, Y.; Sun, M.; Chang, Y.; Lin, X.; Yu, L.; Hu, S.; Zhang, X.; Zheng, S.; Cui, Z.; et al. Molecular epidemiology of Cryptosporidium spp. in dairy cattle in Guangdong Province, South China. Parasitology 2019, 146, 28–32. [Google Scholar] [CrossRef]
- Xue, N.Y.; Liu, F.; Tao, W.F.; Zhao, Q.; Qiu, H.Y.; Hu, Y.; Chen, Y.; Wei, X.Y.; Wang, W.; Gao, D.; et al. Molecular detection of Cryptosporidium spp. and Enterocytozoon bieneusi in Longjiang Wagyu cattle in Northeastern China. Microb. Pathog. 2020, 149, 104526. [Google Scholar] [CrossRef]
- Li, S.; Zou, Y.; Wang, P.; Qu, M.R.; Zheng, W.B.; Wang, P.; Chen, X.Q.; Zhu, X.Q. Prevalence and multilocus genotyping of Cryptosporidium spp. in cattle in Jiangxi Province, southeastern China. Parasitol. Res. 2021, 120, 1281–1289. [Google Scholar] [CrossRef]
- Zhong, Z.; Dan, J.; Yan, G.; Tu, R.; Tian, Y.; Cao, S.; Shen, L.; Deng, J.; Yu, S.; Geng, Y.; et al. Occurrence and genotyping of Giardia duodenalis and Cryptosporidium in pre-weaned dairy calves in central Sichuan province, China. Parasite 2018, 25, 45. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Lee, J.Y.; Liu, S.S.; Chen, C.C.; Hsu, H.Y. Cryptosporidium parvum infection and management-based risk factors of dairy calves in Taiwan. J. Vet. Med. Sci. 2021, 83, 1838–1844. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.W.; Shu, F.F.; Pu, L.H.; Zou, Y.; Yang, J.F.; Zou, F.C.; Zhu, X.Q.; Li, Z.; He, J.J. Occurrence and molecular characterization of Cryptosporidium spp. in dairy cattle and dairy buffalo in Yunnan Province, southwest China. Animals 2022, 12, 1031. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, Z.; Yan, F.; Zhang, Z.; Zhang, G.; Zhang, L.; Jian, F.; Zhang, S.; Ning, C.; Wang, R. Zoonotic and host-adapted genotypes of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in dairy cattle in Hebei and Tianjin, China. Vet. Parasitol. 2017, 248, 68–73. [Google Scholar] [CrossRef]
- Qi, M.; Wang, R.; Jing, B.; Jian, F.; Ning, C.; Zhang, L. Prevalence and multilocus genotyping of Cryptosporidium andersoni in dairy cattle and He cattle in Xinjiang, China. Infect. Genet. Evol. 2016, 44, 313–317. [Google Scholar] [CrossRef]
- Ren, M.; Song, J.K.; Yang, F.; Zou, M.; Wang, P.X.; Wang, D.; Zhang, H.J.; Zhao, G.H.; Lin, Q. First genotyping of Blastocystis in yaks from Qinghai Province, northwestern China. Parasites Vectors 2019, 12, 171. [Google Scholar] [CrossRef]
- Wang, J.; Gong, B.; Yang, F.; Zhang, W.; Zheng, Y.; Liu, A. Subtype distribution and genetic characterizations of Blastocystis in pigs, cattle, sheep and goats in northeastern China’s Heilongjiang Province. Infect. Genet. Evol. 2018, 57, 171–176. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Qin, R.L.; Mei, J.J.; Zou, Y.; Zhang, Z.H.; Zheng, W.B.; Liu, Q.; Zhu, X.Q.; Gao, W.W.; Xie, S.C. Molecular detection and genotyping of Enterocytozoon bieneusi in beef cattle in Shanxi Province, north China. Animals 2022, 12, 2961. [Google Scholar] [CrossRef]
