I Like This New Me: Unravelling Population Structure of Mediterranean Electric Rays and Taxonomic Uncertainties within Torpediniformes
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Extraction, Amplification and Sequencing
2.2. Data Analysis
2.2.1. Genetic Diversity of Sardinian Samples
2.2.2. Population Structure in the Mediterranean and Atlantic Ocean
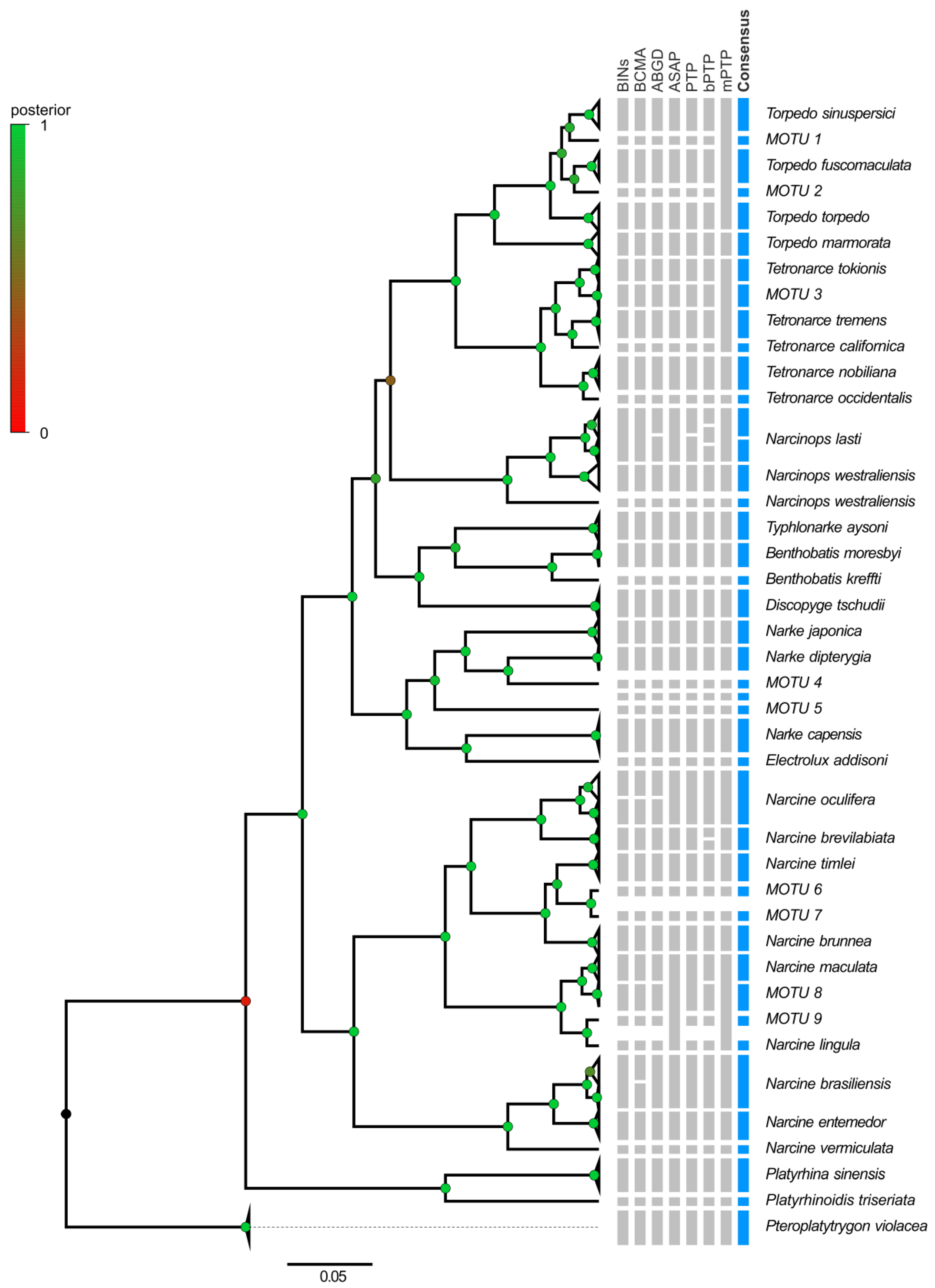
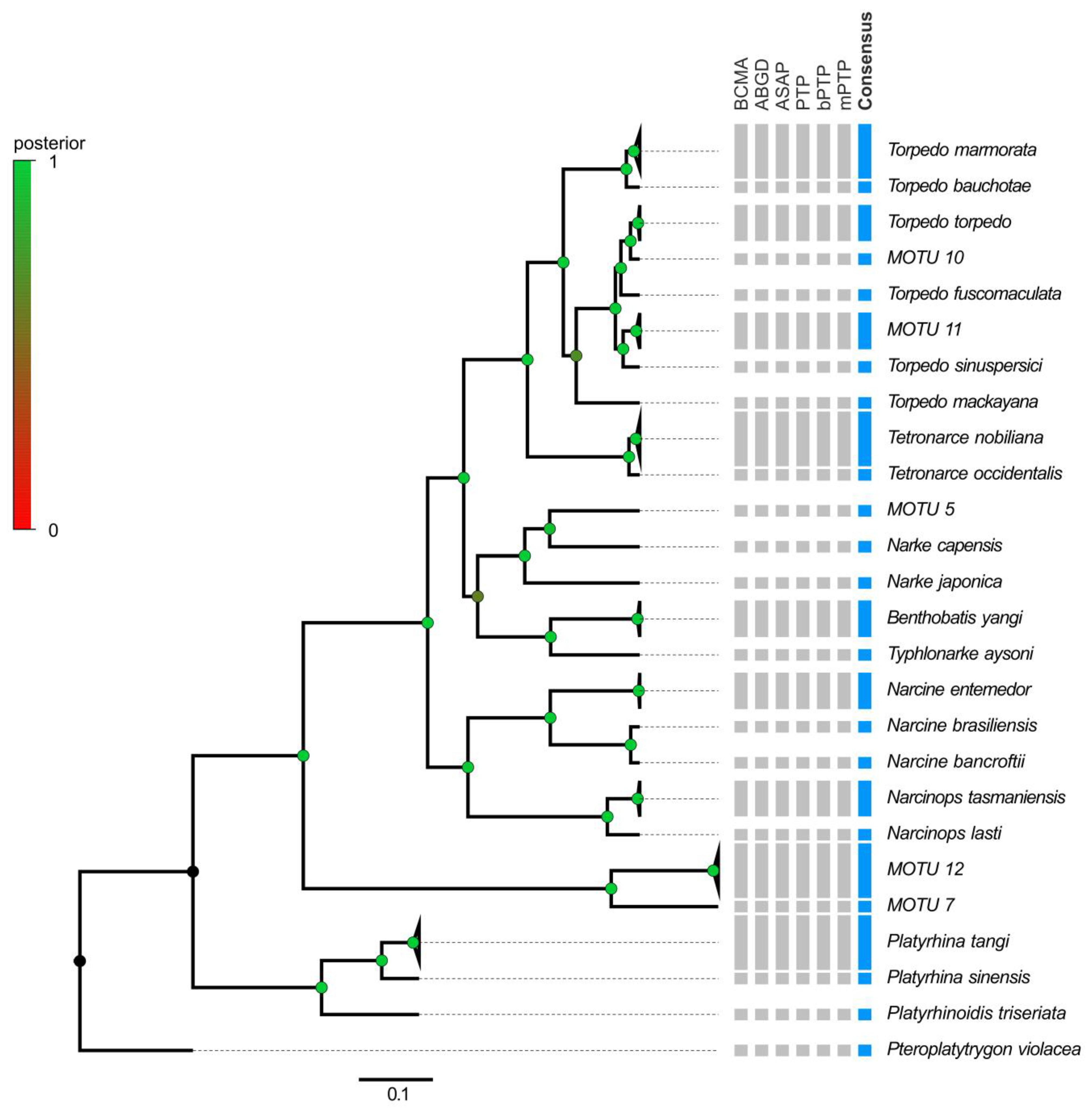
2.2.3. Species Delimitation within the Order Torpediniformes
3. Results
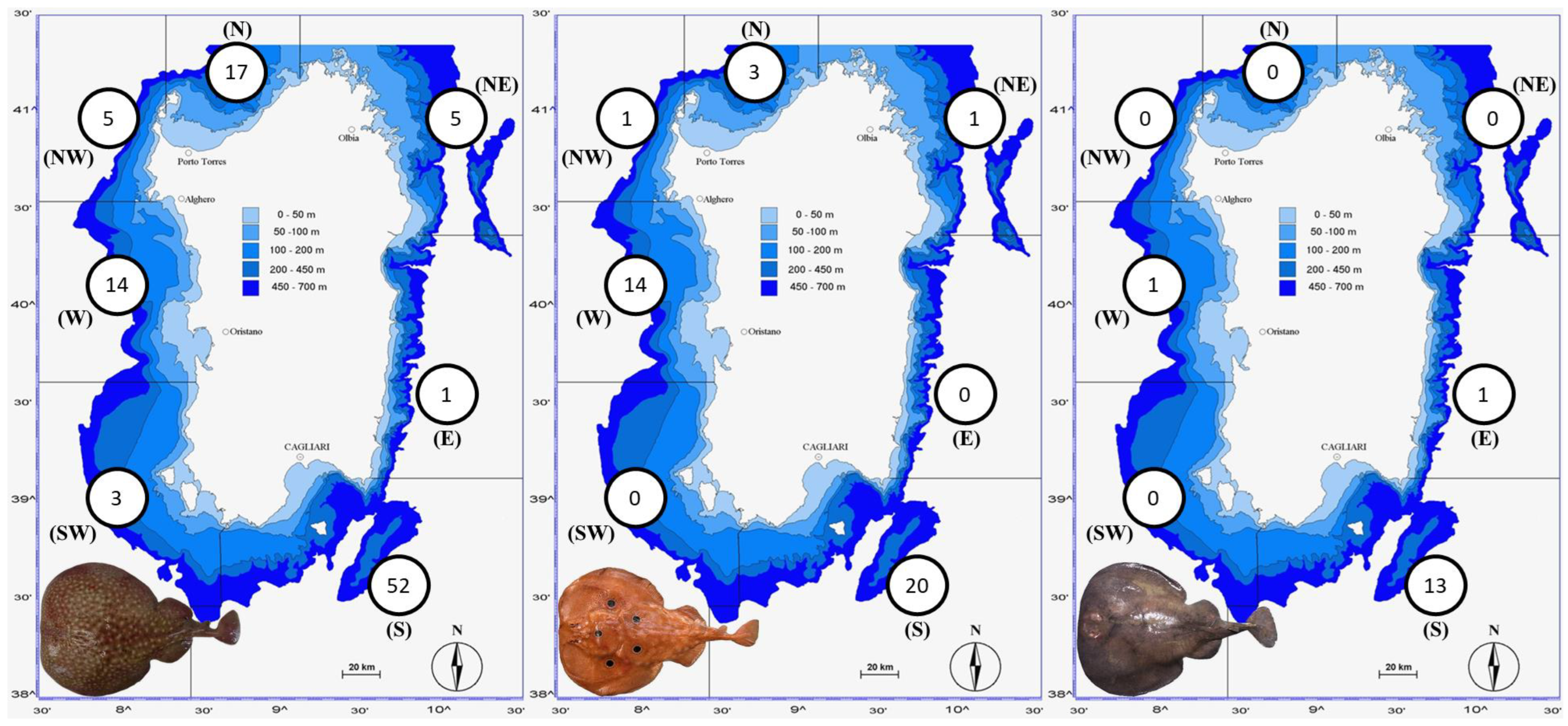
3.1. The Three Electric Rays around Sardinian Coasts
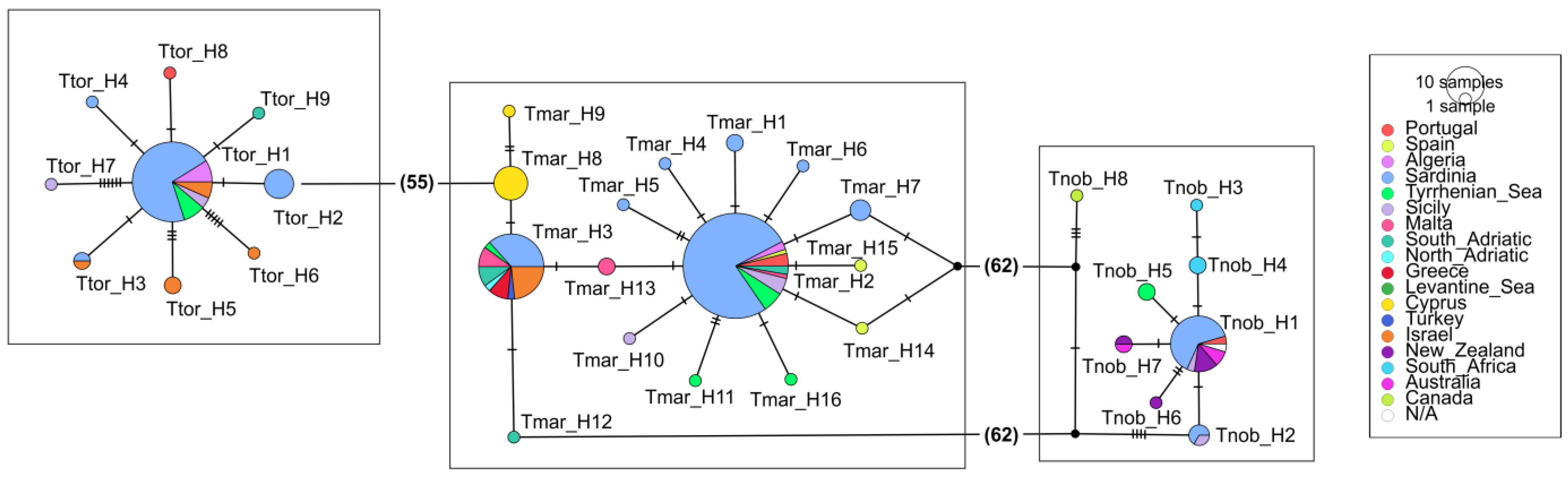
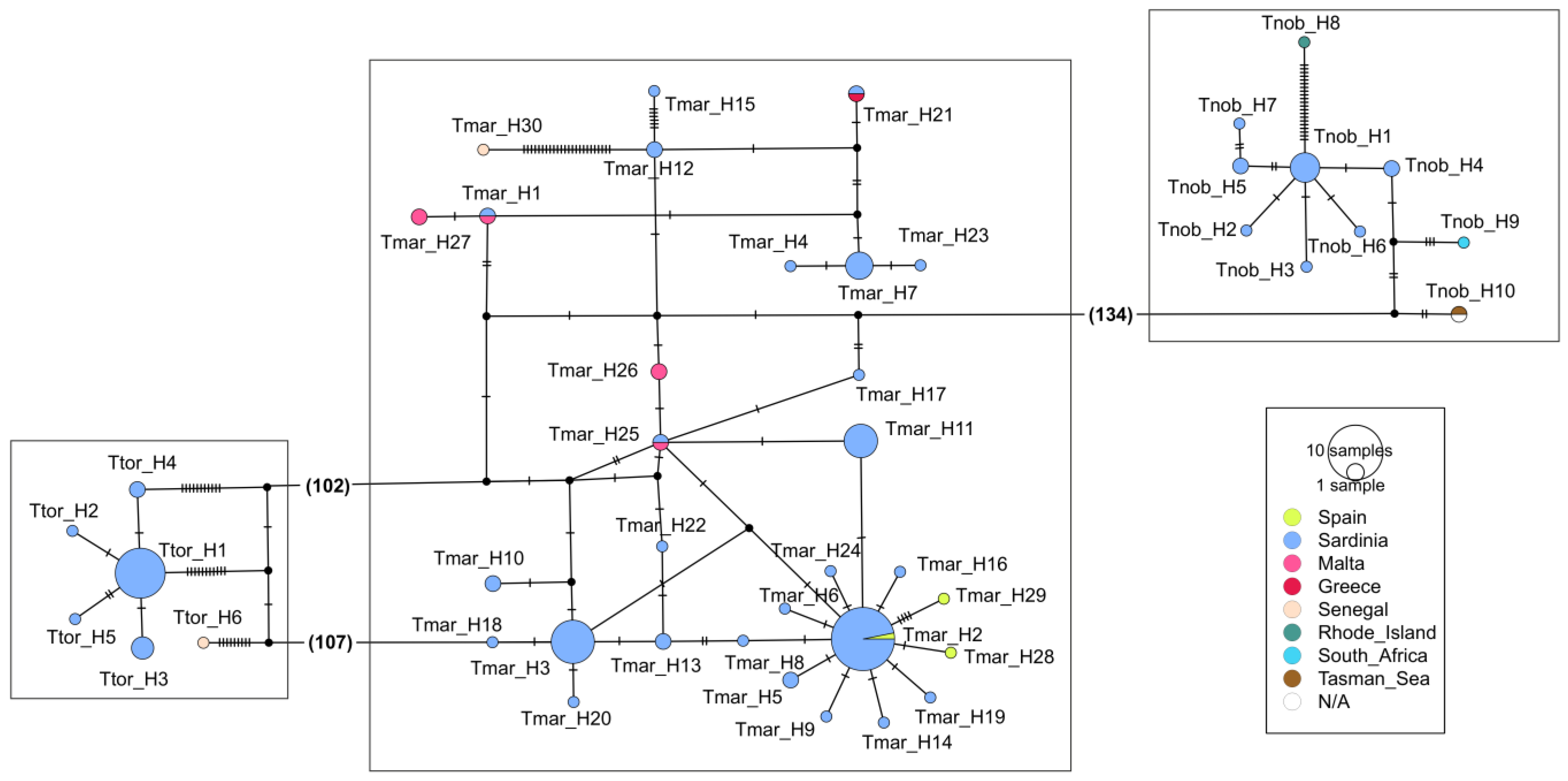
3.2. The Three Electric Rays at a Large Spatial Scale (Mediterranean and Beyond)
3.3. Electric Rays around the World
4. Discussion
4.1. The Three Electric Rays around the Sardinian Coasts
4.2. The Three Electric Rays at a Large Spatial Scale (Mediterranean and Beyond)
4.3. Electric Rays around the World
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cariani, A.; Messinetti, S.; Ferrari, A.; Arculeo, M.; Bonello, J.J.; Bonnici, L.; Cannas, R.; Carbonara, P.; Cau, A.; Charilaou, C.; et al. Improving the Conservation of Mediterranean Chondrichthyans: The ELASMOMED DNA Barcode Reference Library. PLoS ONE 2017, 12, e0170244. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.; Di Crescenzo, S.; Cariani, A.; Crobe, V.; Benvenuto, A.; Piattoni, F.; Mancusi, C.; Bonnici, L.; Bonello, J.J.; Schembri, P.J.; et al. Puzzling over spurdogs: Molecular taxonomy assessment of the Squalus species in the Strait of Sicily. Eur. Zool. J. 2021, 88, 181–190. [Google Scholar] [CrossRef]
