High Prevalence of Microsporidia in the North African Hedgehog (Atelerix algirus) in the Canary Islands, Spain
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Agreement
2.2. Study Area, Sample Collection, and Preparation
2.3. Staining Method
2.4. DNA Extraction
2.5. Nested-PCR Amplification
2.6. Sequencing and Phylogenetic Analysis
3. Results
3.1. Light Microscopy
3.2. Molecular Characterization
3.2.1. Molecular Characterization of Enterocytozoon bieneusi
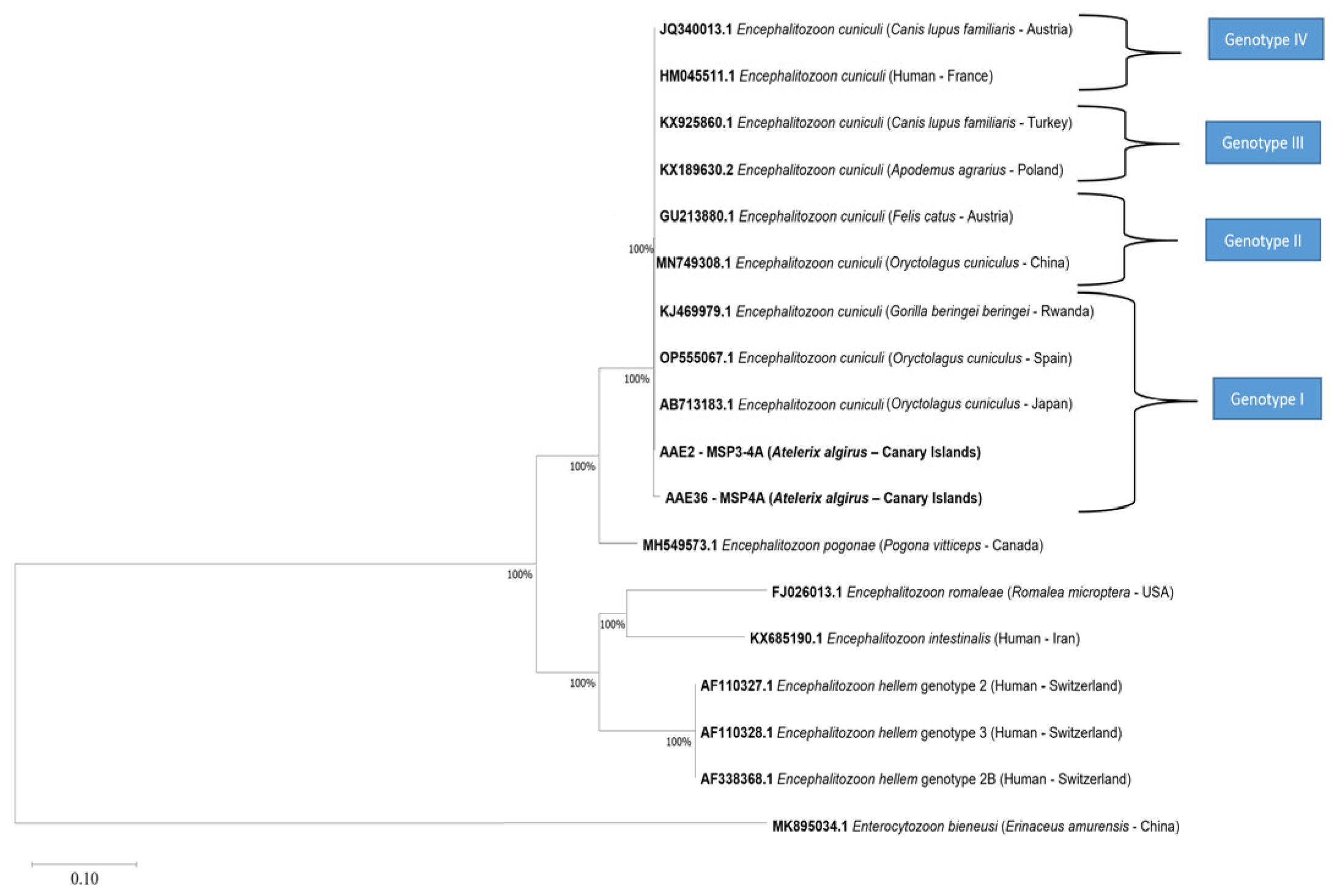
3.2.2. Molecular Characterization of Encephalitozoon cuniculi
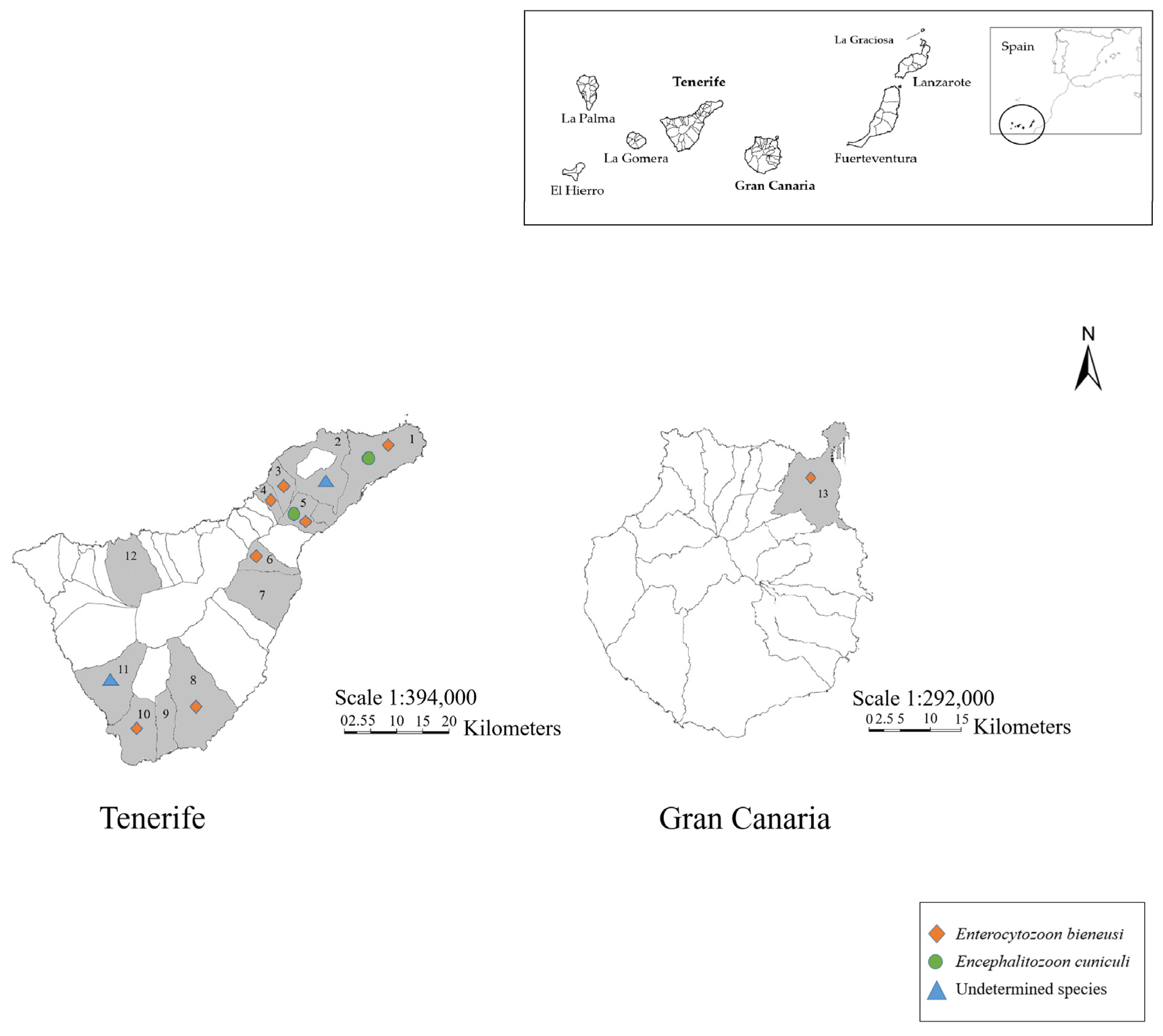
3.2.3. Geographical Distribution
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Han, B.; Pan, G.; Weiss, L.M. Microsporidiosis in Humans. Clin. Microbiol. Rev. 2021, 34, e0001020. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Feng, Y.; Santin, M. Host Specificity of Enterocytozoon bieneusi and Public Health Implications. Trends Parasitol. 2019, 35, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Santaniello, A.; Cimmino, I.; Dipineto, L.; Agognon, A.L.; Beguinot, F.; Formisano, P.; Fioretti, A.; Menna, L.C.; Oriente, F. Zoonotic Risk of Encephalitozoon cuniculi in Animal-Assisted Interventions: Laboratory Strategies for the Diagnosis of Infections in Humans and Animals. Int. J. Environ. Res. Public Health 2021, 18, 9333. [Google Scholar] [CrossRef]
