Simple Summary
With conventional breeding practices, even the best dairy/beef cows will likely produce no more than half-a-dozen calves in their lifetimes; a crime, would you not agree? In contrast, with embryo transfer technologies, a cow can pass her genetics to hundreds of calves through surrogate cows. As such, embryo transfer, particularly involving embryos produced by in vitro fertilization, also known as IVF, is being widely adopted. However, pregnancy rates from the transfer of IVF embryos remain unimpressive. Prior to transfer, embryos are evaluated under a microscope, and only high-quality embryos are transferred. Unfortunately, microscopy cannot accurately distinguish between high-quality and low-quality embryos. Many embryo evaluation methodologies have been tested to date to find a replacement for microscopy-based embryo evaluation, e.g., advanced microscopic systems and the evaluation of genetics and secreted proteins/molecules. The good news: many of them can accurately separate the wheat from the chaff. The bad news: many of them are expensive, kill embryos during evaluation, are too complicated, and/or are laborious and thus useless in the frontline, i.e., thousands of embryo production/transfer facilities. This article reviews the most common (and not so common) techniques that have been tested to date and provides insights about those that have the highest potential to replace microscopy-based embryo evaluation in-field applications.
Abstract
Approximately 80% of the ~1.5 million bovine embryos transferred in 2021 were in vitro produced. However, only ~27% of the transferred IVP embryos will result in live births. The ~73% pregnancy failures are partly due to transferring poor-quality embryos, a result of erroneous stereomicroscopy-based morphological evaluation, the current method of choice for pre-transfer embryo evaluation. Numerous microscopic (e.g., differential interference contrast, electron, fluorescent, time-lapse, and artificial-intelligence-based microscopy) and non-microscopic (e.g., genomics, transcriptomics, epigenomics, proteomics, metabolomics, and nuclear magnetic resonance) methodologies have been tested to find an embryo evaluation technique that is superior to morphologic evaluation. Many of these research tools can accurately determine embryo quality/viability; however, most are invasive, expensive, laborious, technically sophisticated, and/or time-consuming, making them futile in the context of in-field embryo evaluation. However accurate they may be, using complex methods, such as RNA sequencing, SNP chips, mass spectrometry, and multiphoton microscopy, at thousands of embryo production/collection facilities is impractical. Therefore, future research is warranted to innovate field-friendly, simple benchtop tests using findings already available, particularly from omics-based research methodologies. Time-lapse monitoring and artificial-intelligence-based automated image analysis also have the potential for accurate embryo evaluation; however, further research is warranted to innovate economically feasible options for in-field applications.
1. Introduction
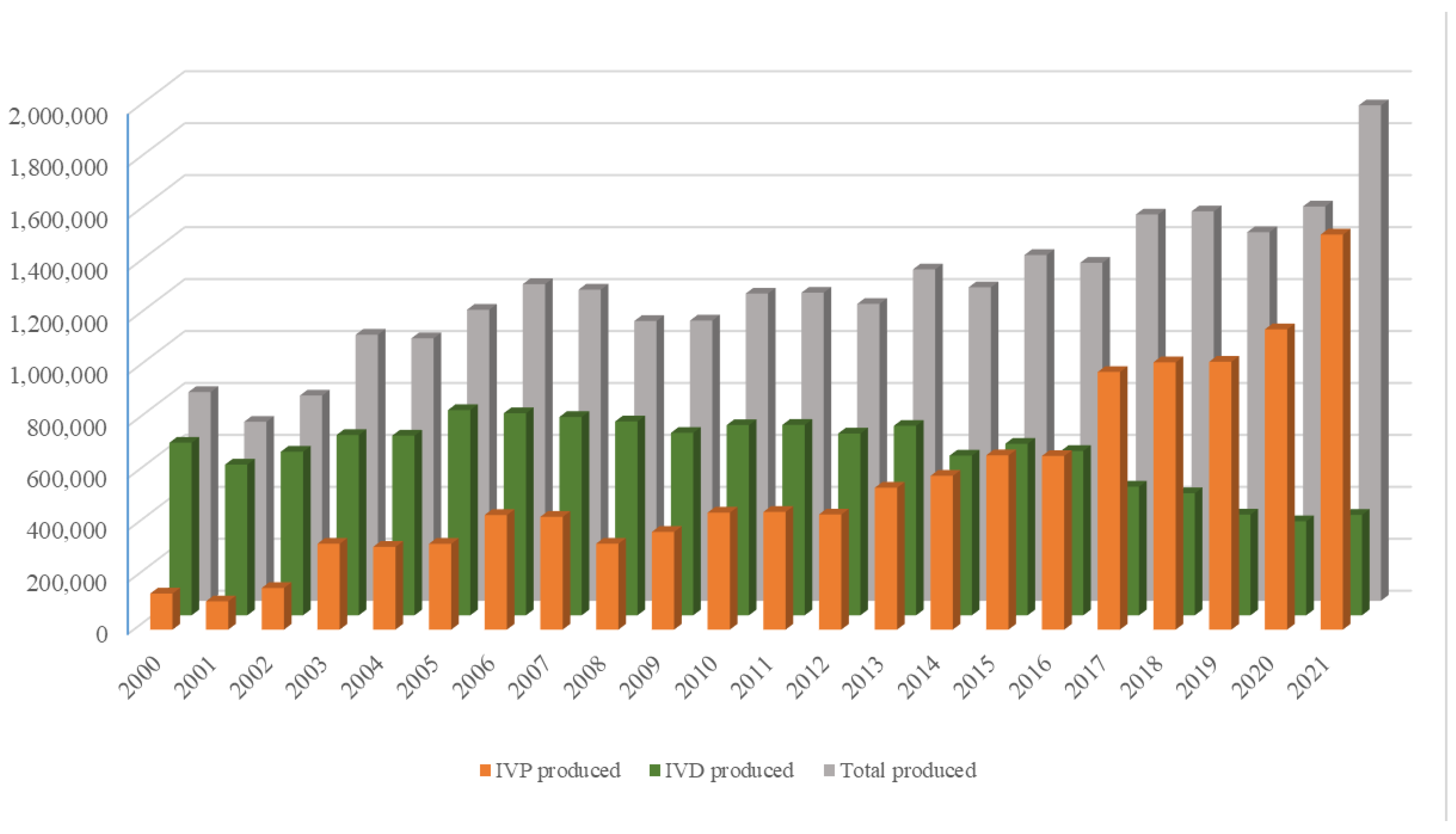
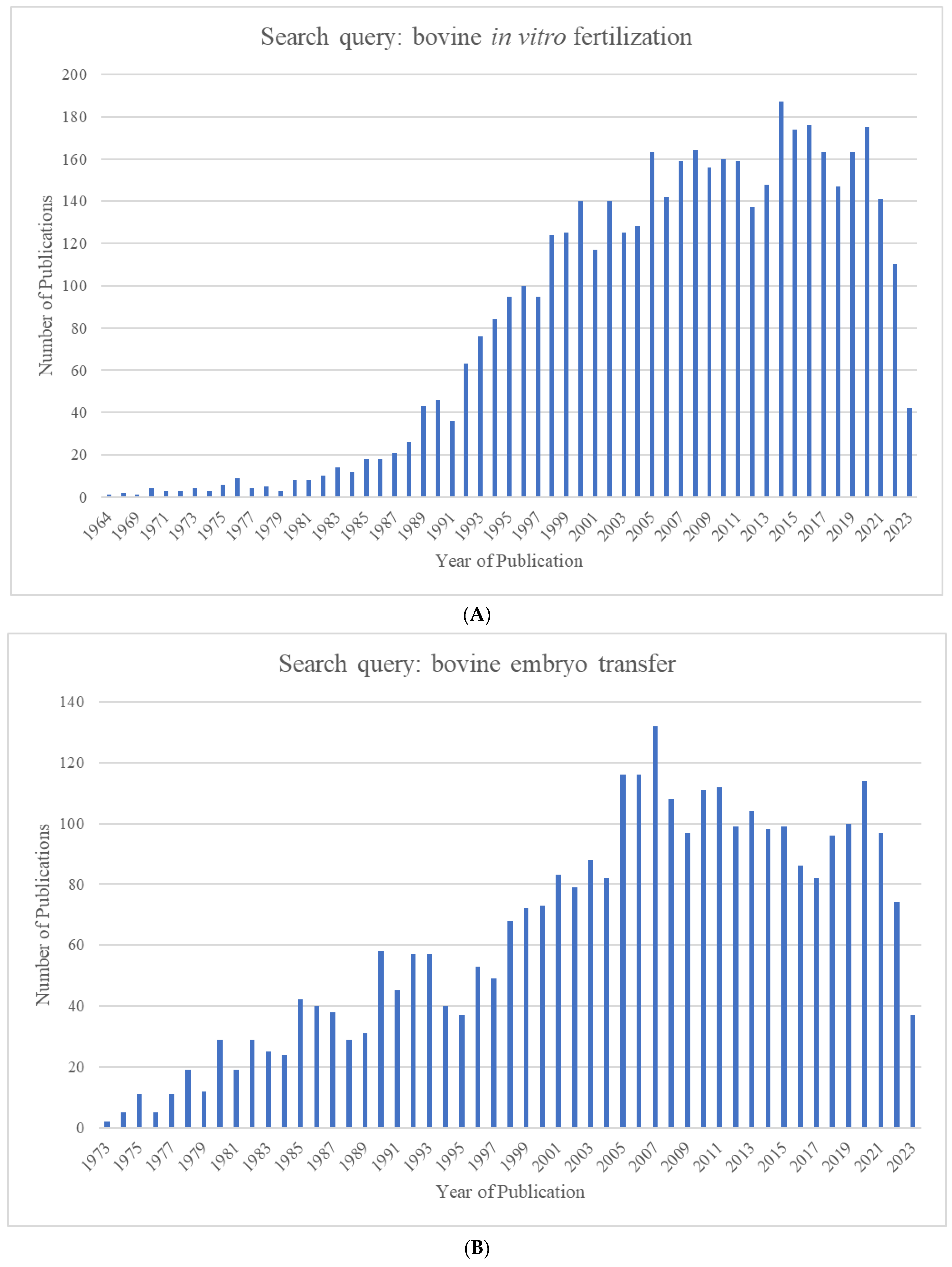
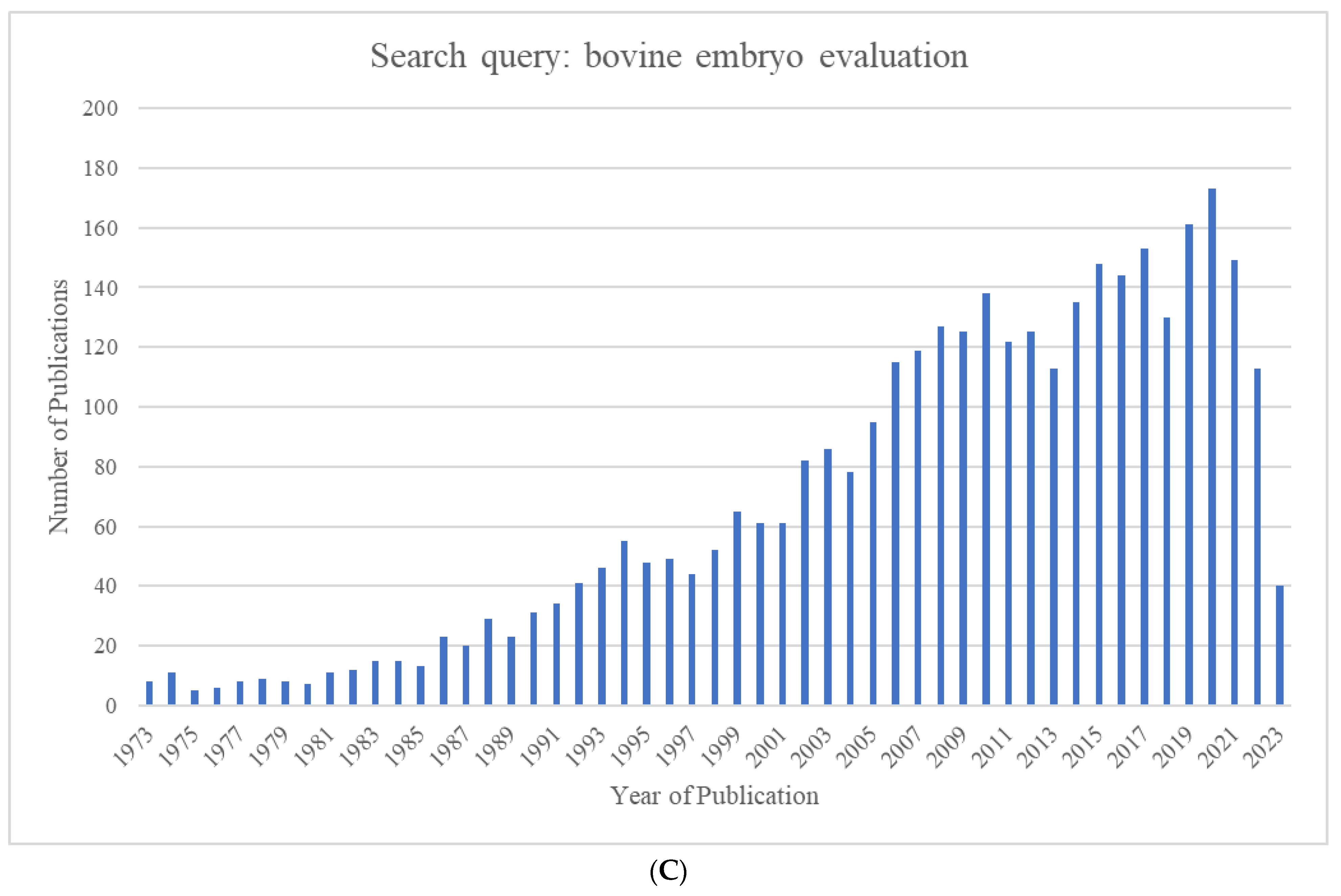
The growing world population has created an ever-increasing demand for animal proteins. The beef and dairy industries play tremendous roles in this regard, annually contributing >72 million metric tons of meat (21% of global meat production) and >500 million metric tons of milk to the global market. Reproduction is a major driver of the bovine meat and milk production industries. As such, Assisted Reproductive Technologies (ARTs) such as in vitro embryo production (IVP), in vivo derived embryo production (IVD), and embryo transfer (ET) have gained enormous importance in the beef and dairy cattle industries [1]. As illustrated in Figure 1, the annual production of transferrable bovine embryos has nearly doubled within the 12-year period from 2009 to 2021. What is even more striking is the shift of embryo production from IVD to IVP, with embryo production by IVP quadrupling over the same period. In 2021, over 1.5 million transferrable IVP embryos were produced (Figure 1), of which nearly 80% (~1.2 million) were transferred [2]. Parallel to this rapid expansion of IVP, the interest from the scientific community and, in turn, research publications have also risen many-fold over the last few decades (Figure 2A–C).
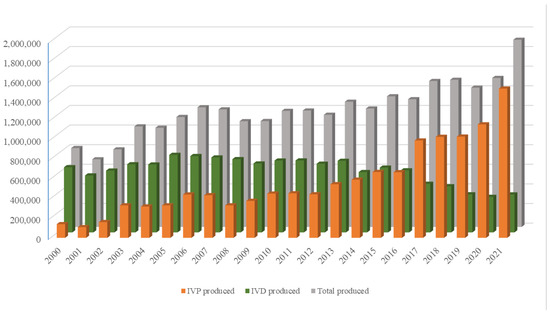
Figure 1.
Annual production of IVP (orange bars), IVD (green bars), and total number (gray bars) of bovine embryos. Compiled from 2001–2022 annual Data Retrieval Committee Reports of the International Embryo Transfer Society.
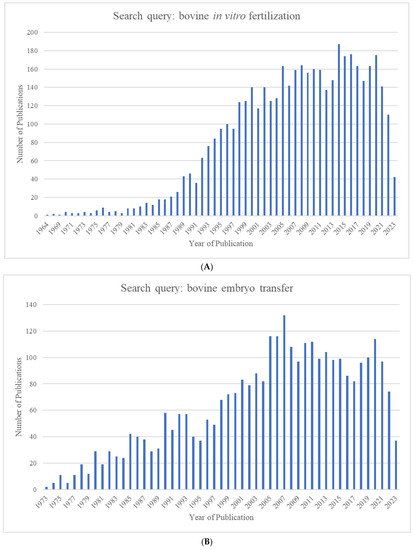
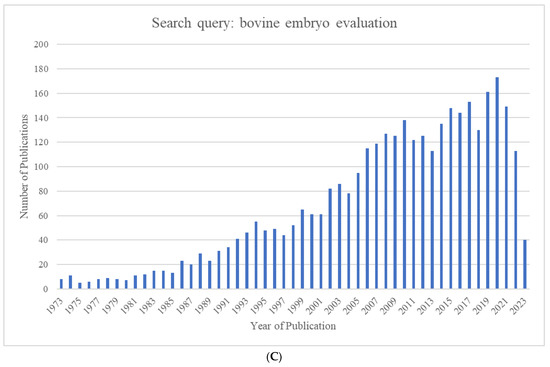
Figure 2.
Number of publications identified in Pubmed.gov by year for the search queries (A) "bovine in vitro fertilization” (1964–2023), (B) “bovine embryo transfer” (1973–2023), and (C) “bovine embryo evaluation” (1973–2023).
It is commonly accepted that the developmental competence, quality, and cryo-survival of IVP embryos are overall lower than those of IVD embryos. Further, pregnancy rates (PRs) resulting from the transfer of IVP embryos are lower than those from the transfer of quality- and stage-matched IVD embryos reviewed by [3,4,5,6]. For both IVP and IVD embryos, it is well established that the transfer of higher-quality blastocysts results in higher PRs compared to the transfer of lower-quality blastocysts [7,8,9]. Therefore, if embryos can be accurately evaluated and the high-quality ones could be selected for ET, the commercial value of selected embryos would increase, with a subsequent decrease in the number of recipients and an overall improvement in the efficiency of ET [10]. Therefore, one of the most important factors associated with the successful application of ARTs is embryo evaluation prior to freezing or transfer to recipients [11,12]. At present, the most widely used method for determining embryo quality is morphological evaluation using stereomicroscopy [6]. However, it is considered to have poor accuracy and poor reproducibility and to be biased by the subjectivity of the evaluator, i.e., intra- and inter-observer variability [1,5].
Together, the current trend favoring IVP and the limitations of current morphology-based embryo evaluation methods demand the in-depth study of pre-implantation bovine embryos (i) to better understand the physiology of embryo development and (ii) to discover embryo viability/quality markers that can be used for the accurate prediction of post-transfer embryo survival and the pregnancy outcome. As a result, a wide variety of methodologies, ranging from optics to omics-based methodologies to gene editing and everything else in between, have been used to evaluate and analyze bovine embryos.
The aim of this review is to provide an overview of the various embryo evaluation methodologies used not only during the post-IVP era but also starting from 1931, the first available record of the microscopic anatomy of pre-implantation bovine embryos from nearly a century ago [13]. Further, we also provide insights about embryo evaluation methodologies that have the highest potential for adoption at the ground level as a replacement for conventional stereomicroscopy-based morphological embryo evaluation.
We conclude that sophisticated embryo evaluation tools such as omics-based methodologies are unfit to be directly used for in-field embryo evaluation. However, findings from these methodologies can be applied indirectly for in-field embryo evaluation if field-friendly, simple, and affordable benchtop tests are developed. Further, findings from time-lapse monitoring (TLM) and artificial-intelligence-based automated image analysis look very promising. With further improvements and the innovation of economically affordable options, they may be directly used for in-field applications.
2. Evaluation of Pre-Implantation Bovine Embryos
Embryo evaluation is two-fold and is typically performed prior to cryopreservation. They are evaluated for the developmental stage (grade) and quality. Currently, only embryos satisfying minimum criteria in both these facets are selected for freezing or direct transfer.
2.1. Evaluation for Embryo Quality
The microscopic anatomy of the bovine embryo was first described in 1931 for a two-celled ovum [13], and subsequent stages up to the blastocyst stage were characterized in 1946 [14]. However, it was not until the mid-1970s, when bovine embryo transfer, carried out at an experimental scale thus far, was transformed into a commercial-scale venture [15], that the first attempts at embryo evaluation were made [16]. The authors rated 8–12-celled embryos and morulae on a scale of 1 to 5 (Table 1) “on the basis of compactness, symmetry and density of the blastomeres”. They observed that pregnancy rates resulting from the transfer of morulae were higher than those resulting from the transfer of 8–12-celled embryos, and that the highest pregnancy rates were obtained by transferring morulae of the highest quality, i.e., a rating of 5. However, the authors admitted that “This rating, by its very nature, was subjective and hence any rigid analysis of the results would not be truly scientific”.