- Carugati, L.; Melis, R.; Cariani, A.; Cau, A.; Crobe, V.; Ferrari, A.; Follesa, M.C.; Geraci, M.L.; Iglésias, S.P.; Pesci, P.; et al. Combined COI barcode-based methods to avoid mislabelling of threatened species of deep-sea skates. Anim. Conserv. 2021, 25, 38–52. [Google Scholar] [CrossRef]
- Castilho, R.; Freitas, M.; Silva, G.; Fernandez-Carvalho, J.; Coelho, R. Morphological and mitochondrial DNA divergence validates blackmouth, Galeus melastomus, and Atlantic sawtail catsharks, Galeus atlanticus, as separate species. J. Fish Biol. 2007, 70, 346–358. [Google Scholar] [CrossRef]
- Puncher, G.N.; Alemany, F.; Arrizabalaga, H.; Cariani, A.; Tinti, F. Misidentification of bluefin tuna larvae: A call for caution and taxonomic reform. Rev. Fish Biol. Fish. 2015, 25, 485–502. [Google Scholar] [CrossRef]
- Bellodi, A.; Benvenuto, A.; Melis, R.; Mulas, A.; Barone, M.; Barría, C.; Cariani, A.; Carugati, L.; Chatzispyrou, A.; Desrochers, M.; et al. Call me by my name: Unravelling the taxonomy of the gulper shark genus Centrophorus in the Mediterranean Sea through an integrated taxonomic approach. Zool. J. Linn. Soc. 2022, 195, 815–840. [Google Scholar] [CrossRef]
- Ferrari, A.; Crobe, V.; Cannas, R.; Leslie, R.W.; Serena, F.; Stagioni, M.; Costa, F.O.; Golani, D.; Hemida, F.; Zaera-Perez, D.; et al. To Be, or Not to Be: That Is the Hamletic Question of Cryptic Evolution in the Eastern Atlantic and Mediterranean Raja miraletus Species Complex. Animals 2023, 13, 2139. [Google Scholar] [CrossRef]
- Daly-Engel, T.S.; Baremore, I.E.; Grubbs, R.D.; Gulak, S.J.B.; Graham, R.T.; Enzenauer, M.P. Resurrection of the sixgill shark Hexanchus vitulus Springer & Waller, 1969 (Hexanchiformes, Hexanchidae), with comments on its distribution in the northwest Atlantic Ocean. Mar. Biodivers. 2019, 49, 759–768. [Google Scholar] [CrossRef]