- Haro, M.; Del Águila, C.; Fenoy, S.; Henriques-Gil, N. Intraspecies genotype variability of the microsporidian parasite Encephalitozoon hellem. J. Clin. Microbiol. 2003, 41, 4166–4171. [Google Scholar] [CrossRef] [PubMed]
- Graczyk, T.K.; Bosco-Nizeyi, J.; da Silva, A.J.; Moura, I.N.; Pieniazek, N.J.; Cranfield, M.R.; Lindquist, H.D. A single genotype of Encephalitozoon intestinalis infects free-ranging gorillas and people sharing their habitats in Uganda. Parasitol. Res. 2002, 88, 926–931. [Google Scholar] [CrossRef] [PubMed]
- Didier, E.S.; Stovall, M.E.; Green, L.C.; Brindley, P.J.; Sestak, K.; Didier, P.J. Epidemiology of microsporidiosis: Sources and modes of transmission. Vet. Parasitol. 2004, 126, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Stentiford, G.D.; Becnel, J.J.; Weiss, L.M.; Keeling, P.J.; Didier, E.S.; Williams, B.A.P.; Bjornson, S.; Kent, M.L.; Freeman, M.A.; Brown, M.J.F.; et al. Microsporidia—Emergent Pathogens in the Global Food Chain. Trends Parasitol. 2016, 32, 336–348. [Google Scholar] [CrossRef]
- Koehler, A.V.; Zhang, Y.; Gasser, R.B. A Perspective on the Molecular Identification, Classification, and Epidemiology of Enterocytozoon bieneusi of Animals. Exp. Suppl. 2022, 114, 389–415. [Google Scholar]
