Genome-Wide Association Study of Body Conformation Traits in a Three-Way Crossbred Commercial Pig Population
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Phenotypic Data
2.3. Genotypes and Quality Control
2.4. Pearson’s Correlation Coefficient and Estimation of Genetic Parameters
2.5. Population Structure Analysis
2.6. Association Analyses
2.6.1. MLM-Based GWAS
2.6.2. FarmCPU-Based GWAS
2.7. Identification of Significant Single Nucleotide Polymorphisms Associated with Body Conformation Traits
2.8. Haplotype Block Analysis
2.9. Candidate Gene Search and Functional Annotation
3. Results and Discussion
3.1. Phenotype Statistics and Correlations among the Traits
3.2. Genome-Wide Association Studies for Body Conformation Traits
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, C.; Wu, Z.; Ren, J.; Huang, Z.; Liu, D.; He, X.; Prakapenka, D.; Zhang, R.; Li, N.; Da, Y.; et al. Genome-wide association study and accuracy of genomic prediction for teat number in Duroc pigs using genotyping-by-sequencing. Genet. Sel. Evol. 2017, 49, 35. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Ye, J.; Dong, L.; Li, Y.; Yan, L.; Cai, G.; Liu, D.; Tan, C.; Wu, Z. Genome-Wide Association Study for Body Length, Body Height, and Total Teat Number in Large White Pigs. Front. Genet. 2021, 12, 650370. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ji, J.; Peng, S.; Zhang, Z.; Fang, S.; Li, L.; Zhu, Y.; Huang, L.; Chen, C.; Ma, J. A GWA study reveals genetic loci for body conformation traits in Chinese Laiwu pigs and its implications for human BMI. Mamm. Genome 2016, 27, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Hoge, M.D.; Bates, R.O. Developmental factors that influence sow longevity. J. Anim. Sci. 2011, 89, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Le, T.H.; Madsen, P.; Lundeheim, N.; Nilsson, K.; Norberg, E. Genetic association between leg conformation in young pigs and sow longevity. J. Anim. Breed. Genet. 2016, 133, 283–290. [Google Scholar] [CrossRef]
- Nikkilä, M.T.; Stalder, K.J.; Mote, B.E.; Rothschild, M.F.; Gunsett, F.C.; Johnson, A.K.; Karriker, L.A.; Boggess, M.V.; Serenius, T.V. Genetic associations for gilt growth, compositional, and structural soundness traits with sow longevity and lifetime reproductive performance. J. Anim. Sci. 2013, 91, 1570–1579. [Google Scholar] [CrossRef]
- Hu, Z.L.; Park, C.A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the future: Meeting new challenges and providing updated services. Nucleic Acids Res. 2022, 50, D956–D961. [Google Scholar] [CrossRef]