- Daly-Engel, T.S.; Koch, A.; Anderson, J.M.; Cotton, C.F.; Grubbs, R.D. Description of a new deep-water dogfish shark from Hawaii, with comments on the Squalus mitsukurii species complex in the West Pacific. ZooKeys 2018, 798, 135–157. [Google Scholar] [CrossRef]
- Whitaker, J.M.; Brower, A.L.; Janosik, A.M. Invasive lionfish detected in estuaries in the northern Gulf of Mexico using environmental DNA. Environ. Biol. Fishes 2021, 104, 1475–1485. [Google Scholar] [CrossRef]
- Nester, G.M.; De Brauwer, M.; Koziol, A.; West, K.M.; DiBattista, J.D.; White, N.E.; Power, M.; Heydenrych, M.J.; Harvey, E.; Bunce, M. Development and evaluation of fish eDNA metabarcoding assays facilitate the detection of cryptic seahorse taxa (family: Syngnathidae). Environ. DNA 2020, 2, 614–626. [Google Scholar] [CrossRef]
- Bellodi, A.; Mulas, A.; Carbonara, P.; Cau, A.; Cuccu, D.; Marongiu, M.F.; Mura, V.; Pesci, P.; Zupa, W.; Porcu, C.; et al. New insights into life-history traits of Mediterranean Electric rays (Torpediniformes: Torpedinidae) as a contribution to their conservation. Zoology 2021, 146, 125922. [Google Scholar] [CrossRef] [PubMed]
- Chatzispyrou, A.; Gubili, C.; Touloumis, K.; Karampetsis, D.; Kioulouris, S.; Anastasopoulou, A.; Christidis, A.; Peristeraki, P.; Batjakas, I.E.; Koutsikopoulos, C. Life-history traits of the marbled electric ray, Torpedo marmorata Risso, 1810, from the Greek Seas, north-eastern Mediterranean Sea. Environ. Biol. Fishes 2021, 104, 1575–1592. [Google Scholar] [CrossRef]