- Mathis, A.; Weber, R.; Deplazes, P. Zoonotic potential of the microsporidia. Clin. Microbiol. Rev. 2005, 18, 423–425. [Google Scholar] [CrossRef]
- Gong, X.Q.; Xiao, X.; Gu, X.L.; Han, H.J.; Yu, X.J. Detection of Intestinal Parasites and Genetic Characterization of Enterocytozoon bieneusi in Hedgehogs from China. Vector Borne Zoonotic Dis. 2021, 21, 63–66. [Google Scholar] [CrossRef]
- Meng, X.; Chu, W.; Tang, Y.; Wang, W.; Chen, Y.; Li, N.; Feng, Y.; Xiao, L.; Guo, Y. High zoonotic potential and heavy environmental burden of Cryptosporidium spp. and Enterocytozoon bieneusi in farmed and pet African pygmy hedgehogs (Atelerix albiventris). One Health 2023, 16, 100532. [Google Scholar] [CrossRef]
- Banco de Datos de Biodiversidad de Canarias (EXOS). Available online: https://www.biodiversidadcanarias.es/exos/especie/V00149 (accessed on 22 March 2023).
- Khaldi, M.; Ribas, A.; Barech, G.; Hugot, J.P.; Benyettou, M.; Albane, L.; Arrizabalaga, A.; Nicolas, V. Molecular evidence supports recent anthropogenic introduction of the Algerian hedgehog Atelerix algirus in Spain, Balearic and Canary islands from North Africa. Mammalia 2016, 80, 313–320. [Google Scholar] [CrossRef]
- Boletín Oficial de Canarias (BOC). Available online: http://www.gobiernodecanarias.org/boc/2020/248/016.html (accessed on 22 March 2023).