- Li, C.; Duan, D.; Xue, Y.; Han, X.; Wang, K.; Qiao, R.; Li, X.L.; Li, X.J. An association study on imputed whole-genome resequencing from high-throughput sequencing data for body traits in crossbred pigs. Anim. Genet. 2022, 53, 212–219. [Google Scholar] [CrossRef]
- Zhou, S.; Ding, R.; Zhuang, Z.; Zeng, H.; Wen, S.; Ruan, D.; Wu, J.; Qiu, Y.; Zheng, E.; Cai, G.; et al. Genome-Wide Association Analysis Reveals Genetic Loci and Candidate Genes for Chest, Abdominal, and Waist Circumferences in Two Duroc Pig Populations. Front. Vet. Sci. 2021, 8, 807003. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.P.; Tribout, T.; Iannuccelli, N.; Bouffaud, M.; Servin, B.; Tenghe, A.; Dehais, P.; Muller, N.; Del Schneider, M.P.; Mercat, M.J.; et al. A genome-wide association study of production traits in a commercial population of Large White pigs: Evidence of haplotypes affecting meat quality. Genet. Sel. Evol. 2014, 46, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Kaler, A.S.; Ray, J.D.; Schapaugh, W.T.; King, C.A.; Purcell, L.C. Genome-wide association mapping of canopy wilting in diverse soybean genotypes. Theor. Appl. Genet. 2017, 130, 2203–2217. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Li, Y.Q.; Wu, H.Y.; Hu, B.; Zheng, J.J.; Zhai, H.; Lv, S.X.; Liu, X.L.; Chen, X.; Qiu, H.M.; et al. Genotyping of Soybean Cultivars with Medium-Density Array Reveals the Population Structure and QTNs Underlying Maturity and Seed Traits. Front. Plant Sci. 2018, 9, 610. [Google Scholar] [CrossRef]
- Zhang, H.; Zhuang, Z.; Yang, M.; Ding, R.; Quan, J.; Zhou, S.; Gu, T.; Xu, Z.; Zheng, E.; Cai, G.; et al. Genome-Wide Detection of Genetic Loci and Candidate Genes for Body Conformation Traits in Duroc × Landrace × Yorkshire Crossbred Pigs. Front. Genet. 2021, 12, 664343. [Google Scholar] [CrossRef]
- Zhuang, Z.; Ding, R.; Peng, L.; Wu, J.; Ye, Y.; Zhou, S.; Wang, X.; Quan, J.; Zheng, E.; Cai, G.; et al. Genome-wide association analyses identify known and novel loci for teat number in Duroc pigs using single-locus and multi-locus models. BMC Genom. 2020, 21, 344. [Google Scholar] [CrossRef]
- Özşensoy, Y.; Şahin, S. Comparison of different DNA isolation methods and use of dodecyle trimethyl ammonium bromide (DTAB) for the isolation of DNA from meat products. J. Adv. Vet. Anim. Res. 2016, 3, 368–374. [Google Scholar] [CrossRef]