- Naylor, G.J.P.; Yang, L.; Corrigan, S.; de Carvalho, M.R. Phylogeny and classification of rays. In Rays of the Word; Last, P.R., White, T.W., de Carvalho, M.R., Séret, B., Stehmann, M.F.W., Naylor, G.J.P., Eds.; CISRO: Canberra, Australia, 2016; pp. 10–15. [Google Scholar]
- Naylor, G.J.P.; Ryburn, J.A.; Fedrigo, O.; Lopez, A. Phylogenetic Relationships among the Major Lineages of Modern Elasmobranchs. Reprod. Biol. Phylogeny Chondrichthyes 2005, 3, 25. [Google Scholar]
- Naylor, G.J.P.; Caira, J.N.; Jensen, K.; Rosana, K.A.M.; White, W.T.; Last, P.R. A DNA Sequence–Based Approach To the Identification of Shark and Ray Species and Its Implications for Global Elasmobranch Diversity and Parasitology. Bull. Am. Mus. Nat. Hist. 2012, 367, 1–262. [Google Scholar] [CrossRef]
- Moreira, R.A.; Carvalho, M.R. Phylogenetic significance of clasper morphology of electric rays (Chondrichthyes: Batoidea: Torpediniformes). J. Morphol. 2021, 282, 438–448. [Google Scholar] [CrossRef]
- Villalobos-Segura, E.; Marramà, G.; Carnevale, G.; Claeson, K.M.; Underwood, C.J.; Naylor, G.J.P.; Kriwet, J. The Phylogeny of Rays and Skates (Chondrichthyes: Elasmobranchii) Based on Morphological Characters Revisited. Diversity 2022, 14, 456. [Google Scholar] [CrossRef]
- Last, P.R.; White, W.T.; de Carvalho, M.R.; Séret, B.; Stehmann, M.F.W.; Naylor, G.J.P. Rays of the World; Cornell University Press: Ithaca, NY, USA, 2016. [Google Scholar]
- Ebert, D.A.; Dando, M. Field Guide to Sharks, Rays, and Chimaeras of Europe and the Mediterranean; Princeton University Press: Princeton, NJ, USA, 2020. [Google Scholar]
- Fricke, R.; Eschmeyer, W.N.; Van der Laan, R. Eschmeyer’s Catalog of Fishes: Genera, Species, References. Electronic Version. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 17 July 2023).