A set of relatively less subjective criteria were used in a 1978 report that categorized embryos into four classes, namely, excellent, good, fair, and poor ([17]; Table 1). They observed that, upon transfer, embryos categorized as “excellent” and “good” gave the highest pregnancy rates (PRs), while those categorized as “poor” gave the lowest PRs. A 1981 study adopted a classification system where embryos were given ratings of 2, 3, or 4 ([18]; Table 1). Similar to previous studies, embryos with a higher rating gave higher pregnancy rates compared to embryos with lower ratings. A 1983 study ([7]; Table 1) used the same “excellent”, “good”, “fair”, and “poor” classification system used by Elsden, Nelson, 1978 [17], but with slightly modified defining criteria. Consistent with previous studies, excellent and good embryos gave the best pregnancy rates. They went on to conclude that embryo quality, but not the stage of development (16-cell through hatched blastocysts), is the most accurate predictor of an embryo’s competency to establish a pregnancy. This finding contradicted the previous observations of Shea, Hines, 1976 [16], and Newcomb and Rowson, 1975 [19], where the authors observed higher PRs when transferring morulae compared to 8–12-celled embryos and higher PRs when transferring 5–7-day-old embryos compared to <5-day-old embryos, respectively. These observations highlight the need to devise strategies for embryo grading, as very early stage embryos with as few as 4–8 blastomeres as well as very late stage embryos as late as hatched blastocysts were being transferred during this period of time.
In addition to the compactness, symmetry, and density that were considered by Shea, Hines, 1976 [16], for embryo evaluation, the later classifications also considered criteria such as the color and shape of blastomeres, signs of degeneration such as vesiculation, the extrusion of blastomeres from the main cell mass, the retardation of embryos, and the integrity of the zona pellucida.

Table 1.
Evaluation of bovine embryo quality based on morphological characteristics.
Table 1.
Evaluation of bovine embryo quality based on morphological characteristics.
| Source | Criteria |
|---|---|
| [16] | “5: excellent appearing 4 3: average appearing 2 1: very poor appearing” rated “on the basis of compactness, symmetry and density of the blastomeres” |
| [17] | “Excellent—judged to be at the normal stage of development at the time of examination; embryos were symmetrical, and blastomeres were polygonal in shape forming a tight mass at the morula stage. Good—similar to excellent embryos but were asymmetrical, contained blastomeres excluded from the main morula mass, or were slightly retarded relative to other embryos recovered from the same donor Fair—embryos were retarded 1 to 2 days in development, had spherical rather than polygonal blastomeres at the morula stage, contained blastomeres of varying sizes, had signs of degeneration such as large vesicles in the cells, and/or were darker or lighter than normal Poor—embryos were retarded 2 or more days in development, had indistinct cell membranes, and/or had more severe faults than the fair embryos” |
| [18] | “4—embryo is above average in appearance (perfectly symmetrical, even granulation, no deformations in the zona pellucida, no blastomeres extruded) 3—embryo is average in appearance 2—embryo is below average in appearance (uneven blastomere size, extensive blastomere extrusion, evidence of membrane rupture)” |
| [7] | “Excellent—an ideal embryo, spherical, symmetrical with cells of uniform size, color and texture Good—trivial imperfections such as a few extruded blastomeres, irregular shape, few vesicles Fair—definite but not severe problems, presence of extruded blastomeres, vesiculation, few degenerated cells Poor—severe problems, numerous extruded blastomeres, degenerated cells, cells of varying sizes, large numerous vesicles but a viable-appearing embryo mass” |
| [20] | “Code 1: Excellent or good Code 2: Fair Code 3: Poor Code 4: Degenerated” |
2.2. Evaluation for Embryo Staging/Grading
None of the embryo evaluation systems shown in Table 1 recognized the developmental stages of embryos; i.e., an embryo categorized as “excellent” could be a morula, early blastocyst, expanded blastocyst, etc. Even though a coding system that identifies embryo developmental stages had already been published ([21]; Table 2), it was not incorporated into embryo evaluation criteria until the International Embryo Transfer Society (IETS) introduced a two-digit coding system that would uniformly and systematically describe embryos by their stage of development as well as their quality [20] (Supplementary Figures S1–S19). The first digit identified the stage of embryonic development, ranging from 1 (an unfertilized oocyte) to 9 (an expanding hatched blastocyst; Table 2), while the second digit identified embryo quality (based on morphological characteristics), ranging from 1 (excellent/good) to 4 (degenerated; Table 2). For example, an embryo with a 7–1 coding would refer to an expanded blastocyst of excellent/good quality. This two-digit coding system, introduced in 1998, remains the gold standard for embryo evaluation and grading within the bovine embryo production and transfer industry.

Table 2.
Coding based on developmental stages of bovine embryos.
3. Embryo Evaluation/Analysis—Tools of the Trade
According to the currently accepted norms of bovine embryo evaluation [20], the ideal unhatched bovine embryo should have the following characteristics: the embryo should be compact, spherical, and 150 to 190 µm in diameter (including the 12 to 15 µm thickness of the zona pellucida); blastomeres should be uniform in size, color, and texture; the blastomere cytoplasm should be free of granules and vesicles; perivitelline spaces should be clear and free of cellular debris; and the zona pellucida should be uniform and free of cracks and surface debris (Supplementary Figures S1–S19).
Embryo evaluation is typically performed using a stereomicroscope at 50 to 100× magnification. Embryos are held in a holding dish with holding media and evaluated by rolling it so that the entire embryo and zona pellucida can be evaluated from different angles. However, this type of morphological assessment is considered biased by the subjectivity of the evaluator, i.e., intra- and inter-observer variability [5], and therefore not considered 100% reliable and trustworthy [22,23]. Supporting this notion, differences in ultrastructure [24] and the transcriptome [25] have been demonstrated in morphologically similar, stage- and quality-matched bovine blastocysts. As mentioned earlier, a wide variety of tools have been used to study embryos to understand pre-implantation development and to search for that perfect embryo evaluation method. The following sections address these tools and techniques in detail.
3.1. Microscopic Analyses
3.1.1. Light Microscopy
Light microscopy has been one of the most fundamental and widely used tools in biology for nearly two centuries. It is ubiquitously used to analyze both fixed and living cells and is widely regarded as the least invasive method to obtain biological information from living cells [26]. This feature makes it particularly useful, to the point of being absolutely required, for analyzing embryos in research and commercial enterprises that produce bovine embryos for breeding. As stated previously, the standard method for evaluating embryo quality utilizes light microscopy: in typical cases, the direct observation of embryo morphology under a stereomicroscope. However, magnification is limited when using stereomicroscopes, and morphological details can be more readily discerned by observation with compound microscopes utilizing higher magnifications. Indeed, a study conducted in 2002 comparing embryo quality, as evaluated by observation under a stereomicroscope, standard compound microscope, and electron microscope, found that observation under a stereomicroscope leads to the overestimation of embryo quality, as signs of embryonic degeneration are more readily apparent at the higher magnifications available with compound microscopes or the electron microscope [27]. Unfortunately, specimen preparation requirements for observation at higher magnifications under a compound microscope or electron microscope are not compatible with embryo viability.
3.1.2. Differential Interference Contrast (DIC) Microscopy
For most living cells observed under standard light microscopy, the contrast is poor, and they appear almost transparent. It is difficult to discern cellular structures, no matter the ability of the magnifying and resolving power of the lenses in use, unless the cells are fixed and stained with chemical agents that provide contrast. These procedures almost invariably kill living samples. Because of this, microscopists have endeavored to develop optical techniques that provide contrast to cellular structure without requiring the use of fixatives and stains, thus allowing better visualization of living cells. These techniques are commonly used in biological microscopy today and include phase-contrast microscopy, darkfield microscopy, polarized light microscopy, Hoffman modulation contrast, and differential interference contrast (DIC) microscopy.
Developed by Georges Nomarski in the early 1950s [28], DIC utilizes polarized light that is passed through a prism, through the specimen, and then through another prism and depolarized to produce interference that results in one side of the structure that the light has passed through appearing bright and the other side appearing dark. Overall, imaging cells using this technique produces a shadowing effect that gives cells a 3D-relief-like appearance [29].
DIC microscopy is a good option for imaging embryos and live cells. DIC imaging of embryos has been used to create the qualitative scoring systems used today for bovine embryo grading/staging (personal communication, Prof. Matthew Wheeler, UIUC). DIC has been used to study bovine embryos, particularly to study pre-implantation-stage embryo development, to study subcellular components such as lipid droplets and pronuclei, to study the effects on different culture media, and to visualize microinjection procedures (Table 3). As a standalone procedure, DIC use for the analysis of embryos is declining, but its presence continues in combination with other techniques and more complex imaging systems.

Table 3.
Applications of DIC microscopy in bovine embryo evaluation and analysis.
3.1.3. Electron Microscopy
The maximum magnification that can be achieved with standard light microscopy is generally no more than 1000×. In contrast, electron microscopy allows magnifications up to around 160,000×. This allows cellular ultrastructure to be clearly visualized. In terms of resolution, while standard light microscopy can resolve structures at about 250 nm in size, the resolving power of electron microscopes provides almost a 1000-fold increase in resolution, getting down to around the 0.2 nm range [40].
The very first study that examined the ultrastructure of pre-implantation bovine embryos was carried out in 1978 [41]. The authors specifically used scanning electron microscopy to study hatching blastocysts that were recovered unhatched from cows 7–10 days after estrus and then allowed to hatch during in vitro culture. Many studies have been carried out since then to study various aspects of pre-implantation bovine embryos, including organelle structures in blastomeres [42,43], the nucleologenesis of blastomeres [44,45,46,47], the embryonic cell cycle [48,49], events associated with fertilization (in vitro) and zygote formation [50], the effects of vitrification [51], the effects of different culture media on IVP blastocysts [52,53,54], and the effects of cryopreservation on IVD and IVP blastocysts [55], and to compare IVD and IVP embryos [43,56,57].
A study that compared the accuracy of bovine embryo grading using stereomicroscopy, light microscopy, and transmission electron microscopy (TEM) found that TEM has a better accuracy in identifying “good”- and “fair”-quality blastocysts compared to the other two types [24]. However, this finding has minimal value from a practical application point of view in the dairy industry because fixing embryos for TEM makes them unviable. Therefore, although TEM can be used as a research tool to learn about pre-implantation embryo development, it has little value for routine IVF procedures in the dairy industry [58].
3.1.4. Fluorescence Microscopy
Another modification of light microscopy that is widely used today in biological research is fluorescence microscopy [59]. Fluorescence is the emission of light by a substance that has absorbed light or other electromagnetic radiation. This form of microscopy takes advantage of the ability of different compounds, referred to as fluorophores, to emit light with a monochromatic wavelength when they absorb light with a higher-energy wavelength. By coupling fluorophores with molecular probes that can recognize different proteins or other types of biological molecules, researchers can visualize and quantify the presence of different biological molecules of interest within cells, and they can observe the location and distribution of different molecules within cells, embryos, and tissues in order to gain insights on molecular and cellular functions. As with standard light microscopy, fluorescence microscopy is limited in resolving power to about 200 nm, and techniques for labeling cells with fluorophore-coupled probes are not usually compatible with cell viability. However, there are some techniques available for the fluorescent imaging of living cells and embryos [60].
Fluorescent microscopy has been used to study various aspects of pre-implantation bovine embryo development for simple tasks, such as counting blastomere numbers using DAPI or Hoechst nuclear staining [61,62] or detecting de novo DNA synthesis [63], as well as for more complicated procedures, such as fluorescent in situ hybridization (FISH) for detecting chromosomal abnormalities [63] or the immunodetection of the epigenetic markers H3K9ac and H3K9m2 [62] and the transcription factor POU5F1 [64].
The fluorescent microscopic techniques outlined above produce flat, two-dimensional (2D) images of embryos. In contrast, confocal microscopy and multiphoton laser scanning microscopy (MPLSM) can be used to produce 3D images of embryos, which can be more informative than 2D images. As with standard fluorescence microscopy, confocal microscopy and MPLSM are typically used in conjunction with molecule-specific fluorescence labeling, which allows the identification and study of not only subcellular organelles and components (e.g., lipid droplets and mitochondria) but also the intraembryonic and intracellular localization of proteins/gene products (e.g., transcription factors and lineage-specific markers). Additionally, a relatively new non-fluorescent microscopic technique, Gradient Light Interference Microscopy (GLIM), allows the production of three-dimensional (3D) images. The following sections discuss how these 3D microscopic techniques have been used in the study of pre-implantation bovine embryos.
3.1.5. Confocal Microscopy
The widespread use of fluorescent confocal microscopy on cells and tissues started in the late 1980s [65,66] because of its great applicability to quantitative analysis [67]. In conventional fluorescence microscopy, the entire sample is exposed to the excitation light source, and the entire sample emits fluorescence. When observed under the microscope and images are acquired, some out-of-focus fluorescent signals interfere with the in-focus light. Additionally, the exposure of whole live cells and embryos to this light source can cause phototoxicity, resulting in decreased cell viability. Through the use of optical sectioning technology, the confocal microscope allows the exposure and gathering of the fluorescent signal from a relatively narrow focal plane of the sample. By gathering images of focal planes throughout the sample (commonly referred to as z-stacks), three-dimensional images can be created. This can be applied to either fixed or live embryos [67,68]. There is still some risk of phototoxicity due to the power of the excitation source and the length of the scanning time, but it is possible to minimize this challenge by properly selecting the microscope, settings, and sample processing method [68]. Additionally, bovine embryos are able to maintain viability by successfully overcoming certain levels of phototoxicity [69]. Of the different confocal microscopic optical sectioning techniques available, the spinning-disk confocal system is considered the preferred technique for live imaging [68,70].
Confocal laser scanning has been widely applied to fixed and live samples to visualize specific cellular structures using fluorescent labeling or a label-free approach [68]. In pre-implantation bovine embryos, confocal microscopy has been used for a variety of applications, as summarized in Table 4.

Table 4.
Applications of confocal microscopy in bovine embryo evaluation and analysis.
3.1.6. Multiphoton Laser Scanning Microscopy (MPLSM)
Even though confocal microscopy has been used successfully for imaging the subcellular components, transcription factors, lineage-specific markers, and metabolites of bovine embryos, it can still be phototoxic to live cells/embryos and potentially compromise their viability. In contrast, MPLSM can be used for imaging fluorescent markers in relatively thick specimens, such as mammalian embryos, without compromising their viability [84]. Further, MPLSM is considered superior to confocal microscopy for scanning thick specimens, such as mammalian embryos, that exhibit significant light-scattering properties. These beneficial characteristics of MPLSM are primarily due to multiphoton systems using infrared lasers as their excitation source. Infrared light is lower in energy and scatters much less compared to light with excitation wavelengths used in confocal or standard fluorescence microscopy [85]. MPLSM has been used to image cellular organelles such as mitochondria in bovine blastomeres [85]; however, its use for studying pre-implantation bovine embryos has been very limited, possibly due to the high expenditure on equipment and the need for technical expertise [69].
Our group has begun preliminary studies using simultaneous label-free autofluorescence-multiharmonic (SLAM) microscopy [86], which is a single-excitation source nonlinear imaging platform to analyze the metabolic pathways in bovine embryos (Tu and Wheeler, 2023, personal communication).
3.1.7. Gradient Light Interference Microscopy (GLIM)
There are two major deficiencies in many of the microscopic techniques discussed earlier. First, the imaging of optically dense specimens such as embryos with conventional microscopy results in multiple scattering that scrambles the optical field, giving rise to low-contrast micrographs that often lack cellular-level detail [11,87]. Secondly, microscopic techniques such as TEM and fluorescent microscopy cannot be used to image live embryos.
GLIM can be used to overcome both of the above deficiencies. On the one hand, it can be used for the real-time imaging of thick embryos, overcoming multiple scattering. On the other hand, as a label-free method, GLIM can be used to image embryos without affecting their viability, thus enabling the technology to be used in the evaluation of in vitro produced bovine embryos prior to freezing and/or transfer [88]. Additionally, GLIM is a relatively rapid technique in that it takes ~1 min for image acquisition and >5 min for the virtual reconstruction of the image. GLIM can also be used to track and follow lipid droplet movement, which could be a possible marker of embryo quality [29], as lipid movement has been linked to protein and lipid exchange between cellular compartments in embryos of certain other species [89].
GLIM is a type of quantitative phase imaging (QPI) that combines DIC microscopy with low-coherence interferometry and holography. It is considered superior to other QPI methods, such as Spatial Light Interference Microscopy (SLIM), for imaging optically thick samples such as embryos. GLIM can be used to obtain high-contrast three-dimensional, white-light, tomographic imaging of both thin samples, such as single cells, and relatively thick specimens, such as bovine embryos. GLIM has been used to obtain 3D stacks of bovine embryos at different development stages over several days using time-resolved tomography. Using GLIM tomography, researchers were able to study individual blastomeres, their plasma membranes, gaps between plasma membranes of different blastomeres, lipid droplets in blastomeres, etc. [58]. Considering these favorable properties, GLIM has the potential to become a valuable tool for the evaluation of in vitro produced bovine embryos.
3.1.8. Time-Lapse Monitoring (TLM)
The previously described microscopic techniques allow the spatial evaluation of embryos. However, embryo development is a dynamic process, and critical stages of development may go unnoticed with such traditional, one-time morphologic assessments [90]. In contrast to these one-time morphological evaluations, time-lapse monitoring (TLM) allows embryos to be evaluated temporally as well. TLM facilitates the study of morphokinetics, allowing the correlation of early events with subsequent development and even pregnancy outcomes. Embryos could be inspected multiple times with conventional microscopy too; however, such frequent evaluations can be detrimental to embryos owing to the frequent handling and exposure to changes in temperature and gas concentrations. In contrast, TLM allows the frequent and non-invasive imaging of developing embryos while maintaining incubator conditions [91].
The first reports on the TLM of bovine embryos were published in the early 1980s [92,93]. Morulae collected from uterine horns were allowed to grow and hatch in vitro while being continuously monitored. These authors observed that pulsatile expansion and contraction was not a necessary condition for the hatching of the bovine blastocyst, an observation that could not have been made with conventional one-time microscopy. Since then, TLM has been used extensively to study pre-implantation bovine embryo morphokinetics, as summarized in Table 5. Some of these studies observed that morphokinetic indicators (MKIs), such as the timing of the first cleavage, the number of blastomeres at the first cleavage, and the number of blastomeres at the onset of the lag phase, could be used as markers for predicting blastocyst quality and the pregnancy outcome [94,95]. Some have even proposed MKIs as a superior substitute to the IETS’ morphology-based grading system [81].
TLM studies from the 1980s–2000s mostly used a cinematographic chamber placed on an inverted microscope. However, with the advancements in technology, high-resolution imaging equipment has been integrated into fully functional incubators, enabling embryo-safe recording, i.e., TLM (e.g., Primo Vision, Vitrolife, Goteborg, Sweden, and Real-Time Cultured Cell Monitoring System with Multiple-Point Imaging Capture (CCM-MULTI), Astec, Fukuoka, Japan). These systems have been successfully used to study pre-implantation bovine embryo development. Taking TLM a step further, several recent studies have used confocal laser microscopy for time-lapse live-cell imaging [81] to shed light on aspects of bovine embryo development in both temporal and spatial dimensions. The advantages of four-dimensional (4D) imaging over other microscopic techniques are numerous and have been reviewed elsewhere [96].

Table 5.
Applications of time-lapse monitoring in bovine embryo evaluation and analysis.
Table 5.