- Zhuang, Z.; Wu, J.; Xu, C.; Ruan, D.; Qiu, Y.; Zhou, S.; Ding, R.; Quan, J.; Yang, M.; Zheng, E.; et al. The Genetic Architecture of Meat Quality Traits in a Crossbred Commercial Pig Population. Foods 2022, 11, 3143. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Li, H.; Xu, C.; Meng, F.; Yao, Z.; Fan, Z.; Yang, Y.; Meng, X.; Zhan, Y.; Sun, Y.; Ma, F.; et al. Genome-Wide Association Studies for Flesh Color and Intramuscular Fat in (Duroc × Landrace × Large White) Crossbred Commercial Pigs. Genes 2022, 13, 2131. [Google Scholar] [CrossRef] [PubMed]
- Luan, M.; Ruan, D.; Meng, F.; Qiu, Y.; Ye, Y.; Zhou, S.; Yang, J.; Sun, Y.; Ma, F.; Wu, Z.; et al. Genome-wide association study for loin muscle area of commercial crossbred pigs. Anim. Biosci. 2023, 36, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Ding, R.; Meng, F.; Wang, X.; Zhuang, Z.; Quan, J.; Geng, Q.; Wu, J.; Zheng, E.; Wu, Z.; et al. A meta-analysis of genome-wide association studies for average daily gain and lean meat percentage in two Duroc pig populations. BMC Genom. 2021, 22, 12. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Zhao, H.; Zhu, S.; Guo, T.; Han, M.; Chen, B.; Qiao, G.; Wu, Y.; Yuan, C.; Liu, J.; Lu, Z.; et al. Whole-genome re-sequencing association study on yearling wool traits in Chinese fine-wool sheep. J. Anim. Sci. 2021, 99, skab210. [Google Scholar] [CrossRef]
- Glickman, M.E.; Rao, S.R.; Schultz, M.R. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J. Clin. Epidemiol. 2014, 67, 850–857. [Google Scholar] [CrossRef]
- Wang, Y.; Ding, X.; Tan, Z.; Ning, C.; Xing, K.; Yang, T.; Pan, Y.; Sun, D.; Wang, C. Genome-Wide Association Study of Piglet Uniformity and Farrowing Interval. Front. Genet. 2017, 8, 194. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M. The structure of haplotype blocks in the human genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Bergsma, R.; Mathur, P.K.; Kanis, E.; Verstegen, M.W.; Knol, E.F.; Van Arendonk, J.A. Genetic correlations between lactation performance and growing-finishing traits in pigs. J. Anim. Sci. 2013, 91, 3601–3611. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Gong, H.; Cui, L.; Zhang, W.; Ma, J.; Chen, C.; Ai, H.; Xiao, S.; Huang, L.; et al. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations. Meat Sci. 2019, 150, 47–55. [Google Scholar] [CrossRef]
- Cuyàs, E.; Corominas-Faja, B.; Joven, J.; Menendez, J.A. Cell cycle regulation by the nutrient-sensing mammalian target of rapamycin (mTOR) pathway. Methods Mol. Biol. 2014, 1170, 113–144. [Google Scholar]