- Burgos-Vázquez, M.I.; Cruz-Escalona, V.H.; González-Acosta, A.F. Biological Notes and Distribution of Tetronarce californica (Ayres 1855) (Elasmobranchii: Torpediniformes: Torpedinidae) from the Gulf of California, Mexico. Thalass. Int. J. Mar. Sci. 2019, 35, 369–374. [Google Scholar] [CrossRef]
- Psomadakis, P.N.; Giustino, S.; Vacchi, M. Mediterranean fish biodiversity: An updated inventory with focus on the Ligurian and Tyrrhenian seas. Zootaxa 2012, 3263, 1–46. [Google Scholar] [CrossRef]
- Akel, E.H.; Karachle, P. The marine ichthyofauna of Egypt. Egyptian. J. Aquat. Biol. Fish. 2017, 21, 81–116. [Google Scholar] [CrossRef]
- Mazhar, F.M.M. The elasmobranchs of the Mediterranean. 6. Three torpedos. Bull. Inst. Oceanogr. Fish. 1987, 8, 109–135. [Google Scholar]
- Saad, A.; Séret, B.; Ali, M. Annotated list of Chondrichthyans Diversity of Mediterranean and Black Sea Chondrichthyes from the coast of Syria. Rapp. Comm. Int. L’exploration Sci. Méditerranée 2005, 37, 430. [Google Scholar]
- Giovos, I.; Aga-Spyridopoulou, R.N.; Serena, F.; Soldo, A.; Barash, A.; Doumpas, N.; Gkafas, G.A.; Katsada, D.; Katselis, G.; Kleitou, P.; et al. An Updated Greek National Checklist of Chondrichthyans. Fishes 2022, 7, 199. [Google Scholar] [CrossRef]
- Mutlu, E.; Deval, M.C.; de Meo, I.; Saygu, İ.; Miglietta, C. Spatiotemporal Density and Ecology of Batoids (Elasmobranchii) Along a Turkish Shelf-upper Slope of the Mediterranean Sea During Years 2009–2015. Thalass. Int. J. Mar. Sci. 2022, 38, 57–69. [Google Scholar] [CrossRef]
- Soldo, A.; Lipej, L. An Annotated Checklist and the Conservation Status of Chondrichthyans in the Adriatic. Fishes 2022, 7, 245. [Google Scholar] [CrossRef]
- Leonetti, F.L.; Giglio, G.; Leone, A.; Coppola, F.; Romano, C.; Bottaro, M.; Reinero, F.R.; Micarelli, C.; Tripepi, S.; Sperone, E. An updated checklist of chondrichthyans of Calabria (Central Mediterranean, southern Italy), with emphasis on rare species. Mediterr. Mar. Sci. 2020, 21, 794–807. [Google Scholar] [CrossRef]
- Olfa, E.; Mnasri, N.; Jamila, B.S.; Boumaiza, M.; Mourad, M.; Amor, M.M.; Capapé, C. Inventory of elasmobranch species caught in the Lagoon of Bizerte (North-eastern Tunisia, central Mediterranean). Pan-Am. J. Aquat. Sci. 2009, 4, 383–412. [Google Scholar]
- Serena, F.; Abella, A.J.; Bargnesi, F.; Barone, M.; Colloca, F.; Ferretti, F.; Fiorentino, F.; Jenrette, J.; Moro, S. Species diversity, taxonomy and distribution of Chondrichthyes in the Mediterranean and Black Sea. Eur. Zool. J. 2020, 87, 497–536. [Google Scholar] [CrossRef]
- Geraci, M.L.; Ragonese, S.; Scannella, D.; Falsone, F.; Gancitano, V.; Mifsud, J.; Gambin, M.; Said, A.; Vitale, S. Batoid Abundances, Spatial Distribution, and Life History Traits in the Strait of Sicily (Central Mediterranean Sea): Bridging a Knowledge Gap through Three Decades of Survey. Animals 2021, 11, 2189. [Google Scholar] [CrossRef]
- Follesa, M.C.; Marongiu, M.F.; Zupa, W.; Bellodi, A.; Cau, A.; Cannas, R.; Colloca, F.; Djurovic, M.; Isajlovic, I.; Jadaud, A.; et al. Spatial variability of Chondrichthyes in the northern Mediterranean. Sci. Mar. 2019, 83, 80. [Google Scholar] [CrossRef]
- Vella, A.; Vella, N.; Schembri, S. A molecular approach towards taxonomic identification of elasmobranch species from Maltese fisheries landings. Mar. Genom. 2017, 36, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Spedicato, M.T.; Massutí, E.; Mérigot, B.; Tserpes, G.; Jadaud, A.; Relini, G. The MEDITS trawl survey specifications in an ecosystem approach to fishery management. Sci. Mar. 2020, 83, 9. [Google Scholar] [CrossRef]
- Milisenda, G.; Vitale, S.; Massi, D.; Enea, M.; Gancitano, V.; Giusto, G.B.; Badalucco, C.; Gristina, M.; Garofalo, G.; Fiorentino, F. Discard composition associated with the deep water rose shrimp fisheries (Parapenaeus longirostris, Lucas 1846) in the south-central Mediterranean Sea. Mediterr. Mar. Sci. 2017, 18, 53. [Google Scholar] [CrossRef]
- Serena, F. Field Identification Guide to the Sharks and Rays of the Mediterranean and Black Sea; Food and Agriculture Organization of the United Nations: Rome, Italy, 2005. [Google Scholar]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D.N. DNA barcoding Australia’s fish species. Phil. Trans. R. Soc. B 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Song, H.; Buhay, J.E.; Whiting, M.F.; Crandall, K.A. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl. Acad. Sci. USA 2008, 105, 13486–13491. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Federhen, S. The NCBI Taxonomy database. Nucleic Acids Res. 2012, 40, D136–D143. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 2005, 1, 117693430500100. [Google Scholar] [CrossRef]