Applications of time-lapse monitoring in bovine embryo evaluation and analysis.
| Applications of TLM in Bovine Embryo Analysis | Source |
|---|---|
| To compare cleavage intervals of healthy embryos and degenerate embryos | [97] |
| To study development kinetics of embryos derived from calf oocytes | [98] |
| To study the effects of activin A and follistatin on developmental kinetics | [99] |
| To study effects of glucose on developmental kinetics of male and female embryos | [100] |
| To compare developmental kinetics of IVD and IVP embryos | [101] |
| To study dynamics of the 4th cell cycle coincident with EGA | [102] |
| To compare kinetics of initial cleavage patterns in viable and non-viable embryos | [103] |
| To study embryo development in well-of-the-well (WOW) systems | [104] |
| To identify morphokinetics predictive of blastocyst quality and pregnancy outcome | [94] |
| To study effects of abnormal cleavage patterns on morphokinetics and growth potential | [105] |
| To study development kinetics of IVP embryos fertilized with sex-sorted semen | [106] |
| To study pronuclear morphokinetics | [107] |
| To study effects of first zygotic cleavage dysmorphisms on the metabolomic profile | [108] |
| To correlate embryo morphokinetics with their transcriptomic profiles | [109] |
| To study morphokinetics of embryos derived from vitrified bovine oocytes | [90] |
In summary, every microscopic technique that has been used to study bovine embryos to date has its unique pros and cons. Conventional light microscopic methods are simple to use, relatively inexpensive, and non-invasive and therefore can be used without affecting embryo viability, even in locations with limited infrastructural facilities. However, morphological evaluation by conventional light microscopy may be limited by its magnification, resolution, and contrast. On the other hand, technologically advanced microscopic methods such as TEM and fluorescent microscopy are not compatible with maintaining the viability of embryos. Therefore, even though these techniques can be used to study embryo physiology, even at the molecular level, they are invasive methods that have no real-world applications in evaluating live embryos prior to freezing or transfer. GLIM can overcome many of the limitations discussed above for other microscopic techniques. However, it involves the use of highly sophisticated and expensive equipment, as this technology is still relatively new. Similarly, the imaging equipment used in TLM is also sophisticated and expensive. Therefore, it is unlikely that these techniques will be adopted by the dairy and beef industries at the national and global scales at the current time.
3.2. Non-Microscopic Analyses
3.2.1. Pre-Implantation Genetic Diagnosis (PGD)
The term Pre-implantation Genetic Diagnosis (PGD) was coined in the late 1980s [110] to describe a procedure where a biopsy of embryonic cells is tested for diagnosing a genetic trait. However, similar procedures took place as early as 1968 when 5–6-day-old pre-implantation-stage rabbit blastocysts were sexed by the microscopic identification of sex chromatin in biopsied trophoblast cells [111]. In fact, until the early 1990s, most genetic diagnoses of pre-implantation embryos were aimed at sex determination because transferring a known female embryo would lead to numerous benefits to livestock industries [112]. This was especially important in the pre-sexed semen era, where embryo sexing was the only viable option if IVD or IVP embryos were to be sexed before being used in various livestock embryo transfer programs.
Initial studies on embryo sexing of pre-implantation bovine embryos involved in situ hybridization with a Y-chromosome-specific DNA probe [113,114]. However, with the advent of the polymerase chain reaction (PCR; [115]), it became the method of choice for sex determination and other pre-implantation genetic analyses. The first PCR-based sexing of a pre-implantation bovine embryo took place in the early 1990s using DNA extracted from a single biopsied blastomere from a 16–32-cell morula [116]. Similar studies followed, with blastomere biopsies from 8- to 16-cell [117] and 16- to 32-cell bovine morulae [118] and trophoblast cell biopsies from bovine blastocysts [119]. The first bovine-embryo-related study to utilize PCR for a non-sexing purpose studied the temporal expression dynamics of the gap junction gene Cx43 [120]. Since then, PCR (especially quantitative real-time PCR) has been used extensively to study various aspects of pre-implantation bovine embryos, including EGA [80,121], early lineage specification [80,122], and the effects of superovulation on embryos [123], and to validate gene expression data from dozens, if not hundreds, of microarray and RNA-seq studies.
An important prerequisite for successful PGD is the need to perform embryo biopsy without affecting embryo viability/competence. As reviewed elsewhere, pregnancy rates of 31–63% have been obtained by different groups after transferring biopsied pre-implantation bovine embryos [124]. Only a handful of studies have directly compared pregnancy rates resulting from transferring biopsied and intact embryos. One of them observed a ~15% reduction in the pregnancy rate when transferring biopsied blastocysts, while others did not (Table 6). This lack of agreement between studies could be attributed to varying conditions between the studies, such as the number of biopsied blastomeres, the gestation length at the time of pregnancy testing, and variations among recipients.

Table 6.
Comparison of pregnancy rates resulting from transfer of biopsied and non-biopsied bovine embryos.
3.2.2. Omics-Based Embryo Analyses
PGD is carried out to diagnose a specific trait of interest (e.g., sex, chromosomal aberration, or a genetic disease, esp. in humans). In contrast, with the advancements in the “omics”, pre-implantation embryos can be screened for entire genomes (also known as pre-implantation genomic/genetic screening: PGS), transcriptomes, epigenomes, proteomes, and metabolomes. These tools can provide quantitative and statistically accurate insights about not just a handful of molecules but hundreds, if not thousands, of potential biomarkers. This massive amount of data can also be used to understand functional relationships among identified markers, i.e., in cellular pathways/networks [128,129]. As such, omics-related studies have been carried out extensively over the last three decades in search of that gold standard test for the pre-transfer evaluation of embryos and in an attempt to further understand pre-implantation embryo development. The following sections examine the various types of studies that have taken place in the “omics” related to pre-implantation bovine embryos.
Genomic Studies
Genomic selection is widely used in dairy [130] and beef [131] breeding programs today. Genomic estimated breeding values (GEBVs) are currently calculated after the birth of calves [132]. This practice creates some degree of wastage in the form of calves that do not meet the expected minimum genetic merit/GEBV and all the resources that have gone into bringing such calves to life, e.g., the dams/surrogates that gestated them. These wastages can be minimized by using pre-implantation genomic screening (PGS) to screen blastocysts for favorable health, production, and reproduction traits prior to their transfer to a recipient [125,133,134]. Such an approach will enable the transfer of only the best embryos, not only avoiding the previously mentioned wastages but also improving the selection intensity and shortening the generation interval, leading to faster genetic gains [124,125,132]. Large-scale commercial applications of PGS are possible today due to the availability of affordable single-nucleotide polymorphism (SNP) chips [124,132,135] and GEBVs with reliabilities approaching 70% [125].
Genomic studies using SNP genotyping have been carried out in pre-implantation bovine embryos mainly to test for aneuploidy [132,135,136,137], chromosomal aberrations [136,138], and karyomapping [135,139] to date. After identifying euploid embryos for transfer (from karyomapped IVP embryos), live calves were born, demonstrating that genomic/SNP studies indeed have ground-level applications [135].
Microarray for Transcriptome Profiling
Genetic evaluations of pre-implantation embryos that started in the form of PGD evolved to high-throughput methods that could rapidly screen entire transcriptomes around the dawn of the 21st century. These transcriptome-wide studies not only allow the study of many thousands of genes simultaneously but also help make biological sense of the underlying pathophysiology by identifying biological pathways or networks that are differentially affected between groups of samples [129].
The very first transcriptome-wide studies involving bovine embryos were carried out using suppression-subtractive hybridization (SSH) in 2002–2004 [140,141]. The first microarray study on bovine embryos was reported in 2004 [142]. The research group compared the transcriptomes of IVD and IVP blastocysts using a custom-made bovine microarray. The next decade saw several dozen microarray studies carried out to study various aspects of pre-implantation bovine embryo development using several commercial and homemade cDNA and oligonucleotide microarray platforms (Table 7).

Table 7.
Applications of microarray technology in bovine embryo evaluation and analysis.
In some of these studies, RNA was extracted from whole embryos [142,143,144], making it a useless approach for pre-transfer embryo evaluation. Nevertheless, such studies are still useful for further studying pre-implantation embryo development from a research perspective. However, many other microarray studies have been carried out by extracting RNA from biopsies of ~30–40% of the intact embryo. During one such study, the remaining 60–70% of the embryos were allowed to expand in vitro, and the resulting demi-blastocysts were transferred to recipients. A 30% pregnancy rate was achieved in this study. Further, by comparing the transcriptomes of blastocysts that succeeded in a live birth vs. those that did not, they found potential candidate genes that are associated with the post-transfer pregnancy outcome [145,146]. Such gene expression signatures may serve as markers for evaluating pre-transfer embryos. Therefore, microarray technology indeed has the potential to be used in the pre-transfer evaluation of embryos in the dairy and beef industries.
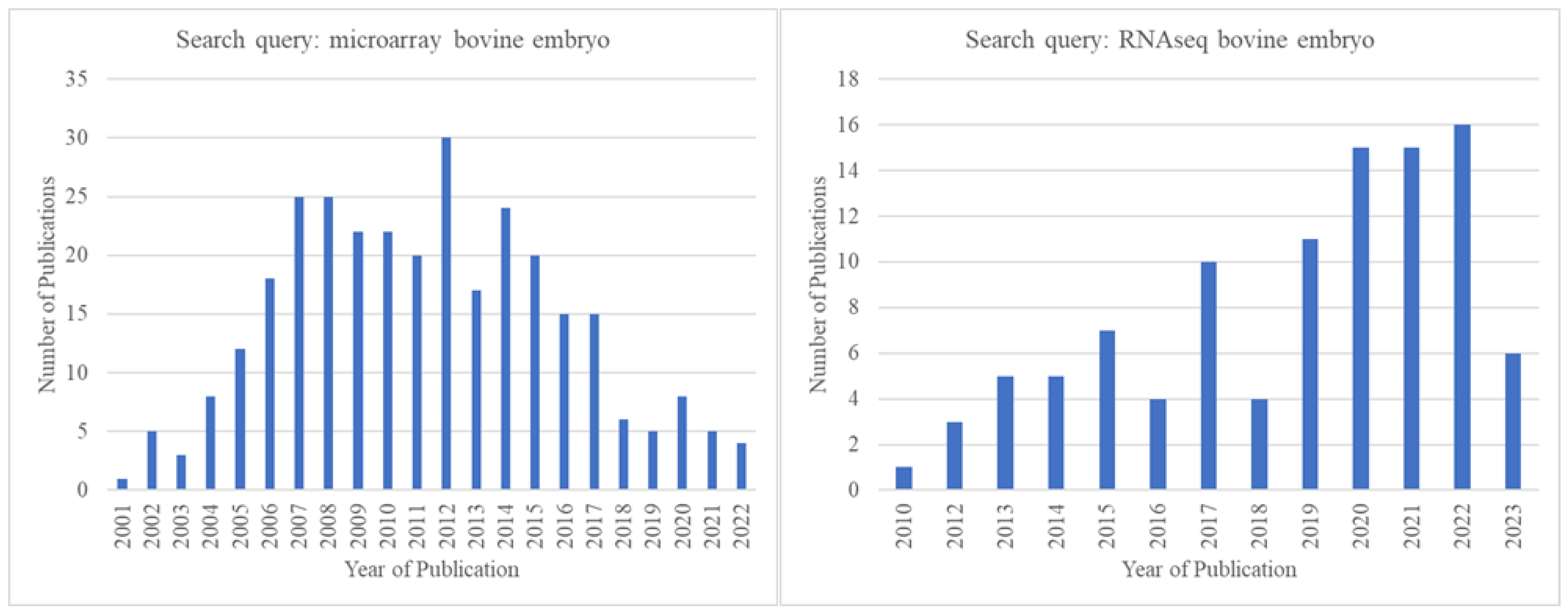
With the advent of RNA-sequencing (RNA-seq) technology, a transcription profiling approach superior to microarrays in many ways [176,177], the popularity of microarrays gradually started decreasing. There seemed to be an obvious shift to RNA-seq from microarrays starting in ~2012, even among those engaged in pre-implantation bovine embryo research. This is clearly reflected in the publication record on Pubmed.gov (Figure 3).
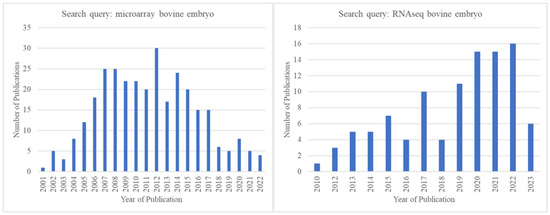
Figure 3.
Comparison of the number of publications identified in Pubmed.gov (https://pubmed.ncbi.nlm.nih.gov/, accessed on 15 May 2023) by year for the search queries “microarray bovine embryo” and “RNA-seq bovine embryo” showing that after the year 2012, microarray-related publications declined, and RNA-seq-related publications increased.
RNA Sequencing (RNA-Seq) for Transcriptome Profiling
The very first RNA-seq study of bovine embryos was carried out in 2010 to compare the transcriptomes of healthy-looking and degenerated IVP blastocysts using the Genome Analyzer platform by Illumina [178]. Since then, several dozen RNA-seq studies have been carried out to study various aspects of pre-implantation bovine embryo development using several commercially available platforms (Table 8). Some of these studies used pooled RNA [25,178,179,180], some used RNA from individual embryos [181,182], and others used RNA from embryo biopsies [183,184], including from those as little as single blastomeres [185,186]. However, extracting minute quantities of RNA from single cells will require the RNA to be amplified, which can lead to undesirable complications.

Table 8.
Applications of RNA-sequencing technology in bovine embryo evaluation and analysis.
Transferring 12 demi-embryos remaining after biopsy for RNA extraction, Zolini, Block, 2020 [183], achieved 5 pregnancies (41.7%) at 60 days of gestation, demonstrating that blastocysts tested by RNA-seq can indeed be used for the pre-transfer evaluation of bovine embryos.
Proteomics
The embryonic proteome is generally considered a better reflection of the actual embryo dynamics than the transcriptome for two reasons. First, it is proteins, metabolites, and other small molecules that are usually the final players in cellular/physiological events. In contrast, transcripts are usually intermediate players. Second, embryos are transcriptionally quiescent during the first few cleavages until EGA takes place. During this phase, embryo development is mainly controlled by oocyte-derived RNAs and proteins [203,204]. In contrast to the relatively invasive methods of embryo evaluation discussed earlier (e.g., genomics, transcriptomics, and even certain types of microscopy), proteomic studies can be carried out relatively non-invasively by analyzing embryos’ secretomes present in the culture media.
Proteomic analyses of pre-implantation embryos have been carried out to study early lineage specification [204], EGA [203], stage-specific proteome dynamics during pre-implantation development [203,205], differences between IVP and IVD embryos [206], proteomes of the blastocoel fluid [207], primitive yolk sac fluid [208], and trophectoderm [209], and to identify embryotrophic factors in the secretome that can serve as biomarkers of high-quality embryos [210].
Traditionally, proteomic profiling was performed using 2D gel electrophoresis and Western blotting. However, modern studies use a variety of mass-spectrometry-based technologies, such as Matrix-Assisted Laser Desorption Ionization–Time-Of-Flight Mass Spectrometry (MALDI-TOF MS), Liquid Chromatography with Tandem Mass Spectrometry (LC-MS-MS), Nanoliquid Chromatography Coupled with Tandem Mass Spectrometry (nanoLC-MS/MS), and Nano-HPLC Tandem Mass Spectrometry.
Metabolic Profiling
The metabolome is a downstream representation of the transcriptome and proteome because metabolites are the final products of cellular functions mediated by the transcriptome and proteome. As such, a strong association has been observed between embryo physiology and its metabolic profile, prompting researchers to develop metabolomics-based biomarkers. Similar to proteomics, the metabolic profile of pre-implantation embryos can be analyzed in a highly non-invasive fashion, making it a powerful tool for pre-transfer or pre-cryopreservation embryo evaluation. The metabolic profile for a specific metabolite can be measured as the quantity (i) consumed/taken up or (ii) released by the embryo. The evaluation of the embryo microenvironment, i.e., spent culture media, can give a reasonably accurate picture of both of the above measurements for individual metabolites or the entire metabolome [10,23,211].
The very first report on measuring metabolites in bovine blastocysts goes back to 1988 [212] when a team of researchers measured glucose, pyruvate, and glutamine, substrates related to energy metabolism. Initial studies in the 1980s and the first half of the 1990s focused on substrates related to energy metabolism, such as glucose, pyruvate, and glutamine. Later on, researchers studied oxygen, too, as an indicator of overall metabolic activity because the generation of cellular energy via mitochondrial oxidative phosphorylation is oxygen-dependent [213,214]. Over the last decade, many other candidates have been identified and tested by various research groups (Table 9).
During the pre-2000 era, mostly fluorometric or reverse-phase HPLC assays were used to quantify individual metabolites, such as glucose, lactate, pyruvate [215], and amino acids [216]. In addition, tools such as nanorespirometers (to measure oxygen consumption; [217]), embryoscopes with built-in oxygen microsensors (to measure oxygen consumption; [218]), REDOX sensors (to measure the production of ROS; [218]), and multi-sensor systems that can measure oxygen, glucose, and lactate concentrations simultaneously [219] had also been developed/used to evaluate the metabolic profiles of bovine embryos.
With massive developments in biomedical technologies, these techniques were replaced by mass spectrometry (MS)-based metabolomics, which allows the mass detection of metabolites, i.e., the metabolome, concurrently (e.g., >10,000 metabolites; [220]). These processes typically consist of two steps: separation using chromatography, followed by detection using MS. Examples of such methodologies used to date are Ultra-High-Performance Liquid Chromatography–Time-of-Flight Mass Spectrometry (UHPLC-TOF MS; UHPLC-TOF MS; [220,221]), Gas Chromatography Quadrupole Time-of-Flight Mass Spectrometry (GC-qTOF; [12]), Fourier Transform Infrared Spectroscopy (FTIR; [222]), and Raman Spectroscopy [223]. In addition to MS-based techniques, high-resolution nuclear magnetic resonance (1H NMR) spectroscopy [224,225] has also been used for metabolomics studies of pre-implantation bovine embryos.
Metabolomics has been used not only to identify embryonic viability markers of bovine embryos but also to determine the sex of the embryo [220,221,222,224,226].

Table 9.
Metabolites analyzed in bovine embryos.
Table 9.
Metabolites analyzed in bovine embryos.
| Analyzed Metabolite or Indicator of Metabolism | Source/s |
|---|---|
| Glucose | [212,215,227,228,229,230] |
| Pyruvate | [212,215,225,228,229,231] |
| Lactate | [215,224,225,229,231] |
| Amino acids | [212,216,220,221,224,225,226,227,228,229,231,232,233] |
| Fatty acids | [221,234,235] |
| Oxygen | [217,218,229,236] |
| Reactive oxygen species (ROS) | [218] |
| Myo-inositol | [224,231] |
| Citrate | [224,231,233] |
| Formate | [224,231] |
| Prostaglandins | [221] |
| Biotin | [220] |
3.2.3. Nuclear Magnetic Resonance (NMR)
Nuclear magnetic resonance (NMR) is used extensively in hospitals and research centers for non-invasive in vivo diagnostic procedures (e.g., commonly known as MRI scanners). In contrast to the ionizing radiation of X-ray and computed tomography (CT scanning), which can be harmful to live cells, the non-ionizing radiation of NMR is considered harmless or minimally harmful to live cells and therefore can be used to identify and quantify biologically important markers in intact cells, embryos, extracts, or the secretome [237,238]. In fact, several research groups have already used NMR spectroscopy to discover metabolome-based biomarkers that can predict fetal sex and the future development of pre-implantation bovine embryos [224,225,231,238]. Once these biomarkers are identified by NMR, simple enzymatic assays can be developed to identify/quantify such markers without the need for sophisticated NMR equipment [237]. As such, there is great potential for metabolomic biomarkers, identified by NMR or other means, to have practical applications in non-invasive embryo evaluation in the bovine ET industry.
ART-related applications of NMR, including those used for the evaluation of embryos, have been reviewed in detail elsewhere [237].
3.2.4. Less Commonly Used Tools
In addition to the methods described above, pre-implantation bovine embryos have been studied using a variety of other tools too (Table 10).

Table 10.
Miscellaneous tools/techniques used to analyze bovine embryos.
4. Discussion
Nearly 1.5 million bovine embryos were transferred in 2021. Of these transfers, nearly 80% (~1.2 million) involved IVP embryos [2]. Despite the massive advancements in every aspect of IVP, only ~27% of them can be expected to produce a live calf [4]. While the causes of pregnancy failures are multi-factorial, part of the problem is the inadvertent transfer of low-quality embryos due to our incompetence in accurately distinguishing high-quality embryos from low-quality ones. The current method of choice for embryo evaluation, i.e., morphological evaluation using a stereomicroscope, is considered to be subjective and to have poor accuracy and reproducibility [1,5]. Over two decades of research has gone into discovering a replacement tool for manual morphological evaluation, yet nothing concrete has come out that is adoptable at the ground level.