- Wang, X.; Zhang, H.; Huang, M.; Tang, J.; Yang, L.; Yu, Z.; Li, D.; Li, G.; Jiang, Y.; Sun, Y.; et al. Whole-genome SNP markers reveal conservation status, signatures of selection, and introgression in Chinese Laiwu pigs. Evol. Appl. 2021, 14, 383–398. [Google Scholar] [CrossRef]
- Falker-Gieske, C.; Blaj, I.; Preuß, S.; Bennewitz, J.; Thaller, G.; Tetens, J. GWAS for Meat and Carcass Traits Using Imputed Sequence Level Genotypes in Pooled F2-Designs in Pigs. G3 Genes Genomes Genet. 2019, 9, 2823–2834. [Google Scholar] [CrossRef]
- Zhou, L.; Zhao, W.; Fu, Y.; Fang, X.; Ren, S.; Ren, J. Genome-wide detection of genetic loci and candidate genes for teat number and body conformation traits at birth in Chinese Sushan pigs. Anim. Genet. 2019, 50, 753–756. [Google Scholar] [CrossRef]
- Li, J.; Peng, S.; Zhong, L.; Zhou, L.; Yan, G.; Xiao, S.; Ma, J.; Huang, L. Identification and validation of a regulatory mutation upstream of the BMP2 gene associated with carcass length in pigs. Genet. Sel. Evol. 2021, 53, 94. [Google Scholar] [CrossRef]
- Zhou, N.; Li, Q.; Lin, X.; Hu, N.; Liao, J.Y.; Lin, L.B.; Zhao, C.; Hu, Z.M.; Liang, X.; Xu, W.; et al. BMP2 induces chondrogenic differentiation, osteogenic differentiation and endochondral ossification in stem cells. Cell Tissue Res. 2016, 366, 101–111. [Google Scholar] [CrossRef]
- Fan, B.; Onteru, S.K.; Du, Z.Q.; Garrick, D.J.; Stalder, K.J.; Rothschild, M.F. Genome-wide association study identifies Loci for body composition and structural soundness traits in pigs. PLoS ONE 2011, 6, e14726. [Google Scholar] [CrossRef] [PubMed]
- Bryk, B.; Hahn, K.; Cohen, S.M.; Teleman, A.A. MAP4K3 regulates body size and metabolism in Drosophila. Dev. Biol. 2010, 344, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Vahedi, S.M.; Salek Ardestani, S.; Karimi, K.; Banabazi, M.H. Weighted Single-Step GWAS for Body Mass Index and Scans for Recent Signatures of Selection in Yorkshire Pigs. J. Hered. 2022, 113, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, P.; Fritz, N.; Smedler, E.; Malmersjö, S.; Kanatani, S. Calcium signaling in neocortical development. Dev. Neurobiol. 2015, 75, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Tuduce, I.L.; Schuh, K.; Bundschu, K. Spred2 expression during mouse development. Dev. Dyn. 2010, 239, 3072–3085. [Google Scholar] [CrossRef]