- Villalobos-Segura, M.D.C.; Garcia-Prieto, L.; Rico-Chavez, O. Effects of latitude, host body size, and host trophic guild on patterns of diversity of helminths associated with humans, wild and domestic mammals of Mexico. Int. J. Parasitol. Parasites Wildl. 2020, 13, 221–230. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, Y.; Jing, B.; Xu, C.; Chen, Y.; Yu, F.; Wei, Z.; Zhang, Y.; Cui, Z.; Qi, M.; et al. Seasonal monitoring of Cryptosporidium species and their genetic diversity in neonatal calves on two large-scale farms in Xinjiang, China. J. Eukaryot. Microbiol. 2022, 69, e12878. [Google Scholar] [CrossRef]
- Paik, S.; Jung, B.Y.; Lee, H.; Hwang, M.H.; Han, J.E.; Rhee, M.H.; Kim, T.H.; Kwon, O.D.; Kwak, D. Molecular detection and subtyping of Blastocystis in Korean pigs. Korean J. Parasitol. 2019, 57, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, D.A.; Ola-Fadunsin, S.D.; Ruviniyia, K.; Gimba, F.I.; Chandrawathani, P.; Lim, Y.a.L.; Jesse, F.F.A.; Sharma, R.S.K. Molecular detection and epidemiological risk factors associated with Cryptosporidium infection among cattle in Peninsular Malaysia. Food Waterborne Parasitol. 2019, 14, e00035. [Google Scholar] [CrossRef] [PubMed]
- Elmahallawy, E.K.; Sadek, H.A.; Aboelsoued, D.; Aloraini, M.A.; Alkhaldi, A.a.M.; Abdel-Rahman, S.M.; Bakir, H.Y.; Arafa, M.I.; Hassan, E.A.; Elbaz, E.; et al. Parasitological, molecular, and epidemiological investigation of Cryptosporidium infection among cattle and buffalo calves from Assiut Governorate, upper Egypt: Current status and zoonotic implications. Front. Vet. Sci. 2022, 9, 899854. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Yang, W.B.; Zou, F.C.; Lin, R.Q.; Zhu, X.Q.; Hou, J.L. Molecular detection and subtype distribution of Blastocystis in farmed pigs in southern China. Microb. Pathog. 2021, 151, 104751. [Google Scholar] [CrossRef]
- Naguib, D.; El-Gohary, A.H.; Mohamed, A.A.; Roellig, D.M.; Arafat, N.; Xiao, L. Age patterns of Cryptosporidium species and Giardia duodenalis in dairy calves in Egypt. Parasitol. Int. 2018, 67, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Tao, W.; Gong, B.; Yang, H.; Li, Y.; Song, M.; Lu, Y.; Li, W. First report of Blastocystis infections in cattle in China. Vet. Parasitol. 2017, 246, 38–42. [Google Scholar] [CrossRef]
- Lee, H.; Lee, S.H.; Seo, M.G.; Kim, H.Y.; Kim, J.W.; Lee, Y.R.; Kim, J.H.; Kwon, O.D.; Kwak, D. Occurrence and genetic diversity of Blastocystis in Korean cattle. Vet. Parasitol. 2018, 258, 70–73. [Google Scholar] [CrossRef]
- Beyhan, Y.E.; Yilmaz, H.; Cengiz, Z.T.; Ekici, A. Clinical significance and prevalence of Blastocystis hominis in Van, Turkey. Saudi Med. J. 2015, 36, 1118–1121. [Google Scholar] [CrossRef]
- Wang, P.; Li, S.; Zou, Y.; Hong, Z.W.; Wang, P.; Zhu, X.Q.; Song, D.P.; Chen, X.Q. Prevalence and subtype distribution of Blastocystis sp. in diarrheic pigs in Southern China. Pathogens 2021, 10, 1189. [Google Scholar] [CrossRef]
- Li, J.; Dong, H.; Karim, M.R.; Yang, X.; Chao, L.; Liu, S.; Song, H.; Zhang, L. Molecular identification and subtyping of Blastocystis sp. in hospital patients in Central China. Eur. J. Protistol. 2021, 79, 125796. [Google Scholar] [CrossRef]
- Song, J.; Yang, X.; Ma, X.; Wu, X.; Wang, Y.; Li, Z.; Liu, G.; Zhao, G. Molecular characterization of Blastocystis sp. in Chinese bamboo rats (Rhizomys sinensis). Parasite 2021, 28, 81. [Google Scholar] [CrossRef]