4.1. Microscopic Techniques
Of the visualization methodologies discussed, stereomicroscopy is the most commonly used tool for the pre-transfer/pre-cryopreservation evaluation of bovine embryo/blastocyst quality. However, the methodology is plagued by human subjectivity and poor accuracy. Electron microscopy and fluorescent microscopy have contributed immensely to the current knowledge about pre-implantation bovine embryo ultrastructure and the intraembryonic/intracellular localization of mRNAs and proteins (through FISH, immunocytochemistry, etc.). However, because sample preparation procedures make the embryos non-viable, they are both invasive techniques of no value for live embryo evaluation.
GLIM is superior to the abovementioned microscopic techniques because it is label-free and non-invasive and captures real-time 3D images of thick embryos, overcoming multiple scattering. Additionally, it takes ~1 min for image acquisition and <5 min for the virtual reconstruction of the image. As such, GLIM can be used for live embryo evaluation without affecting their viability. However, the equipment is expensive and sophisticated and requires a high level of technical expertise to operate. Therefore, it is highly unlikely to have practical applications in the dairy or beef cattle industry in the immediate future.
TLM allows the temporospatial evaluation of embryos and thus can be used to study embryo morphokinetics. Several MKIs predictive of embryo viability/quality have been discovered using TLM; however, the procedure requires embryos to be cultured and tracked individually using expensive imaging equipment. Therefore, however powerful a tool it may be, TLM too is highly unlikely to be adopted by the masses in the immediate future.
4.2. Non-Microscopic Techniques
The most common omics-based methodologies that have been used to study pre-implantation bovine embryo development to date are related to transcriptomics and metabolomics. Relatively few studies have been carried out using SNPs, proteomics, and epigenomics. There is no doubt that many of the discussed “omics” technologies have succeeded in discovering embryo viability/quality markers. However, when it comes to in-field applications of these technologies, they pose more questions than they provide answers. For example, if transcriptomics were to be adopted for in-field embryo evaluations, would it be realistic to biopsy and extract RNA from 2 million embryos (the approximate annual cattle embryo production) and then to run 2 million microarrays/RNA-seq? Would these steps take place at the embryo production/collection facilities? Or would embryos get transported to a different facility? If they are transported out, embryos may need to be frozen for transporting and thawed out before testing. Because there will be an interval of several days between the biopsy and the availability of results after bioinformatic analyses, embryos will need to be refrozen, and the ones that pass the tests will need to be re-thawed before ET. It is well known that freezing and thawing, let alone refreezing and re-thawing, can have undesirable effects on embryo quality/viability [263]. Compared to transcriptomics testing, proteomics and metabolomics are considered non-invasive because it is the culture medium that is analyzed and not the embryo per se. However, these require embryos to be cultured individually for their individual secretomes to be analyzed. If proteomics or metabolomics were to be adopted at the ground level, would it be practically possible to culture two million embryos individually using WOW systems? Additionally, even if this mammoth feat could be accomplished, the same set of challenges as with transcriptomics will need to be dealt with.
Therefore, it is clear that while these tools are extremely powerful and useful for understanding pre-implantation embryo development/physiology, it is almost impossible for them to have direct in-field applications in live embryo evaluation.
4.3. The Ideal Embryo Evaluation Tool
One of the key characteristics of the ideal embryo evaluation tool would be its adoptability by the thousands of embryo-producing/collecting facilities. Such a tool should fulfill most, if not all, of the following seven criteria:
- The method should provide accurate results;
- It should be non-invasive;
- It should be objective (minimize human subjectivity);
- It should be low-cost (initial and ongoing costs);
- It should be technically simple enough to be carried out at the embryo production facility itself;
- The evaluation should be completed by blastulation so that high-quality blastocysts can be immediately identified for direct transfer, without a lag period like in the case of omics-based techniques;
- The results should be available within hours of sample collection so that a second freeze–thaw cycle can be avoided.
Based on Table 11, which compares the common tools of embryo evaluation according to the above criteria, none of the tools fulfill all criteria. However, TLM and AI-based microscopy satisfy the highest number of favorable criteria (green boxes).

Table 11.
Comparison of the different tools available for embryo evaluation (green indicates a favorable feature, whereas orange indicates an unfavorable feature).
TLM fulfills a criterion that others do not: i.e., TLM can complete the embryo evaluation by the time that blastulation takes place by virtue of analyzing the morphokinetics of the first few cleavages. TLM studies have been used to demonstrate that MKIs such as the timing of the first cleavage, the number of blastomeres at the first cleavage, and the number of blastomeres at the onset of the lag phase have a strong correlation with post-transfer pregnancy rates [94,95]. These pre-blastulation observations have even led some to argue that MKIs can replace the IETS’ morphology-based grading system [81]. However, the major obstacle to using MKIs is the need to keep track of individual embryos during culture using WOW systems and sophisticated and expensive TLM imaging equipment. Therefore, we emphasize the need for future research to focus on designing affordable imaging alternatives that can be incorporated into existing incubators. A standalone camera or a smartphone that can be placed inside the incubator and is capable of communicating with a computer via WiFi may suffice for this purpose. For example, de Souza Ciniciato, Takahashi, 2017 [255] and Guilherme, Pronunciate, 2018 [252], used a smartphone to capture digital images of blastocysts with limited success. Considering that a successful embryo evaluation method will need to be adopted by the masses and that everybody has access to smartphones nowadays, we believe that further research needs to be carried out to understand how image capture using smartphones can be used in embryo evaluation, especially in TLM. Smartphone-compatible macro lenses claiming up to 400× magnification and timer-based photography applications are available on the market that may suffice for this type of relatively simple application. Smartphone-based biomedical applications are currently being used in various diagnostic, histopathological, and microbiological applications [264,265,266].
AI-based automated microscopy has been used to process and analyze images in an objective and automated fashion. Using artificial neural networks (ANNs), genetic algorithms (GAs), and automated image processing, embryo quality was classified with a high success rate [1,253]. We believe that this technology has a high potential for in-field embryo evaluation considering its benefits outlined in Table 11. It is well established that subtle variabilities in color, brightness, shape, and features escape the human eye [267] simply due to limitations in the physiological capacity of the human eye and variabilities in the embryologist’s accuracy, experience, and mood [7]. Automated image processing with objective image analysis, aided by machine learning, can overcome these limitations of the human eye.
Certain commercial TLM systems already make use of automatized image processing to a certain degree. We believe that if AI-based evaluation can be combined with TLM in a manner affordable to the masses, it has the potential to be used for in-field embryo evaluation.
4.4. Application of Omics-Based Tools in Field Conditions
In contrast to AI-based microscopy and TLM, omics-based technologies have major limitations that prohibit the direction application of these tools under field conditions, e.g., the need for technical expertise for operation and the need for additional freeze–thaw cycles. However, this does not mean that omics-based methods, GLIM, or NMR cannot be used for in-field embryo evaluation. Even though they cannot be used directly in-field, they can be used indirectly to narrow down embryo viability biomarkers, which may be detected in-field using simpler methodologies. For example, using mass-spectrometry-based proteomics screening of culture media, Raes, Wydooghe, 2023 [210], found that media conditioned by excellent- and good-quality bovine embryos, but not poor-quality embryos, contain cathepsin-L. If cathepsin-L is to be adopted as a bovine embryo viability/quality biomarker for embryo evaluation, mass spectrometry need not be carried out at the ground level. Instead, a simple protein detection methodology such as ELISA, which is non-invasive, simple, affordable, and adoptable by the masses, could be developed for the in-field detection of cathepsin-L. Alternatively, culture media could be developed that change color in the presence of high concentrations of cathepsin-L.
The majority of studies carried out to date have limited their objectives to finding an association between biomarker signatures and blastocyst quality. Only a handful of studies have gone on to correlate biomarker signatures with pregnancy outcomes. We strongly recommend that future studies emphasize investigating correlations between pre-implantation embryo biomarker signatures and pregnancy outcomes. This applies not only to omics-based biomarkers but also to other tools, including microscopic techniques.
5. Conclusions
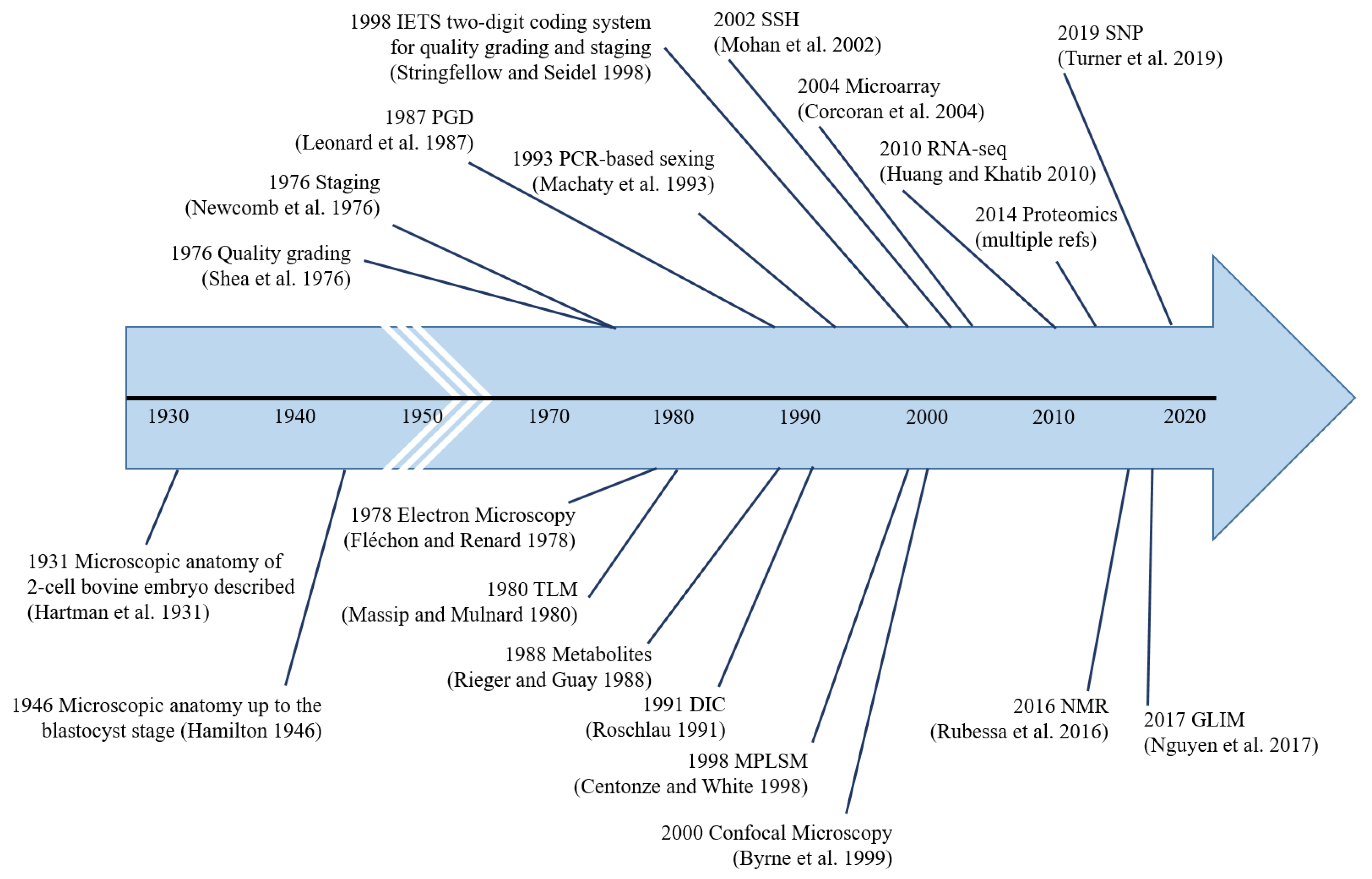
Pre-implantation bovine embryo evaluation has come a long way, from simple microscopic analysis in 1931 to PCR-based analyses in the 1990s to omics-based analyses in the 21st century (Figure 4). The ideal embryo evaluation tool should be not only accurate, objective, non-invasive, and affordable but also simple enough to be adopted by the thousands of embryo production/collection facilities worldwide. As such, under the current circumstances, it is highly unlikely that transcriptomics, proteomics, metabolomics, GLIM, NMR, etc., can be directly used for in-field embryo evaluation. However, biomarkers identified by these methodologies could be used for in-field embryo evaluation by devising simple, affordable benchtop techniques. Further research on TLM and AI-based automated image processing is warranted to make them more affordable (esp. TLM), as we see great potential for these methodologies to be adopted by the masses, provided they can be made affordable.
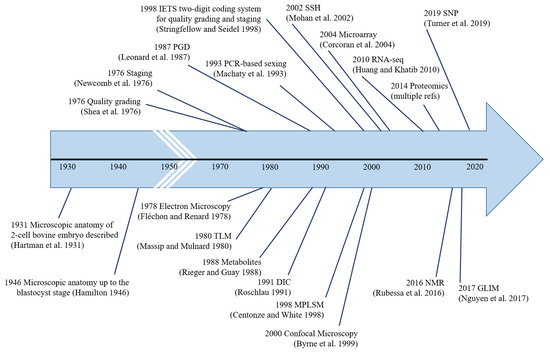
Figure 4.
Major milestones in pre-implantation bovine embryo analysis. The first instance of the use of each methodology is indicated, i.e., microscopic anatomy [13,14], quality grading [16], staging [21], electron microscopy [41], time-lapse microscopy [92], genetic-testing [113], metabolite-testing [212], differential interference contrast microscopy [30], PCR-based sexing [116], IETS two-digit coding system [20], multi-photon laser scanning microscopy [85], confocal microscopy [71], suppression subtractive hybridization [140], microarray [142], RNA-seq [178], proteomics, nuclear magnetic resonance [231], gradient light interference microscopy [58], and single nucleotide polymorphisms [139].
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ani13132102/s1: Figures S1–S19. Figure S1. 100× DIC. Stage 5 Grade 1 embryo. Early blastocyst collected day 7 post onset of estrus. Notice the presence of blastomeres immediately inside the VM causing an irregular bulge, or bumpy edge to the VM. The inner cell mass plus the blastocoele and VM is collectively called the embryo proper. Compare the bulges on the VM of this early blastocyst to the very smooth non-bulging VM of the unfertilized ova (UFO). [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S2. 100× DIC. Same embryo as slide #4 and #5, but arrows point to individual blastomeres. Also, one fused blastomere is outlined in yellow[Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S3. 400× DIC. Stage 4, Grade 1. This is an anatomically near perfect day 7 morula. There are very few, if any, extruded blastomeres outside the VM. The differentiating factor between this and a UFO is the bulging of the blastomeres in this embryo causing the VM to be irregular, yet contiguous and visible. Close inspection of the embryo proper allows one to see a large mass of fused blastomeres. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S4. 400× BF. Stage 4, Grade 2. Arguably, this embryo could be considered a grade 1 instead of a grade 2. The main mass (circled - see next slide) has a very well defined VM covering a healthy group of well fused blastomeres. In the visible plane there are two large extruded blastomeres. Rolling the embryo revealed two more smaller ones on the back side. This is a good example of the subjectivity involved in grading embryos. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S5. 100× DIC. Stage 4, Grade 3. This morula has several extruded unfused cells (arrows) in this plane, plus a few others as it was observed while rolling. The healthy fused mass in the middle represents about half of the total blastomeres. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S6. 200× BF. Stage 5 Grade 1. The arrow points to the "hollow" early developing blastocoele cavity. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S7. 400× DIC. Stage 5, Grade 3. The viable cells in this embryo have differentiated and formed a blastocoele (lower left part of encircled cell mass–see next slide–hollow cavity). The remainder of the cells are unfused and dead. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S8. 200× DIC. Stage 6, Grade 1. The yellow arrow points to the blastocoele cavity. The lighter colored blastocoele has slightly more total volume than the inner cell mass. That is the standard for classifying the embryo as a late-stage blastocyst (stage 6). The blue arrow points to the trophoblast. The red arrow points to the inner cell mass. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S9. 100× DIC. Stage 6, Grade 1. The blastocoele is about twice the size of the inner cell mass. The embryo proper has grown to the point that the VM now contacts the zona, thus eliminating a visible perivitelline space. However, the zona has not begun to expand geometrically and thin, thus this embryo is classified as a Stage 6 instead of Stage 7 (expanded blastocyst). One important point should be noted about embryos that have no perivitelline space–many of these embryos will be classified as Grade 1’s because any extruded blastomeres are flattened between the VM and the zona, and are, therefore often difficult to visualize. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S10. 100× DIC. Stage 6, Grade 2. Although the embryo proper has no imperfections, the zona is severely cracked and torn at 9 o’clock resulting in a down Grade from a 1 to a 2. This embryo would not qualify for export. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S11. 100× DIC. Stage 6 (arguably a stage 5), Grade 3. When this embryo was rolled during evaluation it was easier to see that the blastocoele (circle–see next slide) was slightly larger than the inner cell mass. Also, about half of the total cell mass was dead making the embryo a Grade 3. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S12. 100× DIC. Stage 7, Grade 1. The blastocoele represents about 70% of the embryo proper, and the inner cell mass (yellow arrow) is relatively smaller (about 30% of the embryo proper). The embryo proper has expanded all the way to the zona, and the zona is stretching/thinning to accommodate the growth. A thin layer of trophoblast extends from the corners of the inner cell mass and extends along the entire outer border of the blastocoele and approximates the inner border of the zona. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S13. 400× BF. Hatching blastocyst, Stage 8, Grade 1. The zona has cracked (6 to 8 o’clock) under the pressure of growth by the blastocyst. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S14. 400× BF. Stage 8, Grade 1. This is a hatched blastocyst in almost perfect condition. Normally, the ratio of blastocoele to inner cell mass would be greater than in this embryo. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S15. 100× DIC. #1 is an empty zona. See tear at 9 o’clock. Notice how the zona wall is thinner in #1 as compared to #2 and #3 (both stage 7, grade 1). #4 is a hatched blastocyst (Stage 8, Grade 1) without a zona. #1 is likely the zona that belongs to #4. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S16. 100× DIC. The three embryos with yellow arrows are Stage 7, Grade 2s. Notice the brown colored granular appearance of the blastocoele cavities of those embryos compared to the expanded blastocysts with translucent (green arrows) in this microphotograph. The granulation is an artifact of cellular debris along with dead and degenerated cells trapped tightly between the trophoblast and the zona. The green arrows point to Grade 1 embryos. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S17. 100× DIC. #1 = Stage 7, Grade 1 (7-1). #2 = 6-1, #3 = 5-1, #4 = 6-1. #5 = 6-1, #6 = 6-1,. #7 = 7-1 (notice diameter compared to bordering embryos), #8 = 5-1. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S18. 40× BF. The red arrows point to Stage 7, Grade 1 embryos. The green arrows point to Stage 6, Grade 1 embryos. The yellow arrows represent Stage 5, Grade 1 embryos. The blue arrow points to Stage 4, Grade 1s. The orange dashed arrows point to fragmented UFOs. Any embryo with a cracked zona downgrades from a Grade 1 to a Grade 2. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023]. Figure S19. 40× BF. #1 = Stage 6, Grade 1. #2 and #3 = Stage 7, Grade 1. #4 - Stage 6, Grade 1. The blastocoele appears to be smaller than the inner cell mass (ICM) in this view, but when rolled, the blastocoele is larger than the ICM. #5 = Stage 7, Grade 1. Notice the large diameter, plus the thinning of zona due to physical expansion of growth. #6 = Stage 7, Grade 1 (arguably a stage 6). #7 = Stage 6, Grade 1 (arguably a stage 5). #8 = Stage 7, Grade 1. #9 and #10 = Stage 5, Grade 1. #11 and #12 = Stage 7, Grade 1. #13 = Stage 5, Grade 1 (arguably a stage 6). #14 = Stage 4, Grade 2 (arguably a grade 1). 15 = Stage 5, Grade 1. #16 = Stage 7, Grade 1. #17 = Stage 4, Grade 3. The VM is very ill defined in this embryo. The VM has a "frayed" appearance from about 12 to 6 o’clock. There are most likely a few viable fused cells in the main mass, but many of the peripheral cells are degenerated. #18 = Stage 4, Grade 2. It has dead blastomeres at 6, 7, and 12 o’clock. #19 = Stage 5, Grade 1 (arguably a stage 6). #20 = Stage 6, Grade 1. #21 = Stage 7, Grade 1 (arguably stage 6). #22 = Stage 5, Grade 1. #23, 24, and 25 = Stage 4, Grade 1. #26 = Stage 4, Grade 2. The shape of the embryo proper is geometrically non-spherical. #27 = Stage 4, Grade 2 (arguably a stage 5) due to a cracked zona at 3 o’clock. #28 = Stage 7, Grade 1. #29 = Stage 6, Grade 1. This embryo is slightly oval shaped, but has no other flaws. #30 = Stage 7, Grade 1. #31 = Stage 6, Grade 1. #32 = Stage 7, Grade 1. #33 = Stage 6, Grade 1. #34 = Stage 4, Grade 1. #35 = Stage 7, Grade 1. #36 = Stage 7, Grade 1 (arguably stage 6). #37 = Stage 4, Grade 2 (cracked zona). This embryo could be a Stage 7 with a collapsed blastocoele cavity due to physical forces during the embryo collection procedure. The diameter of the zona is noticeably larger than that of embryo 34. The crack in the zona is at about 7 o’clock. #38 = Stage 7, Grade 1. #39 = Stage 5, Grade 1. #40 = Stage 6, Grade 1. [Figure and caption provided with permission of the International Embryo Technology Society (IETS) 2023].