- Weber, R.; Bryan, R.T.; Owen, R.L.; Wilcox, C.M.; Gorelkin, L.; Visvesvara, G.S. Improved light-microscopical detection of microsporidia spores in stool and duodenal aspirates. The Enteric Opportunistic Infections Working Group. N. Engl. J. Med. 1992, 326, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Katzwinkel-Wladarsch, S.; Lieb, M.; Helse, W.; Löscher, T.; Rinder, H. Direct amplification and species determination of microsporidian DNA from stool specimens. Trop. Med. Int. Health 1996, 1, 373–378. [Google Scholar] [CrossRef]
- Sak, B.; Kváč, M.; Květoňová, D.; Albrecht, T.; Piálek, J. The first report on natural Enterocytozoon bieneusi and Encephalitozoon spp. infections in wild East-European House Mice (Mus musculus musculus) and West-European House Mice (M. m. domesticus) in a hybrid zone across the Czech Republic-Germany border. Vet. Parasitol. 2011, 178, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Candia, A.; Lopez-Velez, R.; Perez-Elias, M.J.; Guerrero, A.; Moreira, V.; Redondo, C. Enterocytozoon bieneusi Infection in an AIDS patient in Spain. Res. Rev. Parasitol. 1994, 54, 269–271. [Google Scholar]
- Miró, O.; Moreno, A.; Valls, M.E.; Miró, J.M.; Piqué, J.M.; Bordas, J.M.; Moreno-Martínez, A.; Bombí, J.A.; Gatell, J.M. Intestinal microsporidiosis in patients with AIDS. Report of three cases. Med. Clin. (Barc) 1995, 104, 96–99. [Google Scholar]
- Subirats, M.; González-Castelao, G.; Aguilera, O.; Moody, A.; Visvesvara, G.; Verdejo, J.; Baquero, M.; del Águila, C. Diagnosis of 4 cases of intestinal microsporidiosis in AIDS patients. Enferm. Infecc. Microbiol. Clin. 1996, 14, 533–537. [Google Scholar] [PubMed]
- Da Silva, A.J.; Bornay-Llinares, F.J.; del Aguila-de la Puente, C.; Moura, H.; Peralta, J.M.; Sobottka, I.; Schwartz, D.A.; Visvesvara, G.S.; Slemenda, S.B.; Pieniazek, N.J. Diagnosis of Enterocytozoon bieneusi (Microsporidia) Infections by Polymerase Chain Reaction in Stool Samples Using Primers Based on the Region Coding for Small-Subunit Ribosomal RNA. Arch. Pathol. Lab. Med. 1997, 121, 874–879. [Google Scholar] [PubMed]
- Moreno-Camacho, A.; Moreno-Martínez, A.; Valls, M.E.; Bordas, J.M.; Piqué, J.M.; Bombí, J.A.; Miró, J.M.; Mallolas, J.; Gatell, J.M.; Soriano, E. Chronic enteropathy of unknown etiology in patients with AIDS. An analysis of 40 cases. Med. Clin. (Barc) 1997, 109, 452–456. [Google Scholar] [PubMed]
- Del Águila, C.; Soriano, V.; Navajas, R.; Subirats, M.; Fenoy, S.; Valencia, E.; Baquero, M.; Pieniazek, N.J. Species identification of intestinal microsporidiosis in HIV-positive patients using the polymerase chain reaction. Enferm. Infecc. Microbiol. Clin. 1997, 15, 456–461. [Google Scholar]
- Del Águila, C.; Navajas, R.; Gurbindo, D.; Ramos, J.T.; Mellado, M.J.; Fenoy, S.; Muñoz-Fernandez, M.A.; Subirats, M.; Ruiz, J.; Pieniazek, N.J. Microsporidiosis in HIV-positive children in Madrid (Spain). J. Eukaryot. Microbiol. 1997, 44, 84S–85S. [Google Scholar] [CrossRef]
- Lores, B.; Arias, C.; Fenoy, S.; Iglesias, I.; Ocampo, A.; Miralles, C.; Garcia-Estevez, J.M.; del Aguila, C. Enterocytozoon bieneusi: A common opportunistic parasite in lungs of HIV-positive patients? J. Eukaryot. Microbiol. 1999, 46, 6S–7S. [Google Scholar]