- Zhao, G.H.; Ren, W.X.; Gao, M.; Bian, Q.Q.; Hu, B.; Cong, M.M.; Lin, Q.; Wang, R.J.; Qi, M.; Qi, M.Z.; et al. Genotyping Cryptosporidium andersoni in cattle in Shaanxi Province, Northwestern China. PLoS ONE 2013, 8, e60112. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Gong, B.; Liu, X.; Jiang, Y.; Cao, J.; Yao, L.; Li, H.; Liu, A.; Shen, Y. Identification of uncommon Cryptosporidium viatorum (a Novel Subtype XVcA2G1c) and Cryptosporidium andersoni as well as common Giardia duodenalis assemblages A and B in humans in Myanmar. Front. Cell Infect. Microbiol. 2020, 10, 614053. [Google Scholar] [CrossRef] [PubMed]
- Tarekegn, Z.S.; Tigabu, Y.; Dejene, H. Cryptosporidium infection in cattle and humans in Ethiopia: A systematic review and meta-analysis. Parasite Epidemiol. Control 2021, 14, e00219. [Google Scholar] [CrossRef]
- Hublin, J.S.Y.; Maloney, J.G.; Santin, M. Blastocystis in domesticated and wild mammals and birds. Res. Vet. Sci. 2021, 135, 260–282. [Google Scholar] [CrossRef] [PubMed]


| Parasite | Loci | Primer ID | Primer Sequences (5′-3′) | Annealing Temperature (°C) | Fragment Length (bp) | Reference |
|---|---|---|---|---|---|---|
| Cryptosporidium | SSU rRNA | F1 | CCCATTTCCTTCGAAACAGGA | 56 | [19] | |
| R1 | TTCTAGAGCTAATACATGCG | |||||
| F2 | AAGGAGTAAGGAACAACCTCCA | 58 | 830 | |||
| R2 | GGAAGGGTTGTATTATTAGATAAAG | |||||
| Blastocystis | SSU rRNA | F1 | ATCTGGTTGATCCTGCCAGT | 65 | 613 | [20] |
| R1 | GAGCTTTTTAACTGCAACAACG |
| Factor | Categories | No. Samples | No. of Positive | Prevalence% (95% CI) | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| Region | Shanyi County | 209 | 0 | - | - | <0.001 |
| Qi County | 313 | 4 | 1.3 (0–2.5) | 1 | ||
| Jishan County | 273 | 19 | 7.0 (3.9–10.0) | 5.8 (1.9–17.2) | ||
| Gender | Female | 525 | 15 | 2.9 (1.4–4.3) | 1 | 0.933 |
| Male | 270 | 8 | 3.0 (0.9–5.0) | 1.0 (0.4–2.5) | ||
| Age | M < 12 | 286 | 7 | 2.4 (0.7–4.2) | 1 | 0.852 |
| 12 ≤ M ≤ 18 | 195 | 6 | 3.1 (0.7–5.5) | 1.3 (0.4–3.8) | ||
| M > 18 | 314 | 10 | 3.2 (1.2–5.1) | 1.3 (0.5–3.5) | ||
| Type | Dairy cattle | 394 | 4 | 1.0 (0–2.0) | 1 | 0.002 |
| Beef cattle | 401 | 19 | 4.7 (2.7–6.8) | 4.8 (1.6–14. 4) | ||
| Total | 795 | 23 | 2.9 (1.7–4.1) |
| Factor | Categories | No. Samples | No. of Positive | Prevalence% (95% CI) | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| Region | Shanyin County | 209 | 8 | 3.8 (1.2–6.4) | 1.7 (0.6–4.9) | <0.001 |
| Qi County | 313 | 7 | 2.2 (0.6–3.9) | 1 | ||
| Jishan County | 273 | 88 | 32.2 (26.7–37.8) | 20.8 (9.4–45.9) | ||
| Gender | Female | 525 | 79 | 15.0 (12.0–18.1) | 1.8 (1.1–2.9) | 0.014 |
| Male | 270 | 24 | 8.9 (5.5–12.3) | 1 | ||
| Age | M < 12 | 286 | 34 | 11.9 (8.1–15.6) | 1 | 0.499 |
| 12 ≤ M ≤ 18 | 195 | 30 | 15.4 (10.3–20.4) | 1.3 (0.8–2.3) | ||
| M > 18 | 314 | 39 | 12.4 (8.8–16.1) | 1.1 (0.6–1.7) | ||
| Type | Beef cattle | 401 | 79 | 19.7 (15.8–23.6) | 3.8 (2.3–6.1) | <0.001 |
| Dairy cattle | 394 | 24 | 6.1 (3.7–8.5) | 1 | ||
| Total | 795 | 103 | 13.0 (10.6–15.3) |
| Categories | Gender | Age | Type | |||||
|---|---|---|---|---|---|---|---|---|
| Female | Male | M < 12 | 12 ≤ M ≤ 18 | M > 18 | Beef Cattle | Dairy Cattle | ||