Author Contributions
Writing—original draft preparation, R.A.C.R., P.V.M., E.A.B., K.W., D.J.M. and M.B.W.; writing—review and editing, R.A.C.R., M.B.W., P.V.M. and D.J.M.; supervision, M.B.W.; project administration, M.B.W.; funding acquisition, M.B.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by the USDA Multistate Project W-4171 #ILLU-538-947 and the Ross Foundation #635148.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
As this is a review article, all data used in the preparation of this manuscript are freely available to the public.
Acknowledgments
This manuscript is dedicated to George E. Seidel, Jr. His vision started an industry, and his ideas helped the ET industry grow and prosper. Seidel’s passion for the pursuit of knowledge and his students has left an everlasting legacy to humankind.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rocha, J.C.; Passalia, F.J.; Matos, F.D.; Takahashi, M.B.; Maserati, M.P., Jr.; Alves, M.F.; de Almeida, T.G.; Cardoso, B.L.; Basso, A.C.; Nogueira, M.F.G. Automatized image processing of bovine blastocysts produced in vitro for quantitative variable determination. Sci. Data 2017, 4, 170192. [Google Scholar] [CrossRef]
- Viana, J. 2021 Statistics of embryo production and transfer in domestic farm animals. Embryo Technol. Newsl. 2022, 40, 19. [Google Scholar]
- Marsico, T.; De Camargo, J.; Valente, R.S.; Sudano, M.J. Embryo competence and cryosurvival: Molecular and cellular features. Anim. Reprod. 2019, 16, 423–439. [Google Scholar] [CrossRef]
- Ealy, A.D.; Wooldridge, L.K.; McCoski, S.R. Board Invited Review: Post-transfer consequences of in vitro-produced embryos in cattle. J. Anim. Sci. 2019, 97, 2555–2568. [Google Scholar] [CrossRef]
- Farin, P.; Britt, J.; Shaw, D.; Slenning, B. Agreement among evaluators of bovine embryos produced in vivo or in vitro. Theriogenology 1995, 44, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Hansen, P.J. The incompletely fulfilled promise of embryo transfer in cattle—Why aren’t pregnancy rates greater and what can we do about it? J. Anim. Sci. 2020, 98, skaa288. [Google Scholar] [CrossRef] [PubMed]
- Lindner, G.M.; Wright, R.W. Bovine embryo morphology and evaluation. Theriogenology 1983, 20, 407–416. [Google Scholar] [CrossRef]
- Schneider, H.; Castleberry, R.; Griffin, J. Commercial aspects of bovine embryo transfer. Theriogenology 1980, 13, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Tervit, H.; Cooper, M.; Goold, P.G.; Haszard, G. Non-surgical embryo transfer in cattle. Theriogenology 1980, 13, 63–71. [Google Scholar] [CrossRef]
- Donnay, I. Metabolic Markers of Embryo Viability. In Assessment of Mammalian Embryo Quality: Invasive and Non-Invasive Techniques; Van Soom, A., Boerjan, M., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 57–94. [Google Scholar]
- Bó, G.; Mapletoft, R. Evaluation and classification of bovine embryos. Anim. Reprod. (AR) 2018, 10, 344–348. [Google Scholar]
- Gomez, E.; Canela, N.; Herrero, P.; Cereto, A.; Gimeno, I.; Carrocera, S.; Martin-Gonzalez, D.; Murillo, A.; Muñoz, M. Metabolites Secreted by Bovine Embryos In Vitro Predict Pregnancies That the Recipient Plasma Metabolome Cannot, and Vice Versa. Metabolites 2021, 11, 162. [Google Scholar] [CrossRef]
- Hartman, C.G.; Lewis, W.H.; Miller, F.W.; Swett, W.W. First findings of tubal ova in the cow, together with notes on oestrus. Anat. Rec. 1931, 48, 267–275. [Google Scholar] [CrossRef]
- Hamilton, W.J.; Laing, J.A. Development of the egg of the cow up to the stage of blastocyst formation. J. Anat. 1946, 80, 194. [Google Scholar] [PubMed]
- Seidel, G.E. Critical Review of Embryo Transfer Procedures with Cattle. In Fertilization and Embryonic Development In Vitro; Mastroianni, L., Biggers, J.D., Eds.; Springer: Boston, MA, USA, 1981; pp. 323–353. [Google Scholar]
- Shea, B.; Hines, D.; Lightfoot, D.; Ollis, G.; Olson, S. The transfer of bovine embryos. In Agriculture Research Seminar; Commission of the European Communities: Brussels, Belgium, 1976; pp. 145–152. [Google Scholar]
- Elsden, R.; Nelson, L.; Seidel, G. Superovulating cows with follicle stimulating hormone and pregnant mare’s serum gonadotrophin. Theriogenology 1978, 9, 17–26. [Google Scholar] [CrossRef]
- Shea, B. Evaluating the bovine embryo. Theriogenology 1981, 15, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Newcomb, R.; Rowson, L.E.A. Conception rate after uterine transfer of cow eggs, in relation to synchronization of oestrus and age of eggs. Reproduction 1975, 43, 539–541. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stringfellow, D.A.; Seidel, S.M. Manual of the International Embryo Transfer Society; The Society: Burlington, ON, Canada, 1998. [Google Scholar]
- Newcomb, R.; Rowson, L.; Trounson, A. The entry of superovulated eggs into the uterus. In Egg Transfer in Cattle; Commission of the European Communities: Brussels, Belgium, 1976; pp. 1–15. [Google Scholar]
- Rocha, J.C.; Passalia, F.J.; Matos, F.D.; Maserati, M.P., Jr.; Alves, M.F.; De Almeida, T.G.; Cardoso, B.L.; Basso, A.C.; Nogueira, M.F.G. Methods for assessing the quality of mammalian embryos: How far we are from the gold standard? JBRA Assist. Reprod. 2016, 20, 150–158. [Google Scholar] [CrossRef]
- Donnay, I.; Partridge, R.J.; Leese, H.J. Can embryo metabolism be used for selecting bovine embryos before transfer? Reprod. Nutr. Dev. 1999, 39, 523–533. [Google Scholar] [CrossRef]
- de México, A. Assessment of Bos taurus embryos comparing stereoscopic microscopy and transmission electron microscopy. J. Cell Anim. Biol. 2008, 2, 072–078. [Google Scholar]
- Driver, A.M.; Peñagaricano, F.; Huang, W.; Ahmad, K.R.; Hackbart, K.S.; Wiltbank, M.C.; Khatib, H. RNA-Seq analysis uncovers transcriptomic variations between morphologically similar in vivo- and in vitro-derived bovine blastocysts. BMC Genom. 2012, 13, 118. [Google Scholar] [CrossRef]
- Antony, P.P.M.A.; Trefois, C.; Stojanovic, A.; Baumuratov, A.S.; Kozak, K. Light microscopy applications in systems biology: Opportunities and challenges. Cell Commun. Signal. 2013, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, M.M.; Galina, C.S.; Merchant, H.; Montiel, F.; Canseco, R.; Márquez, Y.C. Comparison of stereoscopy, light microscopy and ultrastructural methods for evaluation of bovine embryos. Reprod. Domest. Anim. 2002, 37, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Nishiwaki, S.; Narumi, K.; Korenaga, T. Interference phase-contrast imaging technology without beam separation. Sci. Rep. 2019, 9, 1753. [Google Scholar] [CrossRef]
- Rubessa, M.; Wheeler, M.B. Label-free microscopy: A non-invasive new tool to assess gametes and embryo quality. Theriogenology 2020, 150, 241–246. [Google Scholar] [CrossRef]
- Roschlau, K. Gene transfer studies in cattle. J. Reprod. Fertil. 1991, 43, 293–295. [Google Scholar] [CrossRef]
- Bavister, B.D.; Rose-Hellekant, T.A.; Pinyopummintr, T. Development of in vitro matured/in vitro fertilized bovine embryos into morulae and blastocysts in defined culture media. Theriogenology 1992, 37, 127–146. [Google Scholar] [CrossRef]
- Elmileik, A.; Maeda, T.; Terada, T. Higher rates of development into blastocyst following the in vitro fertilization of bovine oocytes matured in a medium supplemented with the fluid from large bovine follicles. Anim. Reprod. Sci. 1995, 38, 85–96. [Google Scholar] [CrossRef]
- Tominaga, K.; Shimizu, M.; Ooyama, S.; Izaike, Y. Effect of lipid polarization by centrifugation at different developmental stages on post-thaw survival of bovine in vitro produced 16-cell embryos. Theriogenology 2000, 53, 1669–1680. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Yamashita, S.; Satoh, T.; Hoshi, H. Accumulation of cytoplasmic lipid droplets in bovine embryos and cryotolerance of embryos developed in different culture systems using serum-free or serum-containing media. Mol. Reprod. Dev. 2001, 61, 57–66. [Google Scholar] [CrossRef]
- Duszewska, A.; Kozikova, L.; Szydlik, H.; Cybulska, M.; Korwin-Kossakowski, M.; Połoszynowicz, J.; Wicińska, K.; Rosochacki, S.; Waś, B. The use of green fluorescent protein (GFP) to select bovine embryos. J. Anim. Feed. Sci. 2003, 12, 71–81. [Google Scholar] [CrossRef][Green Version]
- Rosochacki, S.J.; Kozikova, L.V.; Korwin-Kossakowski, M.; Matejczyk, M.; Połoszynowicz, J.; Duszewska, A. Noninvasive fluorescent screening of microinjected bovine embryos to predict transgene integration. Folia Biol. 2003, 51, 97–104. [Google Scholar]
- Neuber, E.; Luetjens, C.; Chan, A.; Schatten, G. Analysis of DNA fragmentation of in vitro cultured bovine blastocysts using TUNEL. Theriogenology 2002, 57, 2193–2202. [Google Scholar] [CrossRef]
- Pereira, R.; Baptista, M.; Vasques, M.; Horta, A.; Portugal, P.; Bessa, R.; e Silva, J.C.; Pereira, M.S.; Marques, C. Cryosurvival of bovine blastocysts is enhanced by culture with trans-10 cis-12 conjugated linoleic acid (10t,12c CLA). Anim. Reprod. Sci. 2007, 98, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.; Carvalhais, I.; Pimenta, J.; Baptista, M.; Vasques, M.; Horta, A.; Santos, I.; Marques, M.D.R.; Reis, A.; Pereira, M.S.; et al. Biopsied and vitrified bovine embryos viability is improved by trans10, cis12 conjugated linoleic acid supplementation during in vitro embryo culture. Anim. Reprod. Sci. 2007, 106, 322–332. [Google Scholar] [CrossRef]
- Malatesta, M. Transmission Electron Microscopy as a Powerful Tool to Investigate the Interaction of Nanoparticles with Subcellular Structures. Int. J. Mol. Sci. 2021, 22, 12789. [Google Scholar] [CrossRef]
- Flechon, J.-E.; Renard, J.-P. A scanning electron microscope study of the hatching of bovine blastocysts in vitro. Reproduction 1978, 53, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Massip, A. Ultrastructure of the cow blastocyst. J. Submicrose. Cytol. 1981, 13, 31–40. [Google Scholar]
- Crosier, A.E.; Farin, P.W.; Dykstra, M.J.; Alexander, J.E.; Farin, C.E. Ultrastructural Morphometry of Bovine Blastocysts Produced In Vivo or In Vitro. Biol. Reprod. 2001, 64, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Laurincik, J.; Thomsen, P.; Hay-Schmidt, A.; Avery, B.; Greve, T.; Ochs, R.; Hyttel, P. Nucleolar Proteins and Nuclear Ultrastructure in Preimplantation Bovine Embryos Produced In Vitro. Biol. Reprod. 2000, 62, 1024–1032. [Google Scholar] [CrossRef][Green Version]
- King, W.A.; Niar, A.; Chartrain, I.; Betteridge, K.J.; Guay, P. Nucleolus organizer regions and nucleoli in preattachment bovine embryos. Reproduction 1988, 82, 87–95. [Google Scholar] [CrossRef]
- Kopečný, V.; Fléchon, J.E.; Camous, S.; Fulka, J. Nucleologenesis and the onset of transcription in the eight-cell bovine embryo: Fine-structural autoradiographic study. Mol. Reprod. Dev. 1989, 1, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Baran, V.; Fléchon, J.-E.; Pivko, J. Nucleologenesis in the cleaving bovine embryo: Immunocytochemical aspects. Mol. Reprod. Dev. 1996, 44, 63–70. [Google Scholar] [CrossRef]
- Laurincik, J.; Kopecny, V.; Hyttel, P. Detailed analysis of pronucleus development in bovine zygotes in vivo: Ultrastructure and cell cycle chronology. Mol. Reprod. Dev. Inc. Gamete Res. 1996, 43, 62–69. [Google Scholar] [CrossRef]
- Laurincik, J.; Hyttel, P.; Baran, V.; Lucas-Hahn, A.; Eckert, J.; Pivko, J.; Niemann, H.; Brem, G.; Schellander, K. Pronucleus development and intranuclear bodies organization during progress of the first bovine embryonic cell cycle in vitro. Mol. Reprod. Dev. 1998, 50, 192–199. [Google Scholar]
- Crozet, N. Ultrastructural aspects of in vivo fertilization in the cow. Gamete Res. 1984, 10, 241–251. [Google Scholar] [CrossRef]
- Kuwayama, M.; Fujikawa, S.; Nagai, T. Ultrastructure of IVM-IVF Bovine Blastocysts Vitrified after Equilibration in Glycerol 1,2-Propanediol Using 2-Step and 16-Step Procedures. Cryobiology 1994, 31, 415–422. [Google Scholar] [CrossRef]
- Shamsuddin, M.; Rodriguez-Martinez, H. Fine Structure of Bovine Blastocysts Developed Either in Serum-Free Medium or in Conventional Co-Culture With Oviduct Epithelial Cells. J. Vet. Med. Ser. A 1994, 41, 307–316. [Google Scholar] [CrossRef]
- Abe, H.; Hoshi, H. Evaluation of Bovine Embryos Produced in High Performance Serum-Free Media. J. Reprod. Dev. 2003, 49, 193–202. [Google Scholar] [CrossRef]
- Sollecito, N.; Alves, R.; Beletti, M.; Pereira, E.; Miranda, M.; Silva, J.; Borges, A. Morphometry of bovine blastocysts produced in vitro in culture media with antioxidants cysteamine or oily extract of Lippia origanoides. Arq. Bras. Med. Vet. Zootec. 2021, 73, 799–811. [Google Scholar] [CrossRef]
- Fair, T.; Lonergan, P.; Dinnyes, A.; Cottell, D.C.; Hyttel, P.; Ward, F.A.; Boland, M.P. Ultrastructure of bovine blastocysts following cryopreservation: Effect of method of blastocyst production. Mol. Reprod. Dev. 2001, 58, 186–195. [Google Scholar] [CrossRef]
- Abe, H.; Otoi, T.; Tachikawa, S.; Yamashita, S.; Satoh, T.; Hoshi, H. Fine structure of bovine morulae and blastocysts in vivo and in vitro. Anat. Embryol. 1999, 199, 519–527. [Google Scholar] [CrossRef]
- Plante, L.; King, W.A. Light and electron microscopic analysis of bovine embryos derived byin Vitro andin Vivo fertilization. J. Assist. Reprod. Genet. 1994, 11, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Kandel, M.E.; Rubessa, M.; Wheeler, M.B.; Popescu, G. Gradient light interference microscopy for 3D imaging of unlabeled specimens. Nat. Commun. 2017, 8, 210. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, M.J.; Smith, I.; Parker, I.; Bootman, M.D. Fluorescence Microscopy. Cold Spring Harb. Protoc. 2014, 2014, pdb.top071795. [Google Scholar] [CrossRef]
- Specht, E.A.; Braselmann, E.; Palmer, A.E. A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging. Annu. Rev. Physiol. 2017, 79, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Sakatani, M.; Kobayashi, S.-I.; Takahashi, M. Effects of heat shock on in vitro development and intracellular oxidative state of bovine preimplantation embryos. Mol. Reprod. Dev. 2004, 67, 77–82. [Google Scholar] [CrossRef]
- Wang, L.-J.; Zhang, H.; Wang, Y.-S.; Xu, W.-B.; Xiong, X.-R.; Li, Y.-Y.; Su, J.-M.; Hua, S.; Zhang, Y. Scriptaid improves in vitro development and nuclear reprogramming of somatic cell nuclear transfer bovine embryos. Cell. Reprogram. 2011, 13, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Slimane, W.; Heyman, Y.; Lavergne, Y.; Humblot, P.; Renard, J.P. Assessing Chromosomal Abnormalities in Two-Cell Bovine In Vitro-Fertilized Embryos by Using Fluorescent In Situ Hybridization with Three Different Cloned Probes. Biol. Reprod. 2000, 62, 628–635. [Google Scholar] [CrossRef][Green Version]
- Daigneault, B.W.; Rajput, S.; Smith, G.W.; Ross, P.J. Embryonic POU5F1 is Required for Expanded Bovine Blastocyst Formation. Sci. Rep. 2018, 8, 7753. [Google Scholar] [CrossRef]
- Amos, W.B.; White, J.G.; Fordham, M. Use of confocal imaging in the study of biological structures. Appl. Opt. 1987, 26, 3239–3243. [Google Scholar] [CrossRef]
- White, J.G.; Amos, W.B.; Fordham, M. An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. J. Cell Biol. 1987, 105, 41–48. [Google Scholar] [CrossRef]
- Inoué, S. Handbook of Biological Confocal Microscopy; Pawley, J.B., Ed.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Jonkman, J.; Brown, C.M.; Wright, G.D.; Anderson, K.I.; North, A.J. Tutorial: Guidance for quantitative confocal microscopy. Nat. Protoc. 2020, 15, 1585–1611. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.J.; Perez, G.I.; Ko, T.; Yoo, M.S.; Cibelli, J.B. Full developmental potential of mammalian preimplantation embryos is maintained after imaging using a spinning-disk confocal microscope. Biotechniques 2006, 41, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Babbey, C.M.; Dunn, K.W. Performance comparison between the high-speed Yokogawa spinning disc confocal system and single-point scanning confocal systems. J. Microsc. 2005, 218, 148–159. [Google Scholar] [CrossRef]
- Byrne, A.; Southgate, J.; Brison, D.R.; Leese, H.J. Analysis of apoptosis in the preimplantation bovine embryo using TUNEL. Reproduction 1999, 117, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Matwee, C.; Betts, D.H.; King, W.A. Apoptosis in the early bovine embryo. Zygote 1999, 8, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Vanroose, G.; Nauwynck, H.; Van Soom, A.; Ysebaert, M.-T.; Charlier, G.; Van Oostveldt, P.; de Kruif, A. Structural Aspects of the Zona Pellucida of In Vitro-Produced Bovine Embryos: A Scanning Electron and Confocal Laser Scanning Microscopic Study. Biol. Reprod. 2000, 62, 463–469. [Google Scholar] [CrossRef]
- Pennetier, S.; Perreau, C.; Uzbekova, S.; Thélie, A.; Delaleu, B.; Mermillod, P.; Dalbiès-Tran, R. MATER protein expression and intracellular localization throughout folliculogenesis and preimplantation embryo development in the bovine. BMC Dev. Biol. 2006, 6, 26. [Google Scholar] [CrossRef]