- Bundschu, K.; Knobeloch, K.P.; Ullrich, M.; Schinke, T.; Amling, M.; Engelhardt, C.M.; Renné, T.; Walter, U.; Schuh, K. Gene disruption of Spred-2 causes dwarfism. J. Biol. Chem. 2005, 280, 28572–28580. [Google Scholar] [CrossRef]
- Budnik, A.; Heesom, K.J.; Stephens, D.J. Characterization of human Sec16B: Indications of specialized, non-redundant functions. Sci. Rep. 2011, 1, 77. [Google Scholar] [CrossRef]
- Bradfield, J.P.; Vogelezang, S.; Felix, J.F.; Chesi, A.; Helgeland, Ø.; Horikoshi, M.; Karhunen, V.; Lowry, E.; Cousminer, D.L.; Ahluwalia, T.S.; et al. A trans-ancestral meta-analysis of genome-wide association studies reveals loci associated with childhood obesity. Hum. Mol. Genet. 2019, 28, 3327–3338. [Google Scholar] [CrossRef]
- Jiménez-Osorio, A.S.; Aguilar-Lucio, A.O.; Cárdenas-Hernández, H.; Musalem-Younes, C.; Solares-Tlapechco, J.; Costa-Urrutia, P.; Medina-Contreras, O.; Granados, J.; Rodríguez-Arellano, M.E. Polymorphisms in Adipokines in Mexican Children with Obesity. Int. J. Endocrinol. 2019, 2019, 4764751. [Google Scholar] [CrossRef]
- Williams, M.J.; Almén, M.S.; Fredriksson, R.; Schiöth, H.B. What model organisms and interactomics can reveal about the genetics of human obesity. Cell Mol. Life Sci. 2012, 69, 3819–3834. [Google Scholar] [CrossRef]
- León-Mimila, P.; Villamil-Ramírez, H.; Villalobos-Comparán, M.; Villarreal-Molina, T.; Romero-Hidalgo, S.; López-Contreras, B.; Gutiérrez-Vidal, R.; Vega-Badillo, J.; Jacobo-Albavera, L.; Posadas-Romeros, C.; et al. Contribution of common genetic variants to obesity and obesity-related traits in mexican children and adults. PLoS ONE 2013, 8, e70640. [Google Scholar]
- Thorleifsson, G.; Walters, G.B.; Gudbjartsson, D.F.; Steinthorsdottir, V.; Sulem, P.; Helgadottir, A.; Styrkarsdottir, U.; Gretarsdottir, S.; Thorlacius, S.; Jonsdottir, I.; et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009, 41, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Xie, S.; Xu, T.; Wu, X.; Han, M. Rasal2 deficiency reduces adipogenesis and occurrence of obesity-related disorders. Mol. Metab. 2017, 6, 494–502. [Google Scholar] [CrossRef]
- Mouillac, B.; Ibarrondo, J.; Guillon, G. Calcium regulation of hormonal-sensitive phospholipase C. Z. Kardiol. 1991, 80 (Suppl. S7), 79–81. [Google Scholar]
- Kwon, Y.J.; Park, D.H.; Choi, J.E.; Lee, D.; Hong, K.W.; Lee, J.W. Identification of the interactions between specific genetic polymorphisms and nutrient intake associated with general and abdominal obesity in middle-aged adults. Clin. Nutr. 2022, 41, 543–551. [Google Scholar] [CrossRef]