- Chozas, M.; Dashti, A.; Prieto-Pérez, L.; Pérez-Tanoira, R.; Cobo, E.; Bailo, B.; del Palacio, M.; Hernández-Castro, C.; González-Barrio, D.; Carmena, D.; et al. Enterocytozoon bieneusi and Encephalitozoon intestinalis (Microsporidia) in HIV-positive patients in central Spain. Med. Mycol. 2023, 61, myad039. [Google Scholar] [CrossRef]
- Gainzarain, J.C.; Canut, A.; Lozano, M.; Labora, A.; Carreras, F.; Fenoy, S.; Navajas, R.; Pieniazek, N.J.; da Silva, A.J.; del Águila, C. Detection of Enterocytozoon bieneusi in two human immunodeficiency virus-negative patients with chronic diarrhea by polymerase chain reaction in duodenal biopsy specimens and review. Clin. Infect. Dis. 1998, 27, 394–398. [Google Scholar] [CrossRef]
- López-Vélez, R.; Turrientes, M.C.; Garrón, C.; Montilla, P.; Navajas, R.; Fenoy, S.; del Águila, C. Microsporidiosis in travelers with diarrhea from the tropics. J. Travel Med. 1999, 6, 223–227. [Google Scholar] [CrossRef]
- Lores, B.; Arias, C.; López-Miragaya, I.; Torres, J.; Fenoy, S.; del Águila, C. Molecular Diagnosis of Intestinal Microsporidiosis in pediatric patients from Vigo (NW Spain). Res. Rev. Parasitol. 2001, 61, 43–49. [Google Scholar]
- Lores, B.; López-Miragaya, I.; Arias, C.; Fenoy, S.; Torres, J.; del Águila, C. Intestinal microsporidiosis due to Enterocytozoon bieneusi in elderly human immunodeficiency virus-negative patients from Vigo, Spain. Clin. Infect. Dis. 2002, 34, 918–921. [Google Scholar] [CrossRef]
- Lores, B.; del Águila, C.; López-Miragaya, I.; Torres, J.; Arias, C. Detection of Enterocytozoon bieneusi by PCR in Immunocompetent Spanish patients with diarrhea and pneumonia. Res. Rev. Parasitol. 2002, 62, 43–49. [Google Scholar]
- Abreu-Acosta, N.; Lorenzo-Morales, J.; Leal-Guio, Y.; Coronado-Alvarez, N.; Foronda, P.; Alcoba-Florez, J.; Izquierdo, F.; Batista-Díaz, N.; Del Águila, C.; Valladares, B. Enterocytozoon bieneusi (microsporidia) in clinical samples from immunocompetent individuals in Tenerife, Canary Islands, Spain. Trans. R. Soc. Trop. Med. Hyg. 2005, 99, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Galván, A.L.; Sánchez, A.M.; Valentín, M.A.; Henriques-Gil, N.; Izquierdo, F.; Fenoy, S.; del Águila, C. First cases of microsporidiosis in transplant recipients in Spain and review of the literature. J. Clin. Microbiol. 2011, 49, 1301–1306. [Google Scholar] [CrossRef] [PubMed]
- Del Aguila, C.; Lopez-Velez, R.; Fenoy, S.; Turrientes, C.; Cobo, J.; Navajas, R.; Visvesvara, G.S.; Croppo, G.P.; Da Silva, A.J.; Pieniazek, N.J. Identification of Enterocytozoon bieneusi spores in respiratory samples from and AIDS patient with a 2-year history of intestinal microsporidiosis. J. Clin. Microbiol. 1997, 35, 1862–1866. [Google Scholar] [CrossRef]
- Peman, J.; Bornay-Llinares, F.J.; Acosta, B.; Lopez-Aldeguer, J.; Meseguer, I.; Figueras, M.J.; Hernandez, A.; Peset, V.; Gobernado, M.; Visvesvara, G.S. First report of a case of Encephalitozoon sp. microsporidiosis in a Spanish AIDS patient. Res. Rev. Parasitol. 1997, 57, 131–134. [Google Scholar]
- Bornay-Llinares, F.J.; Acosta, B.; Moura, J.H.; Schwartz, D.A.; da Silva, A.J.; Visvesvara, G.S.; Figueras, M.J.; Gobernado, M.; Pieniazek, N.J. Mantenimiento en cultivo y caracterización de un microsporidio (Encephalitozoon hellem) aislado en un paciente con Sida y neumonía. Parasitología 2000, 24, 69–70. [Google Scholar] [CrossRef]