- Clement, M.; Snell, Q.; Walke, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. In Proceedings of the 16th International Parallel and Distributed Processing Symposium, IPDPS 2002, Fort Lauderdale, FL, USA, 15–19 April 2002; p. 7. [Google Scholar]
- Leigh, J.W.; Bryant, D. popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Cheng, L.; Connor, T.R.; Siren, J.; Aanensen, D.M.; Corander, J. Hierarchical and Spatially Explicit Clustering of DNA Sequences with BAPS Software. Mol. Biol. Evol. 2013, 30, 1224–1228. [Google Scholar] [CrossRef]
- Francis, R.M. POPHELPER: An R package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef]
- Collins, R.A.; Trauzzi, G.; Maltby, K.M.; Gibson, T.I.; Ratcliffe, F.C.; Hallam, J.; Rainbird, S.; Maclaine, J.; Henderson, P.A.; Sims, D.W.; et al. Meta-Fish-Lib: A generalised, dynamic DNA reference library pipeline for metabarcoding of fishes. J. Fish Biol. 2021, 99, 1446–1454. [Google Scholar] [CrossRef]
- R_Core_Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Ushey, K. renv: Project Environments. 2021. Available online: https://rstudio.github.io/renv/ (accessed on 5 May 2023).
- Gernhard, T. The conditioned reconstructed process. J. Theor. Biol. 2008, 253, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Kapli, P.; Lutteropp, S.; Zhang, J.; Kobert, K.; Pavlidis, P.; Stamatakis, A.; Flouri, T. Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 2017, 33, 1630–1638. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Meier, R.; Shiyang, K.; Vaidya, G.; Ng, P.K.L. DNA Barcoding and Taxonomy in Diptera: A Tale of High Intraspecific Variability and Low Identification Success. Syst. Biol. 2006, 55, 715–728. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation: ABGD, AUTOMATIC BARCODE GAP DISCOVERY. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.D.J.; Collins, R.A.; Boyer, S.; Lefort, M.C.; Malumbres-Olarte, J.; Vink, C.J.; Cruickshank, R.H. Spider: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Mol. Ecol. Resour. 2012, 12, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Srivathsan, A.; Meier, R. On the inappropriate use of Kimura-2-parameter (K2P) divergences in the DNA-barcoding literature. Cladistics 2012, 28, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Kousteni, V.; Mazzoleni, S.; Vasileiadou, K.; Rovatsos, M. Complete Mitochondrial DNA Genome of Nine Species of Sharks and Rays and Their Phylogenetic Placement among Modern Elasmobranchs. Genes 2021, 12, 324. [Google Scholar] [CrossRef]
- Giovos, I.; Aga Spyridopoulou, R.N.; Doumpas, N.; Glaus, K.; Kleitou, P.; Kazlari, Z.; Katsada, D.; Loukovitis, D.; Mantzouni, I.; Papapetrou, M.; et al. Approaching the “real” state of elasmobranch fisheries and trade: A case study from the Mediterranean. Ocean Coast. Manag. 2021, 211, 105743. [Google Scholar] [CrossRef]
- Giovos, I.; Arculeo, M.; Doumpas, N.; Katsada, D.; Maximiadi, M.; Mitsou, Ε.; Paravas, V.; Naasan Aga-Spyridopoulou, R.; Stoilas, V.-O.; Tiralongo, F.; et al. Assessing multiple sources of data to detect illegal fishing, trade and mislabelling of elasmobranchs in Greek markets. Mar. Policy 2020, 112, 103730. [Google Scholar] [CrossRef]
- Ramírez-Amaro, S.; Ordines, F.; Picornell, A.; Castro, J.A.; Ramon, C.; Massutí, E.; Terrasa, B. The evolutionary history of Mediterranean Batoidea (Chondrichthyes: Neoselachii). Zool. Scr. 2018, 47, 686–698. [Google Scholar] [CrossRef]
- Allendorf, F.; Ryman, N.; Utter, F. Genetics and fishery management: Past, present, and future. In Population Genetics and Fishery Management; University of Washington Press: Seattle, WA, USA, 1987; pp. 1–20. [Google Scholar]
- Martinez, A.S.; Willoughby, J.R.; Christie, M.R. Genetic diversity in fishes is influenced by habitat type and life-history variation. Ecol. Evol. 2018, 8, 12022–12031. [Google Scholar] [CrossRef]
- Frodella, N.; Cannas, R.; Velonà, A.; Carbonara, P.; Farrell, E.D.; Fiorentino, F.; Follesa, M.C.; Garofalo, G.; Hemida, F.; Mancusi, C.; et al. Population connectivity and phylogeography of the Mediterranean endemic skate Raja polystigma and evidence of its hybridization with the parapatric sibling R. montagui. Mar. Ecol. Prog. Ser. 2016, 554, 99–113. [Google Scholar] [CrossRef]
- Melis, R.; Vacca, L.; Bellodi, A.; Cau, A.; Porcu, C.; Follesa, M.C.; Cannas, R. Insights into population genetics, connectivity and demographic history of the longnosed skate Dipturus oxyrinchus (Linnaeus, 1758) in the western Mediterranean Sea. Aquat. Conserv. Mar. Freshw. Ecosyst. 2019, 30, 86–103. [Google Scholar] [CrossRef]