- Merali, Z.; McIntosh, J.; Anisman, H. Role of bombesin-related peptides in the control of food intake. Neuropeptides 1999, 33, 376–386. [Google Scholar] [CrossRef]
- Niu, N.; Liu, Q.; Hou, X.; Liu, X.; Wang, L.; Zhao, F.; Gao, H.; Shi, L.; Wang, L.; Zhang, L. Genome-wide association study revealed ABCD4 on SSC7 and GREB1L and MIB1 on SSC6 as crucial candidate genes for rib number in Beijing Black pigs. Anim. Genet. 2022, 53, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Guo, X.; Zhu, D.; Tan, C.; Bian, C.; Ren, J.; Huang, Z.; Zhao, Y.; Cai, G.; Liu, D.; et al. Accelerated deciphering of the genetic architecture of agricultural economic traits in pigs using a low-coverage whole-genome sequencing strategy. Gigascience 2021, 10, giab048. [Google Scholar] [CrossRef]
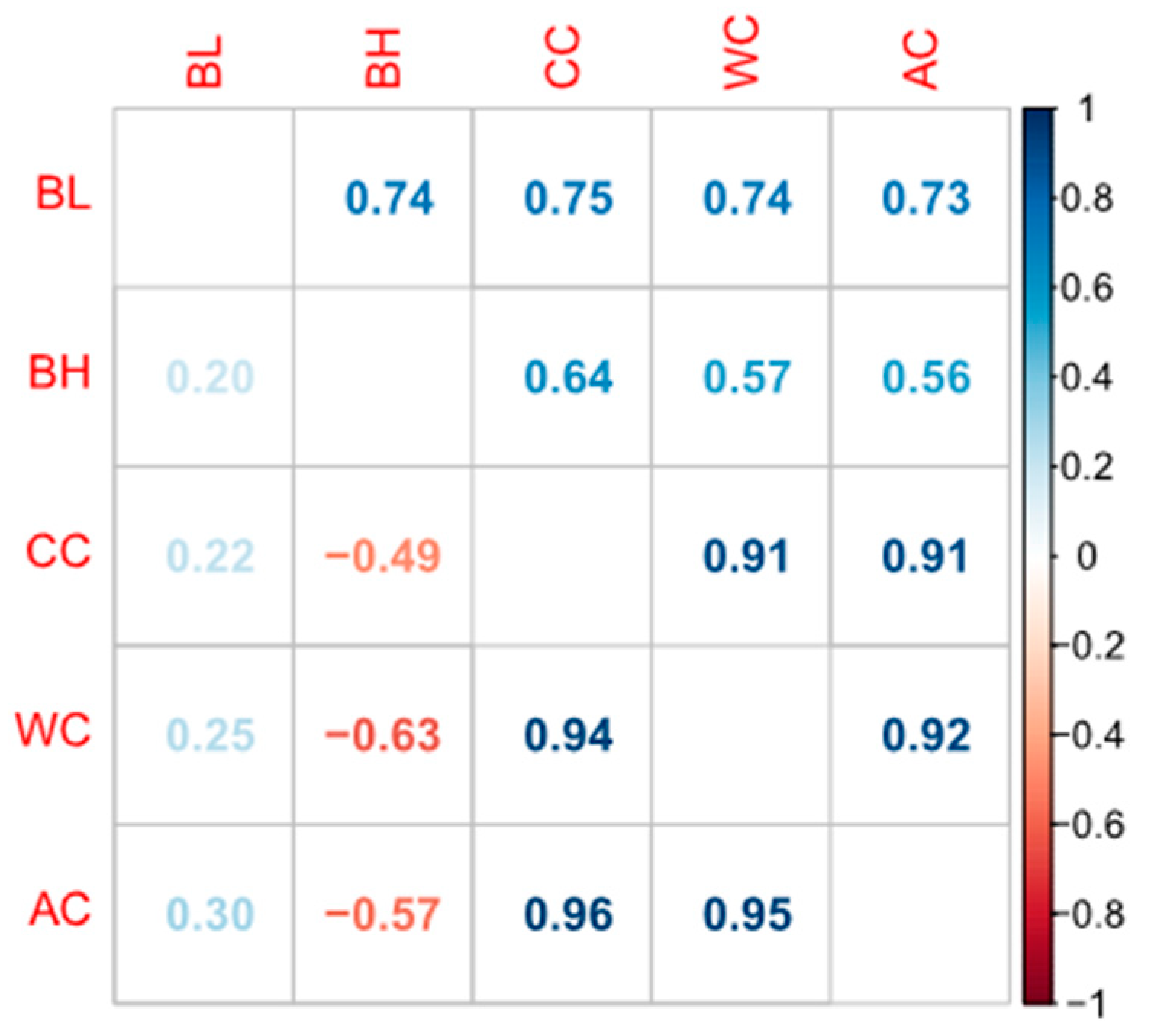



| Traits 1 | Mean ± SD 2 | Min 3 | Max 4 | CV 5 % | h² ± SE 6 |
|---|---|---|---|---|---|
| BL | 123.68 ± 7.15 | 101 | 145 | 5.78 | 0.35 ± 0.04 |
| BH | 64.61 ± 3.64 | 51 | 78 | 5.64 | 0.31 ± 0.05 |
| CC | 112.82 ± 24.24 | 88 | 140 | 7.26 | 0.34 ± 0.04 |
| AC | 121.32 ± 8.58 | 94 | 150 | 7.07 | 0.26 ± 0.04 |
| WC | 110.43 ± 8.78 | 84 | 140 | 7.95 | 0.21 ± 0.04 |
| Trait | SSC 1 | SNP | Location | p-Value | p-Value | R² (%) 3 | Distance (bp) | Nearest Gene |
|---|---|---|---|---|---|---|---|---|
| (bp) 2 | (MLM) | (FarmCPU) | ||||||
| BL | 17 | MARC0030380 | 12,149,145 | 8.19 × 10−5 | 5.77 × 10−7 | 1.91 | 61,261 | INTS10 |
| 9 | ALGA0105578 | 54,113,499 | 9.50 × 10−5 | 1.54 × 10−8 | 1.42 | 50,019 | KIRREL3 | |
| 17 | WU_10.2_17_17479009 | 15,827,454 | 1.05 × 10−4 | 1.18 | 66,239 | BMP2 | ||
| 11 | MARC0052457 | 64,090,448 | 1.06 × 10−4 | 5.75 × 10−8 | 1.36 | 263,981 | SOX21 | |
| 8 | H3GA0024522 | 22,446,465 | 1.51 × 10−7 | 0.57 | NA | NA | ||
| 2 | ALGA0118729 | 131,130,527 | 1.31 × 10−6 | 1.42 | 114,097 | SLC12A2 | ||
| 12 | WU_10.2_12_23896898 | 24,026,238 | 1.77 × 10−6 | 1.14 | 33,694 | OSBPL7 | ||
| 13 | WU_10.2_13_22498141 | 20,661,904 | 2.71 × 10−6 | 1.51 | 272,967 | ARPP21 | ||
| 17 | DRGA0016669 | 24,960,133 | 7.09 × 10−6 | 1.73 | 276,584 | MACROD2 | ||
| 1 | H3GA0002350 | 95,927,556 | 1.53 × 10−5 | 1.15 | 23,858 | RNF165 | ||
| 9 | H3GA0026707 | 16,164,242 | 1.71 × 10−5 | 0.72 | NA | NA | ||
| 12 | WU_10.2_12_4071530 | 4,324,076 | 3.37 × 10−5 | 0.03 | Within | SEPTIN9 | ||
| 16 | WU_10.2_16_67817952 | 62,516,694 | 3.47 × 10−5 | 1.16 | 53,119 | ATP10B | ||