- Del Aguila, C.; Moura, H.; Fenoy, S.; Navajas, R.; Lopez-Velez, R.; Li, L.; Xiao, L.; Leitch, G.J.; da Silva, A.; Pieniazek, N.J.; et al. In vitro culture, ultrastructure, antigenic, and molecular characterization of Encephalitozoon cuniculi from urine and sputum samples from a Spanish patient with AIDS. J. Clin. Microbiol. 2001, 39, 1105–1108. [Google Scholar] [CrossRef]
- Andreu-Ballester, J.C.; Garcia-Ballesteros, C.; Amigo, V.; Ballester, F.; Gil-Borrás, R.; Catalán-Serra, I.; Magnet, A.; Fenoy, S.; del Aguila, C.; Ferrando-Marco, J.; et al. Microsporidia and its relation to Crohn’s disease. A retrospective study. PLoS ONE 2013, 8, e62107. [Google Scholar] [CrossRef]
- Redondo, F.; Hurtado-Marcos, C.; Izquierdo, F.; Cuéllar, C.; Fenoy, S.; Sáez, Y.; Magnet, A.; Galindo-Regal, L.; Uribe, N.; López-Bañeres, M.; et al. Latent Microsporidia Infection Prevalence as a Risk Factor in Colon Cancer Patients. Cancers 2022, 14, 5342. [Google Scholar] [CrossRef]
- Del Aguila, C.; Izquierdo, F.; Navajas, R.; Pieniazek, N.J.; Miró, G.; Alonso, A.I.; da Silva, A.J.; Fenoy, S. Enterocytozoon bieneusi in animals: Rabbits and dogs as new hosts. J. Eukaryot. Microbiol. 1999, 46, 8S–9S. [Google Scholar] [PubMed]
- Lores, B.; del Aguila, C.; Arias, C. Enterocytozoon bieneusi (microsporidia) in faecal samples from domestic animals from Galicia, Spain. Mem. Inst. Oswaldo Cruz 2002, 97, 941–945. [Google Scholar] [CrossRef] [PubMed]
- Galván-Díaz, A.L.; Magnet, A.; Fenoy, S.; Henriques-Gil, N.; Haro, M.; Ponce-Gordo, F.; Millán, J.; Miró, G.; del Águila, C.; Izquierdo, F. Microsporidia detection and genotyping study of human pathogenic E. bieneusi in animals from Spain. PLoS ONE 2014, 9, e92289. [Google Scholar] [CrossRef] [PubMed]
- Dashti, A.; Santín, M.; Cano, L.; de Lucio, A.; Bailo, B.; Hernández-de Mingo, M.; Köster, P.C.; Fernández-Basterra, J.A.; Aramburu-Aguirre, J.; López-Molina, N.; et al. Occurrence and genetic diversity of Enterocytozoon bieneusi (Microsporidia) in owned and sheltered dogs and cats in Northern Spain. Parasitol. Res. 2019, 118, 2979–2987. [Google Scholar] [CrossRef]
- Martínez-Padilla, A.; Caballero-Gómez, J.; Magnet, A.; Gómez-Guillamón, F.; Izquierdo, F.; Camacho-Sillero, L.; Jiménez-Ruiz, S.; del Águila, C.; García-Bocanegra, I. Zoonotic Microsporidia in Wild Lagomorphs in Southern Spain. Animals 2020, 10, 2218. [Google Scholar] [CrossRef]
- Vioque, F.; Dashti, A.; Santín, M.; Ruiz-Fons, F.; Köster, P.C.; Hernández-Castro, C.; García, J.T.; Bailo, B.; Ortega, S.; Olea, P.P.; et al. Wild micromammal host spectrum of zoonotic eukaryotic parasites in Spain. Occurrence and genetic characterisation. Transbound. Emerg. Dis. 2022, 69, e2926–e2942. [Google Scholar] [CrossRef]
- Santín, M.; Calero-Bernal, R.; Carmena, D.; Mateo, M.; Balseiro, A.; Barral, M.; Lima-Barbero, J.F.; Habela, M.A. Molecular Characterization of Enterocytozoon bieneusi in Wild Carnivores in Spain. J. Eukaryot. Microbiol. 2018, 65, 468–474. [Google Scholar] [CrossRef]
- Izquierdo, F.; Ollero, D.; Magnet, A.; Galván-Díaz, A.L.; Llorens, S.; Vaccaro, L.; Hurtado-Marcos, C.; Valdivieso, E.; Miró, G.; Hernández, L.; et al. Microsporidia as a Potential Threat to the Iberian Lynx (Lynx pardinus). Animals 2022, 12, 2507. [Google Scholar] [CrossRef]