- Hou, J.; Liu, L.; Lei, T.; Cui, X.; An, X.; Chen, Y. Genomic DNA methylation patterns in bovine preimplantation embryos derived from in vitro fertilization. Sci. China Life Sci. 2007, 50, 56–61. [Google Scholar] [CrossRef]
- Wang, L.; Feng, H.; Ma, Y.; Cang, M.; Li, H.; Yan, Z.; Zhou, P.; Wen, J.; Bou, S.; Liu, D. Expression of IGF receptors and its ligands in bovine oocytes and preimplantation embryos. Anim. Reprod. Sci. 2009, 114, 99–108. [Google Scholar] [CrossRef]
- Kuijk, E.W.; van Tol, L.T.A.; Van de Velde, H.; Wubbolts, R.; Welling, M.; Geijsen, N.; Roelen, B.A.J. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development 2012, 139, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Madeja, Z.E.; Sosnowski, J.; Hryniewicz, K.; Warzych, E.; Pawlak, P.; Rozwadowska, N.; Plusa, B.; Lechniak, D. Changes in sub-cellular localisation of trophoblast and inner cell mass specific transcription factors during bovine preimplantation development. BMC Dev. Biol. 2013, 13, 32. [Google Scholar] [CrossRef]
- Simmet, K.; Zakhartchenko, V.; Philippou-Massier, J.; Blum, H.; Klymiuk, N.; Wolf, E. OCT4/POU5F1 is required for NANOG expression in bovine blastocysts. Proc. Natl. Acad. Sci. USA 2018, 115, 2770–2775. [Google Scholar] [CrossRef] [PubMed]
- Khan, D.R.; Dubé, D.; Gall, L.; Peynot, N.; Ruffini, S.; Laffont, L.; Le Bourhis, D.; Degrelle, S.; Jouneau, A.; Duranthon, V. Expression of pluripotency master regulators during two key developmental transitions: Ega and early lineage specification in the bovine embryo. PLoS ONE 2012, 7, e34110. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Suzuki, R.; Furuta, N.; Suzuki, Y.; Kabe, K.; Tokoro, M.; Sugawara, A.; Yajima, A.; Nagasawa, T.; Matoba, S.; et al. Live-cell imaging of nuclear–chromosomal dynamics in bovine in vitro fertilised embryos. Sci. Rep. 2018, 8, 7460. [Google Scholar] [CrossRef] [PubMed]
- Mesalam, A.; Lee, K.-L.; Khan, I.; Chowdhury, M.M.R.; Zhang, S.; Song, S.-H.; Joo, M.-D.; Lee, J.-H.; Jin, J.-I.; Kong, I.-K. A combination of bovine serum albumin with insulin–transferrin–sodium selenite and/or epidermal growth factor as alternatives to fetal bovine serum in culture medium improves bovine embryo quality and trophoblast invasion by induction of matrix metalloproteinases. Reprod. Fertil. Dev. 2019, 31, 333–346. [Google Scholar] [CrossRef]
- Monteiro, C.A.S.; Chow, D.J.; Leal, G.R.; Tan, T.C.; Ferreira, A.M.R.; Thompson, J.G.; Dunning, K.R. Optical imaging of cleavage stage bovine embryos using hyperspectral and confocal approaches reveals metabolic differences between on-time and fast-developing embryos. Theriogenology 2020, 159, 60–68. [Google Scholar] [CrossRef]
- Squirrell, J.M.; Wokosin, D.L.; White, J.G.; Bavister, B.D. Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability. Nat. Biotechnol. 1999, 17, 763–767. [Google Scholar] [CrossRef]
- Centonze, V.E.; White, J.G. Multiphoton Excitation Provides Optical Sections from Deeper within Scattering Specimens than Confocal Imaging. Biophys. J. 1998, 75, 2015–2024. [Google Scholar] [CrossRef]
- You, S.; Tu, H.; Chaney, E.J.; Sun, Y.; Zhao, Y.; Bower, A.J.; Liu, Y.-Z.; Marjanovic, M.; Sinha, S.; Pu, Y.; et al. Intravital imaging by simultaneous label-free autofluorescence-multiharmonic microscopy. Nat. Commun. 2018, 9, 2125. [Google Scholar] [CrossRef]
- Yadlowsky, M.J.; Schmitt, J.M.; Bonner, R.F. Multiple scattering in optical coherence microscopy. Appl. Opt. 1995, 34, 5699–5707. [Google Scholar] [CrossRef] [PubMed]
- Rivenson, Y.; Ozcan, A. Deep learning accelerates whole slide imaging for next-generation digital pathology applications. Light. Sci. Appl. 2022, 11, 300. [Google Scholar] [CrossRef]
- Welte, M.A. As the fat flies: The dynamic lipid droplets of Drosophila embryos. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2015, 1851, 1156–1185. [Google Scholar] [CrossRef] [PubMed]
- Angel-Velez, D.; De Coster, T.; Azari-Dolatabad, N.; Fernández-Montoro, A.; Benedetti, C.; Pavani, K.; Van Soom, A.; Pascottini, O.B.; Smits, K. Embryo morphokinetics derived from fresh and vitrified bovine oocytes predict blastocyst development and nuclear abnormalities. Sci. Rep. 2023, 13, 4765. [Google Scholar] [CrossRef] [PubMed]
- Magata, F. Time-lapse monitoring technologies for the selection of bovine in vitro fertilized embryos with high implantation potential. J. Reprod. Dev. 2023, 69, 57–64. [Google Scholar] [CrossRef]
- Massip, A.; Mulnard, J. Time-lapse cinematographic analysis of hatching of normal and frozen--thawed cow blastocysts. Reproduction 1980, 58, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Massip, A.; Mulnard, J.; Vanderzwalmen, P.; Hanzen, C.; Ectors, F. The behaviour of cow blastocyst in vitro: Cinematographic and morphometric analysis. J. Anat. 1982, 134, 399–405. [Google Scholar]
- Sugimura, S.; Akai, T.; Hashiyada, Y.; Somfai, T.; Inaba, Y.; Hirayama, M.; Yamanouchi, T.; Matsuda, H.; Kobayashi, S.; Aikawa, Y.; et al. Promising System for Selecting Healthy In Vitro–Fertilized Embryos in Cattle. PLoS ONE 2012, 7, e36627. [Google Scholar] [CrossRef]
- Sugimura, S.; Akai, T.; Imai, K. Selection of viable in vitro-fertilized bovine embryos using time-lapse monitoring in microwell culture dishes. J. Reprod. Dev. 2017, 63, 353–357. [Google Scholar] [CrossRef]
- Hardin, J. Confocal and Multi-Photon Imaging of Living Embryos. In Handbook Of Biological Confocal Microscopy; Pawley, J.B., Ed.; Springer: Boston, MA, USA, 2006; pp. 746–768. [Google Scholar]
- Holm, P.; Shukri, N.; Vajta, G.; Booth, P.; Bendixen, C.; Callesen, H. Developmental kinetics of the first cell cycles of bovine in vitro produced embryos in relation to their in vitro viability and sex. Theriogenology 1998, 50, 1285–1299. [Google Scholar] [CrossRef]
- Majerus, V.; Lequarré, A.S.; Ferguson, E.M.; Kaidi, S.; Massip, A.; Dessy, F.; Donnay, I. Characterization of embryos derived from calf oocytes: Kinetics of cleavage, cell allocation to inner cell mass, and trophectoderm and lipid metabolism. Mol. Reprod. Dev. 2000, 57, 346–352. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoshioka, K.; Suzuki, C.; Iwamura, S. Effects of activin A and follistatin on developmental kinetics of bovine embryos: Cinematographic analysis in a chemically defined medium. J. Reprod. Fertil. 2000, 118, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Peippo, J.; Kurkilahti, M.; Bredbacka, P. Developmental kinetics of in vitro produced bovine embryos: The effect of sex, glucose and exposure to time-lapse environment. Zygote 2001, 9, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Holm, P.; Booth, P.J.; Callesen, H. Kinetics of early in vitro development of bovine in vivo- and in vitro-derived zygotes produced and/or cultured in chemically defined or serum-containing media. Reproduction 2002, 123, 553–565. [Google Scholar] [CrossRef]
- Lequarre, A.; Marchandise, J.; Moreau, B.; Massip, A.; Donnay, I. Cell Cycle Duration at the Time of Maternal Zygotic Transition for In Vitro Produced Bovine Embryos: Effect of Oxygen Tension and Transcription Inhibition. Biol. Reprod. 2003, 69, 1707–1713. [Google Scholar] [CrossRef][Green Version]
- Somfai, T.; Inaba, Y.; Aikawa, Y.; Ohtake, M.; Kobayashi, S.; Konishi, K.; Imai, K. Relationship between the length of cell cycles, cleavage pattern and developmental competence in bovine embryos generated by in vitro fertilization or parthenogenesis. J. Reprod. Dev. 2010, 56, 200–207. [Google Scholar] [CrossRef]
- Sugimura, S.; Akai, T.; Somfai, T.; Hirayama, M.; Aikawa, Y.; Ohtake, M.; Hattori, H.; Kobayashi, S.; Hashiyada, Y.; Konishi, K.; et al. Time-lapse cinematography-compatible polystyrene-based microwell culture system: A novel tool for tracking the development of individual bovine embryos. Biol. Reprod. 2010, 83, 970–978. [Google Scholar] [CrossRef]
- Magata, F.; Ideta, A.; Okubo, H.; Matsuda, F.; Urakawa, M.; Oono, Y. Growth potential of bovine embryos presenting abnormal cleavage observed through time lapse cinematography. Theriogenology 2019, 133, 119–124. [Google Scholar] [CrossRef]
- Magata, F.; Urakawa, M.; Matsuda, F.; Oono, Y. Developmental kinetics and viability of bovine embryos produced in vitro with sex-sorted semen. Theriogenology 2020, 161, 243–251. [Google Scholar] [CrossRef]
- Suzuki, R.; Okada, M.; Nagai, H.; Kobayashi, J.; Sugimura, S. Morphokinetic analysis of pronuclei using time-lapse cinematography in bovine zygotes. Theriogenology 2021, 166, 55–63. [Google Scholar] [CrossRef]
- Lechniak, D.; Sell-Kubiak, E.; Warzych, E. The metabolic profile of bovine blastocysts is affected by in vitro culture system and the pattern of first zygotic cleavage. Theriogenology 2022, 188, 43–51. [Google Scholar] [CrossRef]
- Yaacobi-Artzi, S.; Kalo, D.; Roth, Z. Association between the morphokinetics of in-vitro-derived bovine embryos and the transcriptomic profile of the derived blastocysts. PLoS ONE 2022, 17, e0276642. [Google Scholar] [CrossRef] [PubMed]
- Verlinsky, Y.; Pergament, E.; Strom, C. The preimplantation genetic diagnosis of genetic diseases. J. Assist. Reprod. Genet. 1990, 7, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Gardner, R.L.; Edwards, R.G. Control of the Sex Ratio at Full Term in the Rabbit by transferring Sexed Blastocysts. Nature 1968, 218, 346–348. [Google Scholar] [CrossRef] [PubMed]
- Herr, C.; Reed, K. Micronanipulation of bovine embryos for sex determination. Theriogenology 1991, 35, 45–54. [Google Scholar] [CrossRef]
- Leonard, M.; Kirszenbaum, M.; Cotinot, C.; Chesne, P.; Heyman, Y.; Stinnakre, M.; Bishop, C.; Delouis, C.; Vaiman, M.; Fellous, M. Sexing bovine embryos using Y chromosome specific DNA probe. Theriogenology 1987, 27, 248. [Google Scholar] [CrossRef]
- Bondioli, K.; Ellis, S.; Pryor, J.; Williams, M.; Harpold, M. The use of male-specific chromosomal DNA fragments to determine the sex of bovine preimplantation embryos. Theriogenology 1989, 31, 95–104. [Google Scholar] [CrossRef]
- Mullis, K.B. The unusual origin of the polymerase chain reaction. Sci. Am. 1990, 262, 56–65. [Google Scholar] [CrossRef]
- Machaty, Z.; Paldi, A.; Csaki, T.; Varga, Z.; Kiss, I.; Bárándi, Z.; Vajta, G. Biopsy and sex determination by PCR of IVF bovine embryos. Reproduction 1993, 98, 467–470. [Google Scholar] [CrossRef]
- Park, J.; Lee, J.; Choi, K.; Joung, S.; Kim, J.; Chung, G.; Jin, D.; Im, K. Rapid sexing of preimplantation bovine embryo using consecutive and multiplex polymerase chain reaction (PCR) with biopsied single blastomere. Theriogenology 2001, 55, 1843–1853. [Google Scholar] [CrossRef]
- Chrenek, P.; Boulanger, L.; Heyman, Y.; Uhrin, P.; Laurincik, J.; Bulla, J.; Renard, J.-P. Sexing and multiple genotype analysis from a single cell of bovine embryo. Theriogenology 2001, 55, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.M.; Hu, C.L.; Wang, C.H.; Hung, C.M.; Wu, H.K.; Choo, K.B.; Cheng, W.T.K. Gender determination in single bovine blastomeres by polymerase chain reaction amplification of sex-specific polymorphic fragments in the amelogenin gene. Mol. Reprod. Dev. 1999, 54, 209–214. [Google Scholar] [CrossRef]
- Wrenzycki, C.; Herrmann, D.; Carnwath, J.W.; Niemann, H. Expression of the gap junction gene connexin43 (Cx43) in preimplantation bovine embryos derived in vitro or in vivo. Reproduction 1996, 108, 17–24. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vigneault, C.; McGraw, S.; Massicotte, L.; Sirard, M.-A. Transcription Factor Expression Patterns in Bovine In Vitro-Derived Embryos Prior to Maternal-Zygotic Transition. Biol. Reprod. 2004, 70, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Perez, V.M.N.; Zhang, Y.; Hansen, P.J. Single-cell gene expression of the bovine blastocyst. Reproduction 2017, 154, 627–644. [Google Scholar] [CrossRef]
- Mundim, T.; Ramos, A.; Sartori, R.; Dode, M.; Melo, E.; Gomes, L.; Rumpf, R.; Franco, M. Changes in gene expression profiles of bovine embryos produced in vitro, by natural ovulation, or hormonal superstimulation. Genet. Mol. Res. 2009, 8, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Ponsart, C.; Le Bourhis, D.; Knijn, H.; Fritz, S.; Guyader-Joly, C.; Otter, T.; Lacaze, S.; Charreaux, F.; Schibler, L.; Dupassieux, D.; et al. Reproductive technologies and genomic selection in dairy cattle. Reprod. Fertil. Dev. 2014, 26, 12–21. [Google Scholar] [CrossRef]
- Mullaart, E.; Wells, D. Embryo biopsies for genomic selection. In Animal Biotechnology 2: Emerging Breeding Technologies; Springer: Berlin, Germany, 2018; pp. 81–94. [Google Scholar]
- de Sousa, R.V.; Cardoso, C.R.D.S.; Butzke, G.; Dode, M.A.N.; Rumpf, R.; Franco, M.M. Biopsy of bovine embryos produced in vivo and in vitro does not affect pregnancy rates. Theriogenology 2017, 90, 25–31. [Google Scholar] [CrossRef]
- Fisher, P.J.; Hyndman, D.L.; Bixley, M.J.; Oback, F.C.; Popovic, L.; McGowan, L.T.; Berg, M.C. Potential for genomic selection of bovine embryos. In Proceedings of the New Zealand Society of Animal Production; New Zealand Society of Animal Production: Christchurch, New Zealand, 2012; pp. 156–158. [Google Scholar]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef]
- Pas, M.F.W.T.; van Hemert, S.; Hulsegge, B.; Hoekman, A.J.W.; Pool, M.H.; Rebel, J.M.J.; Smits, M.A. A pathway analysis tool for analyzing microarray data of species with low physiological information. Adv. Bioinform. 2008, 2008, 719468. [Google Scholar] [CrossRef]
- Scott, B.; Haile-Mariam, M.; Cocks, B.; Pryce, J. How genomic selection has increased rates of genetic gain and inbreeding in the Australian national herd, genomic information nucleus, and bulls. J. Dairy Sci. 2021, 104, 11832–11849. [Google Scholar] [CrossRef]
- Retallick, K.J.; Lu, D.; Garcia, A.; Miller, S.P. Genomic selection in the US: Where it has been and where it is going? In Proceedings of the 12th World Congress on Genetics Applied to Livestock Production (WCGALP), Rotterdam, The Netherland, 3–8 July 2022; pp. 1795–1798. [Google Scholar]
- Silvestri, G.; Canedo-Ribeiro, C.; Serrano-Albal, M.; Labrecque, R.; Blondin, P.; Larmer, S.G.; Marras, G.; Tutt, D.A.; Handyside, A.H.; Farré, M.; et al. Preimplantation Genetic Testing for Aneuploidy Improves Live Birth Rates with In Vitro Produced Bovine Embryos: A Blind Retrospective Study. Cells 2021, 10, 2284. [Google Scholar] [CrossRef]
- Herrera, C. Clinical Applications of Preimplantation Genetic Testing in Equine, Bovine, and Human Embryos. J. Equine Veter. Sci. 2016, 41, 29–34. [Google Scholar] [CrossRef]
- Saadi, H.A.S.; Vigneault, C.; Sargolzaei, M.; Gagné, D.; Fournier, É.; de Montera, B.; Chesnais, J.; Blondin, P.; Robert, C. Impact of whole-genome amplification on the reliability of pre-transfer cattle embryo breeding value estimates. BMC Genom. 2014, 15, 889. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.J.; Silvestri, G.; Black, D.H.; Dobson, G.; Smith, C.; Handyside, A.H.; Sinclair, K.D.; Griffin, D.K. Karyomapping for simultaneous genomic evaluation and aneuploidy screening of preimplantation bovine embryos: The first live-born calves. Theriogenology 2018, 125, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Tutt, D.; Silvestri, G.; Serrano-Albal, M.; Simmons, R.; Kwong, W.; Guven-Ates, G.; Canedo-Ribeiro, C.; Labrecque, R.; Blondin, P.; Handyside, A.; et al. Analysis of bovine blastocysts indicates ovarian stimulation does not induce chromosome errors, nor discordance between inner-cell mass and trophectoderm lineages. Theriogenology 2020, 161, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, A.C.; Mullaart, E. Screening of in vitro-produced cattle embryos to assess incidence and characteristics of unbalanced chromosomal aberrations. JDS Commun. 2023, 4, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Masset, H.; Ding, J.; Dimitriadou, E.; Ardeshirdavani, A.; Debrock, S.; Tšuiko, O.; Smits, K.; Peeraer, K.; Moreau, Y.; Voet, T.; et al. Single-cell genome-wide concurrent haplotyping and copy-number profiling through genotyping-by-sequencing. Nucleic Acids Res. 2022, 50, e63. [Google Scholar] [CrossRef]
- Griffin, D.K.; Jturner, K.; Silvestri, G.; Smith, C.; Dobson, G.; Black, D.H.; Sinclair, K.D.; Handyside, A.H. The use of Karyomapping for genomic evaluation and PGT-A of preimplantation cattle embryos: The first live-born calves. Reprod. Biomed. Online 2019, 38, e54–e55. [Google Scholar] [CrossRef]
- Mohan, M.; Ryder, S.; Claypool, P.; Geisert, R.; Malayer, J. Analysis of Gene Expression in the Bovine Blastocyst Produced In Vitro Using Suppression-Subtractive Hybridization. Biol. Reprod. 2002, 67, 447–453. [Google Scholar] [CrossRef]
- Mohan, M.; Hurst, A.; Malayer, J. Global gene expression analysis comparing bovine blastocysts flushed on day 7 or produced in vitro. Mol. Reprod. Dev. 2004, 68, 288–298. [Google Scholar] [CrossRef]