| 10 | MARC0041569 | 5,298,516 | 3.78 × 10−5 | 0.92 | 455,315 | KCTD3 | ||
| 11 | WU_10.2_11_5570350 | 5,881,250 | 3.88 × 10−5 | 1.41 | 72,493 | POMP | ||
| 15 | WU_10.2_15_136877153 | 123,439,329 | 5.39 × 10−5 | 1.19 | 63,812 | EPHA4 | ||
| BH | 14 | ALGA0081919 | 125,132,825 | 2.08 × 10−5 | 1.13 | 25,823 | FAM160B1 | |
| 8 | WU_10.2_8_18963576 | 18,731,675 | 2.36 × 10−5 | 6.32 × 10−5 | 1.88 | 64,961 | SOD3 | |
| 3 | DIAS0000802 | 101,049,861 | 1.08 × 10−4 | 1.93 | Within | MAP4K3 | ||
| CC | 9 | H3GA0028170 | 119,852,713 | 1.27 × 10−5 | 9.09 × 10−7 | 1.23 | 9783 | SEC16B |
| 3 | H3GA0010240 | 102,073,609 | 2.03 × 10−5 | 2.15 | 43,940 | ATL2 | ||
| 13 | ALGA0119302 | 29,363,145 | 3.50 × 10−5 | 7.21 × 10−6 | 1.75 | 20,158 | CCR5 | |
| 13 | ASGA0055780 | 6,014,822 | 3.90 × 10−5 | 1.95 × 10−6 | 0.81 | 89,715 | KCNH8 | |
| 3 | MARC0004483 | 76,624,951 | 4.32 × 10−5 | 2.06 | Within | SPRED2 | ||
| 3 | ASGA0015185 | 76,651,363 | 4.32 × 10−5 | 2.06 | 1713 | SPRED2 | ||
| 3 | WU_10.2_3_108307418 | 102,136,805 | 6.36 × 10−5 | 5.43 × 10−6 | 2.07 | 58,936 | CYP1B1 | |
| 10 | WU_10.2_10_67005939 | 61,139,023 | 1.33 × 10−6 | 0.31 | NA | NA | ||
| 5 | INRA0019282 | 40,871,511 | 2.18 × 10−6 | 0.69 | 772 | SYT10 | ||
| 18 | ALGA0098775 | 50,846,238 | 3.65 × 10−6 | 1.81 | 15,228 | CAMK2B | ||
| 1 | WU_10.2_1_179575045 | 161,987,727 | 3.74 × 10−6 | 0.7 | 2555 | ZNF532 | ||
| 1 | ASGA0001418 | 16,903,857 | 4.07 × 10−5 | 0.2 | 63,327 | UST | ||
| 2 | WU_10.2_2_21124019 | 19,419,332 | 4.28 × 10−5 | 1.3 | 425,106 | API5 | ||
| AC | 1 | ALGA0009765 | 258,153,534 | 2.98 × 10−5 | 1.26 | 412,557 | ASTN2 | |
| 1 | WU_10.2_1_289532755 | 257,687,154 | 8.53 × 10−5 | 5.04 × 10−9 | 1.73 | Within | ASTN2 | |
| 2 | MARC0066799 | 115,758,230 | 1.59 × 10−6 | 1.71 | 76,874 | WDR36 | ||
| 12 | MARC0115537 | 37,253,607 | 6.72 × 10−6 | 1.34 | 430,372 | C17orf64 | ||
| 13 | ALGA0067602 | 5,297,429 | 7.42 × 10−6 | 1.77 | 16,929 | SATB1 | ||
| 13 | MARC0021524 | 99,647,029 | 1.02 × 10−5 | 0.88 | 278,346 | C3orf80 | ||
| 6 | MARC0000035 | 120,909,790 | 1.63 × 10−5 | 0.78 | Within | KIAA1328 | ||
| 9 | WU_10.2_9_131985977 | 120,299,004 | 2.42 × 10−5 | 0.33 | Within | RASAL2 | ||
| 7 | DRGA0007316 | 20,219,344 | 2.58 × 10−5 | 0.41 | Within | CARMIL1 | ||
| 7 | MARC0033686 | 64,847,978 | 4.51 × 10−5 | 0.13 | 19,104 | SRP54 | ||
| 3 | ASGA0014859 | 59,618,741 | 4.74 × 10−5 | 0.67 | Within | KCMF1 | ||
| 11 | ALGA0124549 | 25,293,190 | 6.91 × 10−5 | 1.59 | Within | VWA8 | ||