- Cannas, R.; Sacco, F.; Cau, A.; Cuccu, D.; Follesa, M.C.; Cau, A. Genetic monitoring of deep-water exploited banks of the precious Sardinia coral Corallium rubrum (L., 1758): Useful data for a sustainable management. Aquat. Conserv. Mar. Freshw. Ecosyst. 2016, 26, 236–250. [Google Scholar] [CrossRef]
- Pascual, M.; Rives, B.; Schunter, C.; Macpherson, E. Impact of life history traits on gene flow: A multispecies systematic review across oceanographic barriers in the Mediterranean Sea. PLoS ONE 2017, 12, e0176419. [Google Scholar] [CrossRef]
- Le Port, A.; Lavery, S. Population structure and phylogeography of the short-tailed stingray, Dasyatis brevicaudata (Hutton 1875), in the southern hemisphere. J. Hered. 2012, 103, 174–185. [Google Scholar] [CrossRef]
- Melis, R.; Vacca, L.; Cariani, A.; Carugati, L.; Cau, A.; Charilaou, C.; Di Crescenzo, S.; Ferrari, A.; Follesa, M.C.; Hemida, F.; et al. Commercial sharks under scrutiny: Baseline genetic distinctiveness supports structured populations of small-spotted catsharks in the Mediterranean Sea. Front. Mar. Sci. 2023, 10, 1050055. [Google Scholar] [CrossRef]
- Di Crescenzo, S.; Ferrari, A.; Barría, C.; Cannas, R.; Cariani, A.; Drewery, J.; Fernández-Peralta, L.; Giordano, D.; Hidalgo, M.; Kousteni, V.; et al. First evidence of population genetic structure of the deep-water blackmouth catshark Galeus melastomus Rafinesque, 1810. Front. Mar. Sci. 2022, 9, 953895. [Google Scholar] [CrossRef]
- Ferrari, A.; Tinti, F.; Bertucci Maresca, V.; Velonà, A.; Cannas, R.; Thasitis, I.; Costa, F.O.; Follesa, M.C.; Golani, D.; Hemida, F.; et al. Natural history and molecular evolution of demersal Mediterranean sharks and skates inferred by comparative phylogeographic and demographic analyses. PeerJ 2018, 6, e5560. [Google Scholar] [CrossRef]
- Catalano, G.; Crobe, V.; Ferrari, A.; Baino, R.; Massi, D.; Titone, A.; Mancusi, C.; Serena, F.; Cannas, R.; Carugati, L.; et al. Strongly structured populations and reproductive habitat fragmentation increase the vulnerability of the Mediterranean starry ray Raja asterias (Elasmobranchii, Rajidae). Aquat. Conserv. Mar. Freshw. Ecosyst. 2022, 32, 66–84. [Google Scholar] [CrossRef]
- Johri, S.; Doane, M.; Allen, L.; Dinsdale, E. Taking Advantage of the Genomics Revolution for Monitoring and Conservation of Chondrichthyan Populations. Diversity 2019, 11, 49. [Google Scholar] [CrossRef]
- Ariza, A.A.; Adachi, A.M.C.L.; Roque, P.; Hazin, F.H.V.; Vianna, M.; Rotundo, M.M.; Delpiani, S.M.; de Astarloa, J.M.D.; Delpiani, G.; Oliveira, C.; et al. DNA Barcoding and Species Delimitation for Dogfish Sharks Belonging to the Squalus Genus (Squaliformes: Squalidae). Diversity 2022, 14, 544. [Google Scholar] [CrossRef]
- van Staden, M.; Ebert, D.A.; Gennari, E.; Leslie, R.W.; McCord, M.E.; Parkinson, M.; Watson, R.G.A.; Wintner, S.; da Silva, C.; Bester-van der Merwe, A.E. Molecular Taxonomy of South Africa’s Catsharks: How Far Have We Come? Diversity 2023, 15, 828. [Google Scholar] [CrossRef]
- Naylor, G.J.; Caira, J.N.; Jensen, K.; Rosana, K.A.M.; Straube, N.; Lakner, C. Elasmobranch phylogeny: A mitochondrial estimate based on 595 species. In Biology of Sharks and Their Relatives; Evans, D.H., Ed.; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Ruiz-Escobar, F.; Oceguera-Figueroa, A.; Islas-Villanueva, V. Northernmost record of the Chilean torpedo Tetronarce tremens from tropical coastal waters of the Mexican Pacific. J. Mar. Biol. Assoc. UK 2023, 103, e6. [Google Scholar] [CrossRef]
- Fernando, D.; Bown, R.M.K.; Tanna, A.; Gobiraj, R.; Ralicki, H.; Jockusch, E.L.; Ebert, D.A.; Jensen, K.; Caira, J.N. New insights into the identities of the elasmobranch fauna of Sri Lanka. Zootaxa 2019, 4585, 201–238. [Google Scholar] [CrossRef] [PubMed]
- Hubert, N.; Meyer, C.P.; Bruggemann, H.J.; Guerin, F.; Komeno, R.J.; Espiau, B.; Causse, R.; Williams, J.T.; Planes, S. Cryptic diversity in Indo-Pacific coral-reef fishes revealed by DNA-barcoding provides new support to the centre-of-overlap hypothesis. PLoS ONE 2012, 7, e28987. [Google Scholar] [CrossRef]
- Bineesh, K.K.; Gopalakrishnan, A.; Akhilesh, K.V.; Sajeela, K.A.; Abdussamad, E.M.; Pillai, N.G.K.; Basheer, V.S.; Jena, J.K.; Ward, R.D. DNA barcoding reveals species composition of sharks and rays in the Indian commercial fishery. Mitochondrial DNA Part A 2017, 28, 458–472. [Google Scholar] [CrossRef]
- Weigmann, S. Annotated checklist of the living sharks, batoids and chimaeras (Chondrichthyes) of the world, with a focus on biogeographical diversity. J. Fish Biol. 2016, 88, 837–1037. [Google Scholar] [CrossRef] [PubMed]
- Sujatha, K.; Iswarya Deepti, V.A.; Ravali, V.; Jha, S. Narcine atzi Carvalho & Randall, 2003 (Pisces: Narcinidae)—An addition to Indian ichthyofauna. Indian J. Fish. 2016, 63. [Google Scholar] [CrossRef]