| Cryptosporidium spp. | Shanyin County | 0 (0/184) | 0 (0/25) | 0 (0/95) | 0 (0/0) | 0 (0/114) | 0 (0/0) | 0 (0/209) |
| Qi County | 1.0% (2/196) | 1.7% (2/117) | 2.6% (3/116) | 1.1% (1/93) | 0 (0/104) | 0 (0/177) | 2.9% (4/136) | |
| Jishan County | 4.1% (6/145) | 10.2% (13/128) | 5.3% (4/75) | 4.9% (5/102) | 10.4% (10/96) | 8.5% (19/224) | 0 (0/49) | |
| Blastocystis sp. | Shanyin County | 4.3% (8/184) | 0 (0/25) | 8.4% (8/95) | 0 (0/0) | 0 (0/114) | 0 (0/0) | 3.8% (8/209) |
| Qi County | 3.6% (7/196) | 0 (0/117) | 2.6% (3/116) | 0 (0/93) | 3.8% (4/104) | 0 (0/177) | 5.1% (7/136) | |
| Jishan County | 44.1% (64/145) | 18.8% (24/128) | 30.7% (23/75) | 29.4% (30/102) | 36.5% (35/96) | 35.3% (79/224) | 18.4% (9/49) | |
| Parasite | Type | Subtypes | Accession Numbers |
|---|---|---|---|
| Cryptosporidium | Beef cattle | - | ON054428, ON054429, ON054430, ON054431 |
| Dairy cattle | - | ON102776, ON102777, ON102778 | |
| Blastocystis | Beef cattle | ST1 | ON062437 |
| ST10 | ON062422, ON062423, ON062424, ON062425, ON062426, ON062427, ON062428, ON062429, ON062430, ON062431 | ||
| ST14 | ON062435 | ||
| ST21 | ON062434 | ||
| ST26 | ON062432, ON062433 | ||
| Unknown subtype | ON062436 | ||
| Dairy cattle | ST1 | ON110348 | |
| ST10 | ON110349 | ||
| ST14 | ON110350 | ||
| ST21 | ON110351 | ||
| ST26 | ON110352 | ||
| Unknown subtype | ON110353 | ||
| Unknown subtype | ON110354 |
| Factors | Categories | No. Samples | No. of Positive | Subtype (n) |
|---|---|---|---|---|
| Region | Shanyi County | 209 | 8 | ST10 (7), ST26 (1) |
| Qi County | 313 | 7 | ST1 (1), ST10 (2), ST14 (2), ST21 (1), ST26 (1) | |
| Jishan County | 273 | 88 | ST1 (3), ST10 (54), ST14 (6), ST21 (3), ST26 (19), unknown subtypes (3) | |
| Type | Beef cattle | 401 | 79 | ST1 (3), ST10 (48), ST14 (6), ST21 (2), ST26 (19), unknown subtype (1) |
| Dairy cattle | 394 | 24 | ST1 (1), ST10 (15), ST14 (2), ST21 (2), ST26 (2), unknown subtypes (2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Y.; Liu, Y.-Y.; Mei, J.-J.; Zheng, W.-B.; Liu, Q.; Gao, W.-W.; Zhu, X.-Q.; Xie, S.-C. Molecular Identification and Genotyping of Cryptosporidium spp. and Blastocystis sp. in Cattle in Representative Areas of Shanxi Province, North China. Animals 2023, 13, 2929. https://doi.org/10.3390/ani13182929
Liang Y, Liu Y-Y, Mei J-J, Zheng W-B, Liu Q, Gao W-W, Zhu X-Q, Xie S-C. Molecular Identification and Genotyping of Cryptosporidium spp. and Blastocystis sp. in Cattle in Representative Areas of Shanxi Province, North China. Animals. 2023; 13(18):2929. https://doi.org/10.3390/ani13182929
Chicago/Turabian StyleLiang, Yao, Ya-Ya Liu, Jin-Jin Mei, Wen-Bin Zheng, Qing Liu, Wen-Wei Gao, Xing-Quan Zhu, and Shi-Chen Xie. 2023. "Molecular Identification and Genotyping of Cryptosporidium spp. and Blastocystis sp. in Cattle in Representative Areas of Shanxi Province, North China" Animals 13, no. 18: 2929. https://doi.org/10.3390/ani13182929
APA StyleLiang, Y., Liu, Y.-Y., Mei, J.-J., Zheng, W.-B., Liu, Q., Gao, W.-W., Zhu, X.-Q., & Xie, S.-C. (2023). Molecular Identification and Genotyping of Cryptosporidium spp. and Blastocystis sp. in Cattle in Representative Areas of Shanxi Province, North China. Animals, 13(18), 2929. https://doi.org/10.3390/ani13182929