| WC | 7 | ALGA0039140 | 18,645,244 | 2.25 × 10−5 | 1.58 | 362,537 | NRSN1 | |
| 7 | WU_10.2_7_103232787 | 97,584,287 | 5.15 × 10−5 | 1.84 | Within | ABCD4 | ||
| 7 | Affx-115258151 | 97,595,573 | 6.36 × 10−5 | 4.58 × 10−7 | 1.63 | 9894 | ABCD4 | |
| 7 | WU_10.2_7_103460706 | 97,617,907 | 7.98 × 10−5 | 1.62 | 32,228 | ABCD4 | ||
| 7 | Affx-114892585 | 97,575,068 | 8.58 × 10−5 | 1.61 | Within | ABCD4 | ||
| 7 | Affx-114687136 | 97,568,284 | 8.99 × 10−5 | 1.62 | Within | ABCD4 | ||
| 14 | ASGA0062816 | 38,090,626 | 1.79 × 10−6 | 1.63 | Within | RBM19 | ||
| 11 | ASGA0049251 | 3,324,036 | 9.00 × 10−6 | 1.12 | Within | ATP8A2 | ||
| 5 | WU_10.2_5_65149069 | 62,312,551 | 1.03 × 10−5 | 0.32 | 54,860 | KLRB1 | ||
| 7 | WU_10.2_7_105348813 | 99,303,783 | 1.22 × 10−5 | 1.71 | 21,786 | GPATCH2L | ||
| 16 | ASGA0072515 | 19,669,219 | 1.42 × 10−5 | 0.82 | Within | ADAMTS12 | ||
| 15 | ALGA0088031 | 131,517,298 | 1.47 × 10−5 | 0.69 | 24,648 | CAB39 | ||
| 1 | DIAS0002061 | 161,757,996 | 2.05 × 10−5 | 2.2 | 36,214 | GRP | ||
| 6 | WU_10.2_6_21220801 | 22,691,873 | 3.09 × 10−5 | 1.02 | 231,267 | CDH8 | ||
| 7 | WU_10.2_7_118076533 | 111,437,802 | 3.68 × 10−5 | 2.48 | Within | FOXN3 | ||
| 12 | ALGA0064332 | 2,354,756 | 4.46 × 10−5 | 1.13 | Within | CCDC40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, S.; Qiu, Y.; Zhuang, Z.; Wu, J.; Li, X.; Ruan, D.; Xu, C.; Zheng, E.; Yang, M.; Cai, G.; et al. Genome-Wide Association Study of Body Conformation Traits in a Three-Way Crossbred Commercial Pig Population. Animals 2023, 13, 2414. https://doi.org/10.3390/ani13152414
Deng S, Qiu Y, Zhuang Z, Wu J, Li X, Ruan D, Xu C, Zheng E, Yang M, Cai G, et al. Genome-Wide Association Study of Body Conformation Traits in a Three-Way Crossbred Commercial Pig Population. Animals. 2023; 13(15):2414. https://doi.org/10.3390/ani13152414
Chicago/Turabian StyleDeng, Shaoxiong, Yibin Qiu, Zhanwei Zhuang, Jie Wu, Xuehua Li, Donglin Ruan, Cineng Xu, Enqing Zheng, Ming Yang, Gengyuan Cai, and et al. 2023. "Genome-Wide Association Study of Body Conformation Traits in a Three-Way Crossbred Commercial Pig Population" Animals 13, no. 15: 2414. https://doi.org/10.3390/ani13152414
APA StyleDeng, S., Qiu, Y., Zhuang, Z., Wu, J., Li, X., Ruan, D., Xu, C., Zheng, E., Yang, M., Cai, G., Yang, J., Wu, Z., & Huang, S. (2023). Genome-Wide Association Study of Body Conformation Traits in a Three-Way Crossbred Commercial Pig Population. Animals, 13(15), 2414. https://doi.org/10.3390/ani13152414