- Hossain, N.; Haque, A.B. Distribution range extension and first record of tonkin numbfish Narcine prodorsalis (Bessednov 1966) (Torpediniformes: Narcinidae) an electric ray in Bangladesh waters. Bangladesh J. Zool. 2018, 46, 147–154. [Google Scholar] [CrossRef]
- de Carvalho, M.R.; White, W.T. Narcine baliensis, a new species of electric ray from southeast Asia (Chondrichthyes: Torpediniformes). Zootaxa 2016, 4127, 149–160. [Google Scholar] [CrossRef]
- de Carvalho, M.; Randall, J. Numbfishes from the Arabian Sea and surrounding gulfs, with the description of a new species from Oman (Chondrichthyes: Torpediniformes: Narcinidae). Ichthyol. Res. 2003, 50, 0059–0066. [Google Scholar] [CrossRef]
- Reich, D.; Thangaraj, K.; Patterson, N.; Price, A.L.; Singh, L. Reconstructing Indian population history. Nature 2009, 461, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Willing, E.-M.; Dreyer, C.; Van Oosterhout, C. Estimates of Genetic Differentiation Measured by FST Do Not Necessarily Require Large Sample Sizes When Using Many SNP Markers. PLoS ONE 2012, 7, e42649. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, S.J.; Micheli, F.; White, T.D.; Van Houtan, K.S.; Alfaro-Shigueto, J.; Andrzejaczek, S.; Arnoldi, N.S.; Baum, J.K.; Block, B.; Britten, G.L.; et al. Emergent research and priorities for shark and ray conservation. Endanger. Species Res. 2022, 47, 171–203. [Google Scholar] [CrossRef]
- Crobe, V.; Ferrari, A.; Hanner, R.; Leslie, R.W.; Steinke, D.; Tinti, F.; Cariani, A. Molecular Taxonomy and Diversification of Atlantic Skates (Chondrichthyes, Rajiformes): Adding More Pieces to the Puzzle of Their Evolutionary History. Life 2021, 11, 596. [Google Scholar] [CrossRef]
- Ball, R.E.; Serra-Pereira, B.; Ellis, J.; Genner, M.J.; Iglésias, S.; Johnson, A.F.; Jones, C.S.; Leslie, R.; Lewis, J.; Mariani, S.; et al. Resolving taxonomic uncertainty in vulnerable elasmobranchs: Are the Madeira skate (Raja maderensis) and the thornback ray (Raja clavata) distinct species? Conserv. Genet. 2016, 17, 565–576. [Google Scholar] [CrossRef]
- Corrigan, S.; Maisano Delser, P.; Eddy, C.; Duffy, C.; Yang, L.; Li, C.; Bazinet, A.L.; Mona, S.; Naylor, G.J.P. Historical introgression drives pervasive mitochondrial admixture between two species of pelagic sharks. Mol. Phylogenet. Evol. 2017, 110, 122–126. [Google Scholar] [CrossRef]
- Kumar, A.; Prakash, S. Mitochondrial Genome of “Spotted Numbfish” Narcine timlei (Bloch & Schneider, 1801) and Phylogenetic Relationships among Order Torpediniformes. J. Appl. Ichthyol. 2023, 2023, 6829359. [Google Scholar] [CrossRef]
| Species | COI | NADH2 | ||||||
|---|---|---|---|---|---|---|---|---|
| N | H | hd | π | N | H | hd | π | |
| T. marmorata | 77 | 7 (5) | 0.419 ± 0.066 | 0.001 ± 0.000 | 86 | 25 (21) | 0.829 ± 0.032 | 0.004 ± 0.000 |
| T. torpedo | 38 | 4 (2) | 0.352 ± 0.088 | 0.001 ± 0.000 | 28 | 5 (5) | 0.781 ± 0.107 | 0.001 ± 0.000 |
| T. nobiliana | 15 | 2 (0) | 0.248 ± 0.131 | 0.001 ± 0.000 | 15 | 7 (7) | 0.479 ± 0.016 | 0.002 ± 0.006 |
| Total | 130 | 13 | 0.736 ± 0.000 | 0.060 ± 0.004 | 129 | 37 | 0.898 ± 0.016 | 0.075 ± 0.006 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melis, R.; Di Crescenzo, S.; Cariani, A.; Ferrari, A.; Crobe, V.; Bellodi, A.; Mulas, A.; Carugati, L.; Coluccia, E.; Follesa, M.C.; et al. I Like This New Me: Unravelling Population Structure of Mediterranean Electric Rays and Taxonomic Uncertainties within Torpediniformes. Animals 2023, 13, 2899. https://doi.org/10.3390/ani13182899
Melis R, Di Crescenzo S, Cariani A, Ferrari A, Crobe V, Bellodi A, Mulas A, Carugati L, Coluccia E, Follesa MC, et al. I Like This New Me: Unravelling Population Structure of Mediterranean Electric Rays and Taxonomic Uncertainties within Torpediniformes. Animals. 2023; 13(18):2899. https://doi.org/10.3390/ani13182899
Chicago/Turabian StyleMelis, Riccardo, Simone Di Crescenzo, Alessia Cariani, Alice Ferrari, Valentina Crobe, Andrea Bellodi, Antonello Mulas, Laura Carugati, Elisabetta Coluccia, Maria Cristina Follesa, and et al. 2023. "I Like This New Me: Unravelling Population Structure of Mediterranean Electric Rays and Taxonomic Uncertainties within Torpediniformes" Animals 13, no. 18: 2899. https://doi.org/10.3390/ani13182899
APA StyleMelis, R., Di Crescenzo, S., Cariani, A., Ferrari, A., Crobe, V., Bellodi, A., Mulas, A., Carugati, L., Coluccia, E., Follesa, M. C., & Cannas, R. (2023). I Like This New Me: Unravelling Population Structure of Mediterranean Electric Rays and Taxonomic Uncertainties within Torpediniformes. Animals, 13(18), 2899. https://doi.org/10.3390/ani13182899