- Corcoran, D.; Fair, T.; Rizos, D.; Smith, G.; Coussens, P.; Patel, O.; Ireland, J.; Boland, M.; Evans, A.; Lonergan, P. 216 Identification of Differentially Expressed Genes in Bovine Embryos Cultured In Vivo or In Vitro. Reprod. Fertil. Dev. 2005, 17, 259. [Google Scholar] [CrossRef]
- Corcoran, D.; Fair, T.; Park, S.; Rizos, D.; Patel, O.V.; Smith, G.W.; Coussens, P.M.; Ireland, J.J.; Boland, M.P.; Evans, A.C.O.; et al. Suppressed expression of genes involved in transcription and translation in in vitro compared with in vivo cultured bovine embryos. Reproduction 2006, 131, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Vallée, M.; Dufort, I.; Desrosiers, S.; Labbe, A.; Gravel, C.; Gilbert, I.; Robert, C.; Sirard, M.-A. Revealing the bovine embryo transcript profiles during early in vivo embryonic development. Reproduction 2009, 138, 95–105. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.; Hoelker, M.; Rings, F.; Salilew, D.; Jennen, D.; Tholen, E.; Sirard, M.-A.; Schellander, K.; Tesfaye, D. Large-scale transcriptional analysis of bovine embryo biopsies in relation to pregnancy success after transfer to recipients. Physiol. Genom. 2006, 28, 84–96. [Google Scholar] [CrossRef]
- Ghanem, N.; Wondim, D.S.; Gad, A.; Tesfaye, D.; Phatsara, C.; Tholen, E.; Looft, C.; Schellander, K.; Hoelker, M. Bovine blastocysts with developmental competence to term share similar expression of developmentally important genes although derived from different culture environments. Reproduction 2011, 142, 551–564. [Google Scholar] [CrossRef]
- Hoelker, M.; Rings, F.; Lund, Q.; Ghanem, N.; Phatsara, C.; Griese, J.; Schellander, K.; Tesfaye, D. Effect of the microenvironment and embryo density on developmental characteristics and gene expression profile of bovine preimplantative embryos cultured in vitro. Reproduction 2009, 137, 415–425. [Google Scholar] [CrossRef]
- Kepková, K.V.; Vodicka, P.; Toralova, T.; Lopatarova, M.; Cech, S.; Dolezel, R.; Havlicek, V.; Besenfelder, U.; Kuzmany, A.; Sirard, M.A.; et al. Transcriptomic analysis of in vivo and in vitro produced bovine embryos revealed a developmental change in cullin 1 expression during maternal-to-embryonic transition. Theriogenology 2011, 75, 1582–1595. [Google Scholar] [CrossRef]
- Labrecque, R.; Sirard, M.-A. Gene expression analysis of bovine blastocysts produced by parthenogenic activation or fertilisation. Reprod. Fertil. Dev. 2011, 23, 591–602. [Google Scholar] [CrossRef]
- Sudano, M.J.; Caixeta, E.S.; Paschoal, D.M.; Rascado, T.S.; Crocomo, L.F.; Maziero, R.R.; Magalhães, L.C.O.; Guastali, M.D.; Vergara, L.E.; Monteiro, B.A.; et al. Microarray Analysis of In Vitro-Produced Bovine Embryos with Phenazine Ethosulfate. Biol. Reprod. 2012, 87, 215. [Google Scholar] [CrossRef]
- Sudano, M.J.; Paschoal, D.M.; Caixeta, E.S.; Maziero, R.R.; Guastali, M.D.; Crocomo, L.F.; Rascado, T.S.; Magalhães, L.C.O.; Monteiro, B.A.; Martins, A.; et al. 82 Effect of High Fetal Calf Serum Concentration in the Gene Expression Pattern of In Vitro Produced Bovine Embryos. Reprod. Fertil. Dev. 2013, 26, 155. [Google Scholar] [CrossRef]
- Misirlioglu, M.; Page, G.P.; Sagirkaya, H.; Kaya, A.; Parrish, J.J.; First, N.L.; Memili, E. Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos. Proc. Natl. Acad. Sci. USA 2006, 103, 18905–18910. [Google Scholar] [CrossRef]
- Kues, W.A.; Sudheer, S.; Herrmann, D.; Carnwath, J.W.; Havlicek, V.; Besenfelder, U.; Lehrach, H.; Adjaye, J.; Niemann, H. Genome-wide expression profiling reveals distinct clusters of transcriptional regulation during bovine preimplantation development in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 19768–19773. [Google Scholar] [CrossRef]
- Carter, F.; Rings, F.; Mamo, S.; Hölker, M.; Kuzmany, A.; Besenfelder, U.; Havlicek, V.; Mehta, J.P.; Tesfaye, D.; Schellander, K.; et al. Effect of elevated circulating progesterone concentration on bovine blastocyst development and global transcriptome following endoscopic transfer of in vitro produced embryos to the bovine oviduct. Biol. Reprod. 2010, 83, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Yandell, B.S.; Khatib, H. Transcriptomic profiling of bovine IVF embryos revealed candidate genes and pathways involved in early embryonic development. BMC Genom. 2010, 11, 23. [Google Scholar] [CrossRef]
- Bermejo-Alvarez, P.; Rizos, D.; Rath, D.; Lonergan, P.; Gutierrez-Adan, A. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc. Natl. Acad. Sci. USA 2010, 107, 3394–3399. [Google Scholar] [CrossRef] [PubMed]
- Salilew-Wondim, D.; Hölker, M.; Rings, F.; Ghanem, N.; Ulas-Cinar, M.; Peippo, J.; Tholen, E.; Looft, C.; Schellander, K.; Tesfaye, D. Bovine pretransfer endometrium and embryo transcriptome fingerprints as predictors of pregnancy success after embryo transfer. Physiol. Genom. 2010, 42, 201–218. [Google Scholar] [CrossRef]
- Gad, A.; Besenfelder, U.; Rings, F.; Ghanem, N.; Salilew-Wondim, D.; Hossain, M.; Tesfaye, D.; Lonergan, P.; Becker, A.; Cinar, U.; et al. Effect of reproductive tract environment following controlled ovarian hyperstimulation treatment on embryo development and global transcriptome profile of blastocysts: Implications for animal breeding and human assisted reproduction. Hum. Reprod. 2011, 26, 1693–1707. [Google Scholar] [CrossRef]
- Clemente, M.; Lopez-Vidriero, I.; O’Gaora, P.; Mehta, J.; Forde, N.; Gutierrez-Adan, A.; Lonergan, P.; Rizos, D. Transcriptome Changes at the Initiation of Elongation in the Bovine Conceptus. Biol. Reprod. 2011, 85, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Nagatomo, H.; Kagawa, S.; Kishi, Y.; Takuma, T.; Sada, A.; Yamanaka, K.-I.; Abe, Y.; Wada, Y.; Takahashi, M.; Kono, T.; et al. Transcriptional Wiring for Establishing Cell Lineage Specification at the Blastocyst Stage in Cattle. Biol. Reprod. 2013, 88, 158. [Google Scholar] [CrossRef] [PubMed]
- Gad, A.; Hoelker, M.; Besenfelder, U.; Havlicek, V.; Cinar, U.; Rings, F.; Held, E.; Dufort, I.; Sirard, M.A.; Schellander, K.; et al. Molecular Mechanisms and Pathways Involved in Bovine Embryonic Genome Activation and Their Regulation by Alternative In Vivo and In Vitro Culture Conditions. Biol. Reprod. 2012, 87, 100. [Google Scholar] [CrossRef] [PubMed]
- Cagnone, G.; Dufort, I.; Vigneault, C.; Sirard, M.-A. Differential Gene Expression Profile in Bovine Blastocysts Resulting from Hyperglycemia Exposure During Early Cleavage Stages. Biol. Reprod. 2012, 86, 50. [Google Scholar] [CrossRef] [PubMed]
- Cagnone, G.L.; Sirard, M.-A. Transcriptomic signature to oxidative stress exposure at the time of embryonic genome activation in bovine blastocysts. Mol. Reprod. Dev. 2013, 80, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.M.; Dufort, I.; Caballero, J.; Moulavi, F.; Ghanaei, H.R.; Sirard, M.A. Transcriptome profiling of bovine inner cell mass and trophectoderm derived from in vivo generated blastocysts. BMC Dev. Biol. 2015, 15, 49. [Google Scholar] [CrossRef] [PubMed]
- Desmet, K.L.J.; Van Hoeck, V.; Gagné, D.; Fournier, E.; Thakur, A.; O’doherty, A.M.; Walsh, C.P.; Sirard, M.A.; Bols, P.E.J.; Leroy, J.L.M.R. Exposure of bovine oocytes and embryos to elevated non-esterified fatty acid concentrations: Integration of epigenetic and transcriptomic signatures in resultant blastocysts. BMC Genom. 2016, 17, 1004. [Google Scholar] [CrossRef]
- Gupta, A.; Singh, J.; Dufort, I.; Robert, C.; Dias, F.; Anzar, M. Transcriptomic difference in bovine blastocysts following vitrification and slow freezing at morula stage. PLoS ONE 2017, 12, e0187268. [Google Scholar] [CrossRef]
- Wu, C.; Blondin, P.; Vigneault, C.; Labrecque, R.; Sirard, M.-A. The age of the bull influences the transcriptome and epigenome of blastocysts produced by IVF. Theriogenology 2019, 144, 122–131. [Google Scholar] [CrossRef]
- Salilew-Wondim, D.; Tesfaye, D.; Rings, F.; Held-Hoelker, E.; Miskel, D.; Sirard, M.-A.; Tholen, E.; Schellander, K.; Hoelker, M. The global gene expression outline of the bovine blastocyst: Reflector of environmental conditions and predictor of developmental capacity. BMC Genom. 2021, 22, 408. [Google Scholar] [CrossRef]
- Hallberg, I.; Persson, S.; Olovsson, M.; Sirard, M.-A.; Damdimopoulou, P.; Rüegg, J.; Sjunnesson, Y.C. Perfluorooctane sulfonate (PFOS) exposure of bovine oocytes affects early embryonic development at human-relevant levels in an in vitro model. Toxicology 2021, 464, 153028. [Google Scholar] [CrossRef]
- Rabaglino, M.B.; Salilew-Wondim, D.; Zolini, A.; Tesfaye, D.; Hoelker, M.; Lonergan, P.; Hansen, P.J. Machine-learning methods applied to integrated transcriptomic data from bovine blastocysts and elongating conceptuses to identify genes predictive of embryonic competence. FASEB J. 2023, 37, e22809. [Google Scholar] [CrossRef]
- Brinkhof, B.; Van Tol, H.T.A.; Koerkamp, M.J.A.G.; Wubbolts, R.W.; Haagsman, H.P.; Roelen, B.A.J. Characterization of bovine embryos cultured under conditions appropriate for sustaining human naïve pluripotency. PLoS ONE 2017, 12, e0172920. [Google Scholar] [CrossRef]
- Ushizawa, K.; Herath, C.B.; Kaneyama, K.; Shiojima, S.; Hirasawa, A.; Takahashi, T.; Imai, K.; Ochiai, K.; Tokunaga, T.; Tsunoda, Y.; et al. cDNA microarray analysis of bovine embryo gene expression profiles during the pre-implantation period. Reprod. Biol. Endocrinol. 2004, 2, 77. [Google Scholar] [CrossRef]
- Smith, S.L.; Everts, R.E.; Tian, X.C.; Du, F.; Sung, L.-Y.; Rodriguez-Zas, S.L.; Jeong, B.-S.; Renard, J.-P.; Lewin, H.A.; Yang, X. Global gene expression profiles reveal significant nuclear reprogramming by the blastocyst stage after cloning. Proc. Natl. Acad. Sci. USA 2005, 102, 17582–17587. [Google Scholar] [CrossRef]
- Pfister-Genskow, M.; Myers, C.; Childs, L.A.; Lacson, J.C.; Patterson, T.; Betthauser, J.M.; Goueleke, P.J.; Koppang, R.W.; Lange, G.; Fisher, P.; et al. Identification of Differentially Expressed Genes in Individual Bovine Preimplantation Embryos Produced by Nuclear Transfer: Improper Reprogramming of Genes Required for Development. Biol. Reprod. 2005, 72, 546–555. [Google Scholar] [CrossRef]
- Somers, J.; Smith, C.; Donnison, M.; Wells, D.; Henderson, H.; McLeay, L.; Pfeffer, P. Gene expression profiling of individual bovine nuclear transfer blastocysts. Reproduction 2006, 131, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gong, B.; Bushel, P.R.; Thierry-Mieg, J.; Thierry-Mieg, D.; Xu, J.; Fang, H.; Hong, H.; Shen, J.; Su, Z.; et al. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat. Biotechnol. 2014, 32, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.S.; Van Vleet, T.R.; Ciurlionis, R.; Buck, W.R.; Mittelstadt, S.W.; Blomme, E.A.G.; Liguori, M.J. Comparison of RNA-Seq and Microarray Gene Expression Platforms for the Toxicogenomic Evaluation of Liver from Short-Term Rat Toxicity Studies. Front. Genet. 2019, 9, 636. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Khatib, H. Comparison of transcriptomic landscapes of bovine embryos using RNA-Seq. BMC Genom. 2010, 11, 711. [Google Scholar] [CrossRef]
- Mamo, S.; Mehta, J.P.; McGettigan, P.; Fair, T.; Spencer, T.E.; Bazer, F.W.; Lonergan, P. RNA Sequencing Reveals Novel Gene Clusters in Bovine Conceptuses Associated with Maternal Recognition of Pregnancy and Implantation. Biol. Reprod. 2011, 85, 1143–1151. [Google Scholar] [CrossRef]
- Graf, A.; Krebs, S.; Zakhartchenko, V.; Schwalb, B.; Blum, H.; Wolf, E. Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, 4139–4144. [Google Scholar] [CrossRef]
- Chitwood, J.L.; Rincon, G.; Kaiser, G.G.; Medrano, J.F.; Ross, P.J. RNA-seq analysis of single bovine blastocysts. BMC Genom. 2013, 14, 350. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Shi, Y.; Wang, H.; Wang, Z.; Dang, Y.; Li, S.; Wang, S.; Zhang, K. Base editing in bovine embryos reveals a species-specific role of SOX2 in regulation of pluripotency. PLoS Genet. 2022, 18, e1010307. [Google Scholar] [CrossRef] [PubMed]
- Zolini, A.M.; Block, J.; Rabaglino, M.B.; Rincon, G.; Hoelker, M.; Bromfield, J.J.; Salilew-Wondim, D.; Hansen, P.J. Genes associated with survival of female bovine blastocysts produced in vivo. Cell Tissue Res. 2020, 382, 665–678. [Google Scholar] [CrossRef]
- Zolini, A.M.; Block, J.; Rabaglino, M.B.; Tríbulo, P.; Hoelker, M.; Rincon, G.; Bromfield, J.J.; Hansen, P.J. Molecular fingerprint of female bovine embryos produced in vitro with high competence to establish and maintain pregnancy†. Biol. Reprod. 2019, 102, 292–305. [Google Scholar] [CrossRef]
- Nõmm, M.; Ivask, M.; Pärn, P.; Reimann, E.; Kõks, S.; Jaakma, Ü. Detecting Embryo Developmental Potential by Single Blastomere RNA-Seq. Genes 2023, 14, 569. [Google Scholar] [CrossRef] [PubMed]
- Lavagi, I.; Krebs, S.; Simmet, K.; Beck, A.; Zakhartchenko, V.; Wolf, E.; Blum, H. Single-cell RNA sequencing reveals developmental heterogeneity of blastomeres during major genome activation in bovine embryos. Sci. Rep. 2018, 8, 4071. [Google Scholar] [CrossRef] [PubMed]
- Kropp, J.; Khatib, H. Characterization of microRNA in bovine in vitro culture media associated with embryo quality and development. J. Dairy Sci. 2015, 98, 6552–6563. [Google Scholar] [CrossRef]
- Simmet, K.; Kurome, M.; Zakhartchenko, V.; Reichenbach, H.-D.; Springer, C.; Bähr, A.; Blum, H.; Philippou-Massier, J.; Wolf, E. The second lineage differentiation of bovine embryos fails in the absence of OCT4/POU5F. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zhao, X.-M.; Cui, L.-S.; Hao, H.-S.; Wang, H.-Y.; Zhao, S.-J.; Du, W.-H.; Wang, D.; Liu, Y.; Zhu, H.-B. Transcriptome analyses of inner cell mass and trophectoderm cells isolated by magnetic-activated cell sorting from bovine blastocysts using single cell RNA-seq. Reprod. Domest. Anim. 2016, 51, 726–735. [Google Scholar] [CrossRef]
- Kropp, J.; Carrillo, J.A.; Namous, H.; Daniels, A.; Salih, S.M.; Song, J.; Khatib, H. Male fertility status is associated with DNA methylation signatures in sperm and transcriptomic profiles of bovine preimplantation embryos. BMC Genom. 2017, 18, 280. [Google Scholar] [CrossRef]
- Gilchrist, G.C.; Tscherner, A.; Nalpathamkalam, T.; Merico, D.; LaMarre, J. MicroRNA Expression during Bovine Oocyte Maturation and Fertilization. Int. J. Mol. Sci. 2016, 17, 396. [Google Scholar] [CrossRef]
- Tríbulo, P.; Rabaglino, M.B.; Bo, M.B.; Carvalheira, L.D.R.; Bishop, J.V.; Hansen, T.R.; Hansen, P.J. Dickkopf-related protein 1 is a progestomedin acting on the bovine embryo during the morula-to-blastocyst transition to program trophoblast elongation. Sci. Rep. 2019, 9, 11816. [Google Scholar] [CrossRef]
- Diaz-Lundahl, S.; Sundaram, A.Y.; Gillund, P.; Gilfillan, G.D.; Olsaker, I.; Krogenæs, A. Gene Expression in Embryos From Norwegian Red Bulls With High or Low Non Return Rate: An RNA-Seq Study of in vivo-Produced Single Embryos. Front. Genet. 2022, 12, 780113. [Google Scholar] [CrossRef] [PubMed]
- Halstead, M.M.; Ma, X.; Zhou, C.; Schultz, R.M.; Ross, P.J. Chromatin remodeling in bovine embryos indicates species-specific regulation of genome activation. Nat. Commun. 2020, 11, 4654. [Google Scholar] [CrossRef] [PubMed]
- Gross, N.; Strillacci, M.G.; Peñagaricano, F.; Khatib, H. Characterization and functional roles of paternal RNAs in 2–4 cell bovine embryos. Sci. Rep. 2019, 9, 20347. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhao, P.; Dang, Y.; Li, S.; Luo, L.; Hu, B.; Wang, S.; Wang, H.; Zhang, K. Functional roles of the chromatin remodeler SMARCA5 in mouse and bovine preimplantation embryos†. Biol. Reprod. 2021, 105, 359–370. [Google Scholar] [CrossRef]
- Li, S.; Shi, Y.; Dang, Y.; Luo, L.; Hu, B.; Wang, S.; Wang, H.; Zhang, K. NOTCH signaling pathway is required for bovine early embryonic development. Biol. Reprod. 2021, 105, 332–344. [Google Scholar] [CrossRef]
- Zhu, L.; Zhou, T.; Iyyappan, R.; Ming, H.; Dvoran, M.; Wang, Y.; Chen, Q.; Roberts, R.M.; Susor, A.; Jiang, Z. High-resolution ribosome profiling reveals translational selectivity for transcripts in bovine preimplantation embryo development. Development 2022, 149, dev.200819. [Google Scholar] [CrossRef]
- Kajdasz, A.; Warzych, E.; Derebecka, N.; Madeja, Z.E.; Lechniak, D.; Wesoly, J.; Pawlak, P. Lipid Stores and Lipid Metabolism Associated Gene Expression in Porcine and Bovine Parthenogenetic Embryos Revealed by Fluorescent Staining and RNA-seq. Int. J. Mol. Sci. 2020, 21, 6488. [Google Scholar] [CrossRef]
- Nix, J.L.; Schettini, G.P.; Biase, F.H. Sexing of cattle embryos using RNA-sequencing data or polymerase chain reaction based on a complete sequence of cattle chromosome Y. Front. Genet. 2023, 14, 569. [Google Scholar] [CrossRef]
- Jiang, Z.; Sun, J.; Dong, H.; Luo, O.; Zheng, X.; Obergfell, C.; Tang, Y.; Bi, J.; O’neill, R.; Ruan, Y.; et al. Transcriptional profiles of bovine in vivo pre-implantation development. BMC Genom. 2014, 15, 756. [Google Scholar] [CrossRef]
- Jiang, Z.; Dong, H.; Zheng, X.; Marjani, S.L.; Donovan, D.M.; Chen, J.; Tian, X. mRNA Levels of Imprinted Genes in Bovine In Vivo Oocytes, Embryos and Cross Species Comparisons with Humans, Mice and Pigs. Sci. Rep. 2015, 5, 17898. [Google Scholar] [CrossRef] [PubMed]
- Banliat, C.; Mahé, C.; Lavigne, R.; Com, E.; Pineau, C.; Labas, V.; Guyonnet, B.; Mermillod, P.; Saint-Dizier, M. Dynamic changes in the proteome of early bovine embryos developed in vivo. Front. Cell Dev. Biol. 2022, 10, 863700. [Google Scholar] [CrossRef]
- Demant, M.; Deutsch, D.R.; Fröhlich, T.; Wolf, E.; Arnold, G.J. Proteome analysis of early lineage specification in bovine embryos. Proteomics 2014, 15, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, D.R.; Fröhlich, T.; Otte, K.A.; Beck, A.; Habermann, F.A.; Wolf, E.; Arnold, G.J. Stage-Specific Proteome Signatures in Early Bovine Embryo Development. J. Proteome Res. 2014, 13, 4363–4376. [Google Scholar] [CrossRef]
- Banliat, C.; Mahé, C.; Lavigne, R.; Com, E.; Pineau, C.; Labas, V.; Guyonnet, B.; Mermillod, P.; Saint-Dizier, M. The proteomic analysis of bovine embryos developed in vivo or in vitro reveals the contribution of the maternal environment to early embryo. BMC Genom. 2022, 23, 839. [Google Scholar] [CrossRef]
- Jensen, P.L.; Grøndahl, M.L.; Beck, H.C.; Petersen, J.; Stroebech, L.; Christensen, S.T.; Andersen, C.Y. Proteomic analysis of bovine blastocoel fluid and blastocyst cells. Syst. Biol. Reprod. Med. 2014, 60, 127–135. [Google Scholar] [CrossRef]
- Jensen, P.L.; Beck, H.C.; Petersen, T.S.; Stroebech, L.; Schmidt, M.; Rasmussen, L.M.; Hyttel, P. Proteomic analysis of the early bovine yolk sac fluid and cells from the day 13 ovoid and elongated preimplantation embryos. Theriogenology 2014, 82, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Talbot, N.C.; Powell, A.M.; Caperna, T.J.; Garrett, W.M. Proteomic analysis of the major cellular proteins of bovine trophectoderm cell lines derived from IVP, parthenogenetic and nuclear transfer embryos: Reduced expression of annexins I and II in nuclear transfer-derived cell lines. Anim. Reprod. Sci. 2010, 120, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Raes, A.; Wydooghe, E.; Pavani, K.C.; Pascottini, O.B.; Van Steendam, K.; Dhaenens, M.; Boel, A.; Heras, S.; Heindryckx, B.; Peelman, L.; et al. Cathepsin-L Secreted by High-Quality Bovine Embryos Exerts an Embryotrophic Effect In Vitro. Int. J. Mol. Sci. 2023, 24, 6563. [Google Scholar] [CrossRef]
- Nel-Themaat, L.; Nagy, Z. A review of the promises and pitfalls of oocyte and embryo metabolomics. Placenta 2011, 32, S257–S263. [Google Scholar] [CrossRef] [PubMed]
- Rieger, D.; Guay, P. Measurement of the metabolism of energy substrates in individual bovine blastocysts. Reproduction 1988, 83, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.G.; McNaughton, C.; Gasparrini, B.; McGowan, L.T.; Tervit, H.R. Effect of inhibitors and uncouplers of oxidative phosphorylation during compaction and blastulation of bovine embryos cultured in vitro. J. Reprod. Fertil. 2000, 118, 47–56. [Google Scholar] [CrossRef]
- Leese, H.J. What does an embryo need? Hum. Fertil. 2003, 6, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Gardner, D.; Pawelczynski, M.; Trounson, A. Nutrient uptake and utilization can be used to select viable day 7 bovine blastocysts after cryopreservation. Mol. Reprod. Dev. 1996, 44, 472–475. [Google Scholar] [CrossRef]
- Partridge, R.; Leese, H. Consumption of amino acids by bovine preimplantation embryos. Reprod. Fertil. Dev. 1996, 8, 945–950. [Google Scholar] [CrossRef]
- Lopes, A.; Madsen, S.; Ramsing, N.; Løvendahl, P.; Greve, T.; Callesen, H. Investigation of respiration of individual bovine embryos produced in vivo and in vitro and correlation with viability following transfer. Hum. Reprod. 2006, 22, 558–566. [Google Scholar] [CrossRef]
- Lopes, A.S.; Lane, M.; Thompson, J.G. Oxygen consumption and ROS production are increased at the time of fertilization and cell cleavage in bovine zygotes. Hum. Reprod. 2010, 25, 2762–2773. [Google Scholar] [CrossRef]
- Obeidat, Y.; Catandi, G.; Carnevale, E.; Chicco, A.J.; DeMann, A.; Field, S.; Chen, T. A multi-sensor system for measuring bovine embryo metabolism. Biosens. Bioelectron. 2018, 126, 615–623. [Google Scholar] [CrossRef]
- Gómez, E.; Muñoz, M.; Simó, C.; Ibáñez, C.; Carrocera, S.; Martín-González, D.; Cifuentes, A. Non-invasive metabolomics for improved determination of embryonic sex markers in chemically defined culture medium. J. Chromatogr. A 2016, 1474, 138–144. [Google Scholar] [CrossRef]
- Gómez, E.; Carrocera, S.; Martin, D.; Herrero, P.; Canela, N.; Muñoz, M. Differential release of cell-signaling metabolites by male and female bovine embryos cultured in vitro. Theriogenology 2018, 114, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, M.; Uyar, A.; Correia, E.; Díez, C.; Fernandez-Gonzalez, A.; Caamaño, J.N.; Trigal, B.; Carrocera, S.; Seli, E.; Gomez, E. Non-invasive assessment of embryonic sex in cattle by metabolic fingerprinting of in vitro culture medium. Metabolomics 2014, 10, 443–451. [Google Scholar] [CrossRef]
- dos Santos, É.; da Silva Martinho, H.; Annes, K.; da Silva, T.; Soares, C.; Leite, R.F.; Milazzotto, M. Raman-based noninvasive metabolic profile evaluation of in vitro bovine embryos. J. Biomed. Opt. 2016, 21, 075002. [Google Scholar] [CrossRef] [PubMed]
- Rubessa, M.; Ambrosi, A.; Gonzalez-Pena, D.; Polkoff, K.M.; Wheeler, M.B. Non-invasive nuclear magnetic resonance analysis of male and female embryo metabolites during in vitro embryo culture. Metabolomics 2018, 14, 113. [Google Scholar] [CrossRef] [PubMed]
- Perkel, K.J.; Madan, P. Spent culture medium analysis from individually cultured bovine embryos demonstrates metabolomic differences. Zygote 2017, 25, 662–674. [Google Scholar] [CrossRef]
- Sturmey, R.; Bermejo-Alvarez, P.; Gutierrez-Adan, A.; Rizos, D.; Leese, H.; Lonergan, P. Amino acid metabolism of bovine blastocysts: A biomarker of sex and viability. Mol. Reprod. Dev. 2010, 77, 285–296. [Google Scholar] [CrossRef]
- Tiffin, G.J.; Rieger, D.; Betteridge, K.J.; Yadav, B.R.; King, W.A. Glucose and glutamine metabolism in pre-attachment cattle embryos in relation to sex and stage of development. Reproduction 1991, 93, 125–132. [Google Scholar] [CrossRef]
- Rieger, D.; Loskutoff, N.; Betteridge, K. Developmentally related changes in the uptake and metabolism of glucose, glutamine and pyruvate by cattle embryos produced in vitro. Reprod. Fertil. Dev. 1992, 4, 547–557. [Google Scholar] [CrossRef]
- Donnay, I.; Leese, H. Embryo metabolism during the expansion of the bovine blastocyst. Mol. Reprod. Dev. 1999, 53, 171–178. [Google Scholar] [CrossRef]
- Khurana, N.K.; Niemann, H. Energy Metabolism in Preimplantation Bovine Embryos Derived In Vitro or In Vivo. Biol. Reprod. 2000, 62, 847–856. [Google Scholar] [CrossRef]
- Rubessa, M.; Ambrosi, A.; Gonzalez-Pena, D.; Polkoff, K.M.; Denmark, S.E.; Wheeler, M.B. Non-invasive analysis of bovine embryo metabolites during in vitro embryo culture using nuclear magnetic resonance. AIMS Bioeng. 2016, 3, 538–551. [Google Scholar] [CrossRef]
- Steeves, T.; Gardner, D. Temporal and Differential Effects of Amino Acids on Bovine Embryo Development in Culture. Biol. Reprod. 1999, 61, 731–740. [Google Scholar] [CrossRef]
- Nõmm, M.; Porosk, R.; Pärn, P.; Kilk, K.; Soomets, U.; Kõks, S.; Jaakma, Ü. In vitro culture and non-invasive metabolic profiling of single bovine embryos. Reprod. Fertil. Dev. 2019, 31, 306. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, E.M.; Leese, H.J. A potential role for triglyceride as an energy source during bovine oocyte maturation and early embryo development. Mol. Reprod. Dev. 2006, 73, 1195–1201. [Google Scholar] [CrossRef]
- Sudano, M.J.; Rascado, T.D.; Tata, A.; Belaz, K.R.; Santos, V.G.; Valente, R.S.; Mesquita, F.S.; Ferreira, C.R.; Araújo, J.P.; Eberlin, M.N.; et al. Lipidome signatures in early bovine embryo development. Theriogenology 2016, 86, 472–484.e1. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, H.; Utsunomiya, H.; Shiga, N.; Takahashi, A.; Ihara, M.; Ishibashi, M.; Nishimoto, M.; Watanabe, Z.; Abe, H.; Kumagai, J.; et al. Development of a new clinically applicable device for embryo evaluation which measures embryo oxygen consumption. Hum. Reprod. 2016, 31, 2321–2330. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Asampille, G.; Cheredath, A.; Joseph, D.; Adiga, S.K.; Atreya, H.S. The utility of nuclear magnetic resonance spectroscopy in assisted reproduction. Open Biol. 2020, 10, 200092. [Google Scholar] [CrossRef]
- Sivelli, G.; Conley, G.M.; Herrera, C.; Marable, K.; Rodriguez, K.J.; Bollwein, H.; Sudano, M.J.; Brugger, J.; Simpson, A.J.; Boero, G.; et al. NMR spectroscopy of a single mammalian early stage embryo. J. Magn. Reson. 2022, 335, 107142. [Google Scholar] [CrossRef]
- Pomar, F.R.; Teerds, K.; Kidson, A.; Colenbrander, B.; Tharasanit, T.; Aguilar, B.; Roelen, B. Differences in the incidence of apoptosis between in vivo and in vitro produced blastocysts of farm animal species: A comparative study. Theriogenology 2005, 63, 2254–2268. [Google Scholar] [CrossRef]
- Sagirkaya, H.; Misirlioglu, M.; Kaya, A.; First, N.L.; Parrish, J.; Memili, E. Developmental and molecular correlates of bovine preimplantation embryos. Reproduction 2006, 131, 895–904. [Google Scholar] [CrossRef]
- Ramos-Ibeas, P.; Gimeno, I.; Cañón-Beltrán, K.; Gutiérrez-Adán, A.; Rizos, D.; Gómez, E. Senescence and Apoptosis during in vitro Embryo Development in a Bovine Model. Front. Cell Dev. Biol. 2020, 8, 619902. [Google Scholar] [CrossRef] [PubMed]
- Wondim, D.S.; Fournier, E.; Hoelker, M.; Saeed-Zidane, M.; Tholen, E.; Looft, C.; Neuhoff, C.; Besenfelder, U.; Havlicek, V.; Rings, F.; et al. Genome-Wide DNA Methylation Patterns of Bovine Blastocysts Developed In Vivo from Embryos Completed Different Stages of Development In Vitro. PLoS ONE 2015, 10, e0140467. [Google Scholar] [CrossRef]
- Dobbs, K.B.; Rodriguez, M.; Sudano, M.J.; Ortega, M.S.; Hansen, P.J. Dynamics of DNA methylation during early development of the preimplantation bovine embryo. PLoS ONE 2013, 8, e66230. [Google Scholar] [CrossRef] [PubMed]
- Morin-Doré, L.; Blondin, P.; Vigneault, C.; Grand, F.; Labrecque, R.; Sirard, M. DNA methylation status of bovine blastocysts obtained from peripubertal oocyte donors. Mol. Reprod. Dev. 2020, 87, 910–924. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, D.; Humblot, P.; Sirard, M.-A.; Sjunnesson, Y.; Jhamat, N.; Båge, R.; Andersson, G. DNA methylation pattern of bovine blastocysts associated with hyperinsulinemia in vitro. Mol. Reprod. Dev. 2018, 85, 599–611. [Google Scholar] [CrossRef]
- Salilew-Wondim, D.; Saeed-Zidane, M.; Hoelker, M.; Gebremedhn, S.; Poirier, M.; Pandey, H.O.; Tholen, E.; Neuhoff, C.; Held, E.; Besenfelder, U.; et al. Genome-wide DNA methylation patterns of bovine blastocysts derived from in vivo embryos subjected to in vitro culture before, during or after embryonic genome activation. BMC Genom. 2018, 19, 424. [Google Scholar] [CrossRef]
- Duan, J.E.; Jiang, Z.C.; Alqahtani, F.; Mandoiu, I.; Dong, H.; Zheng, X.; Marjani, S.L.; Chen, J.; Tian, X.C. Methylome Dynamics of Bovine Gametes and in vivo Early Embryos. Front. Genet. 2019, 10, 512. [Google Scholar] [CrossRef]
- Ming, H.; Sun, J.; Pasquariello, R.; Gatenby, L.; Herrick, J.R.; Yuan, Y.; Pinto, C.R.; Bondioli, K.R.; Krisher, R.L.; Jiang, Z. The landscape of accessible chromatin in bovine oocytes and early embryos. Epigenetics 2020, 16, 300–312. [Google Scholar] [CrossRef]
- Kurosaka, S.; Eckardt, S.; McLaughlin, K.J. Pluripotent Lineage Definition in Bovine Embryos by Oct4 Transcript Localization. Biol. Reprod. 2004, 71, 1578–1582. [Google Scholar] [CrossRef]
- Hyttel, P.; Viuff, D.; Laurincik, J.; Schmidt, M.; Thomsen, P.; Avery, B.; Callesen, H.; Rath, D.; Niemann, H.; Rosenkranz, H.; et al. Risks of in-vitro production of cattle and swine embryos: Aberrations in chromosome numbers, ribosomal RNA gene activation and perinatal physiology. Hum. Reprod. 2000, 15, 87–97. [Google Scholar] [CrossRef]
- Demyda-Peyrás, S.; Dorado, J.; Hidalgo, M.; Anter, J.; De Luca, L.; Genero, E.R.; Moreno-Millán, M. Effects of oocyte quality, incubation time and maturation environment on the number of chromosomal abnormalities in IVF-derived early bovine embryos. Reprod. Fertil. Dev. 2013, 25, 1077–1084. [Google Scholar] [CrossRef]
- Guilherme, V.B.; Pronunciate, M.; dos Santos, P.H.; Ciniciato, D.D.S.; Takahashi, M.B.; Rocha, J.C.; Nogueira, M.F.G. Distinct Sources of a Bovine Blastocyst Digital Image Do not Produce the Same Classification by a Previously Trained Software Using Artificial Neural Network. bioRxiv 2018, 424028. [Google Scholar] [CrossRef]
- Rocha, J.C.; Passalia, F.J.; Matos, F.D.; Takahashi, M.B.; Ciniciato, D.D.S.; Maserati, M.P.; Alves, M.F.; de Almeida, T.G.; Cardoso, B.L.; Basso, A.C.; et al. A method based on artificial intelligence to fully automatize the evaluation of bovine blastocyst images. Sci. Rep. 2017, 7, 7659. [Google Scholar] [CrossRef]
- Melo, D.H.; Nascimento, M.Z.; Oliveira, D.L.; Neves, L.A.; Annes, K. Algorithms for automatic segmentation of bovine embryos produced in vitro. J. Phys. Conf. Ser. 2014, 490, 012125. [Google Scholar] [CrossRef]
- Ciniciato, D.D.S.; Takahashi, M.B.; Nogueira, M.F.G.; Rocha, J.C. Potential Use of Smartphone as a Tool to Capture Embryo Digital Images from Stereomicroscope and to Evaluate Them by an Artificial Neural Network. In Proceedings of the International Conference on Computer-Human Interaction Research and Applications, Funchal, Portugal, 31 October–2 November 2017; pp. 185–189. [Google Scholar] [CrossRef]
- Nogueira, M.F.G.; Guilherme, V.B.; Pronunciate, M.; Dos Santos, P.H.; da Silva, D.L.B.; Rocha, J.C. Artificial Intelligence-Based Grading Quality of Bovine Blastocyst Digital Images: Direct Capture with Juxtaposed Lenses of Smartphone Camera and Stereomicroscope Ocular Lens. Sensors 2018, 18, 4440. [Google Scholar] [CrossRef]
- Workman, A.M.; Heaton, M.P.; Ley, B.L.V.; Webster, D.A.; Sherry, L.; Larson, S.; Kalbfleisch, T.S.; Harhay, G.P.; Jobman, E.E.; Carlson, D.F.; et al. First gene-edited calf with reduced susceptibility to a major viral pathogen. bioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Mellisho, E.; Briones, M.A.; Velasquez, A.E.; Cabezas, J.; Castro, F.O.; Rodriguez-Alvarez, L. Extracellular vesicles secreted during blastulation show viability of bovine embryos. Reproduction 2019, 158, 477–492. [Google Scholar] [CrossRef] [PubMed]
- Mellisho, E.A.; Velásquez, A.E.; Nuñez, M.J.; Cabezas, J.G.; Cueto, J.A.; Fader, C.; Castro, F.O.; Rodríguez-Álvarez, L. Identification and characteristics of extracellular vesicles from bovine blastocysts produced in vitro. PLoS ONE 2017, 12, e0178306. [Google Scholar] [CrossRef]
- Saadeldin, I.M.; Kim, S.J.; Bin Choi, Y.; Lee, B.C. Improvement of cloned embryos development by co-culturing with parthenotes: A possible role of exosomes/microvesicles for embryos paracrine communication. Cell. Reprogram. 2014, 16, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Rabaglino, M.B.; O’doherty, A.; Secher, J.B.-M.; Lonergan, P.; Hyttel, P.; Fair, T.; Kadarmideen, H.N. Application of multi-omics data integration and machine learning approaches to identify epigenetic and transcriptomic differences between in vitro and in vivo produced bovine embryos. PLoS ONE 2021, 16, e0252096. [Google Scholar] [CrossRef]
- Milazzotto, M.P.; Noonan, M.J.; Ferraz, M.D.A.M.M. Mining RNAseq data reveals dynamic metaboloepigenetic profiles in human, mouse and bovine pre-implantation embryos. iScience 2022, 25, 103904. [Google Scholar] [CrossRef] [PubMed]
- Dochi, O. Direct transfer of frozen-thawed bovine embryos and its application in cattle reproduction management. J. Reprod. Dev. 2019, 65, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Banik, S.; Melanthota, S.K.; Arbaaz; Vaz, J.M.; Kadambalithaya, V.M.; Hussain, I.; Dutta, S.; Mazumder, N. Recent trends in smartphone-based detection for biomedical applications: A review. Anal. Bioanal. Chem. 2021, 413, 2389–2406. [Google Scholar] [CrossRef] [PubMed]
- Freeman, K.; Dinnes, J.; Chuchu, N.; Takwoingi, Y.; Bayliss, S.E.; Matin, R.N.; Jain, A.; Walter, F.M.; Williams, H.C.; Deeks, J.J. Algorithm based smartphone apps to assess risk of skin cancer in adults: Systematic review of diagnostic accuracy studies. BMJ 2020, 368, m127. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.K.; Nair, M.R.; Manjunath, A.; Mujib, B.R.A. Evaluation and comparison between smartphone and photomicrography based whole slide imaging. J. Fam. Med. Prim. Care 2020, 9, 2319–2323. [Google Scholar] [CrossRef]
- Russ, J.C.; Matey, J.R.; Mallinckrodt, A.J.; McKay, S. The Image Processing Handbook. Comput. Phys. 1994, 8, 177–178. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).