- Dashti, A.; Rivero-Juarez, A.; Santín, M.; López-López, P.; Caballero-Gómez, J.; Frías-Casas, M.; Köster, P.C.; Bailo, B.; Calero-Bernal, R.; Briz, V.; et al. Enterocytozoon bieneusi (Microsporidia): Identification of novel genotypes and evidence of transmission between sympatric wild boars (Sus scrofa ferus) and Iberian pigs (Sus scrofa domesticus) in Southern Spain. Transbound. Emerg. Dis. 2020, 67, 2869–2880. [Google Scholar] [CrossRef]
- Dashti, A.; Santín, M.; Köster, P.C.; Bailo, B.; Ortega, S.; Imaña, E.; Habela, M.A.; Rivero-Juarez, A.; Vicente, J.; WE&H Group; et al. Zoonotic Enterocytozoon bieneusi genotypes in free-ranging and farmed wild ungulates in Spain. Med. Mycol. 2022, 60, myac070. [Google Scholar] [CrossRef]
- Haro, M.; Izquierdo, F.; Henriques-Gil, N.; Andrés, I.; Alonso, F.; Fenoy, S.; del Aguila, C. First detection and genotyping of human-associated microsporidia in pigeons from urban parks. Appl. Environ. Microbiol. 2005, 71, 3153–3157. [Google Scholar] [CrossRef] [PubMed]
- Haro, M.; Henriques-Gil, N.; Fenoy, S.; Izquierdo, F.; Alonso, F.; Del Aguila, C. Detection and genotyping of Enterocytozoon bieneusi in pigeons. J. Eukaryot. Microbiol. 2006, 53, S58–S60. [Google Scholar] [CrossRef]
- Dashti, A.; Rivero-Juárez, A.; Santín, M.; George, N.S.; Köster, P.C.; López-López, P.; Risalde, M.A.; García-Bocanegra, I.; Gómez-Villamandos, J.C.; Caballero-Gómez, J.; et al. Diarrhoea-causing enteric protest species in intensively and extensively raised pigs (Sus scrofa domesticus) in Southern Spain. Part I: Prevalence and genetic diversity. Transbound. Emerg. Dis. 2022, 69, e1051–e1064. [Google Scholar] [CrossRef] [PubMed]
- Abarca, N.; Santín, M.; Ortega, S.; Maloney, J.G.; George, N.S.; Molokin, A.; Cardona, G.A.; Dashti, A.; Köster, P.C.; Bailo, B.; et al. Molecular Detection and Characterization of Blastocystis sp. and Enterocytozoon bieneusi in Cattle in Northern Spain. Vet. Sci. 2021, 8, 191. [Google Scholar] [CrossRef] [PubMed]
- Köster, P.C.; Martínez-Nevado, E.; González, A.; Abelló-Poveda, M.T.; Fernández-Bellon, H.; de la Riva-Fraga, M.; Marquet, B.; Guéry, J.P.; Knauf-Witzens, T.; Weigold, A.; et al. Intestinal Protists in Captive Non-human Primates and Their Handlers in Six European Zoological Gardens. Molecular Evidence of Zoonotic Transmission. Front. Vet. Sci. 2022, 8, 819887. [Google Scholar] [CrossRef]
- Rego, L.; Castro-Scholten, S.; Cano, C.; Jiménez-Martín, D.; Köster, P.C.; Caballero-Gómez, J.; Bailo, B.; Dashti, A.; Hernández-Castro, C.; Cano-Terriza, D.; et al. Iberian wild leporidae as hosts of zoonotic enteroparasites in Mediterranean ecosystems of Southern Spain. Zoonoses Public Health 2022, 70, 223–237. [Google Scholar] [CrossRef]
- Rosell, J.; Máinez, M.; Didier, E.S.; Bowers, L.C.; Marco, A.; Juan-Sallés, C. Encephalitozoon hellem infection in aviary passerine and psittacine birds in Spain. Vet. Parasitol. 2016, 219, 57–60. [Google Scholar] [CrossRef]
- Sak, B.; Kvác, M.; Petrzelková, K.; Kvetonová, D.; Pomajbíková, K.; Mulama, M.; Kiyang, J.; Modrý, D. Diversity of microsporidia (Fungi: Microsporidia) among captive great apes in European zoos and African sanctuaries: Evidence for zoonotic transmission? Folia Parasitol. 2011, 58, 81–86. [Google Scholar] [CrossRef]
- Baz-González, E.; Martin-Carrillo, N.; García-Livia, K.; Abreu-Acosta, N.; Foronda, P. Molecular Detection of Microsporidia in Rabbits (Oryctolagus cuniculus) in Tenerife, Canary Islands, Spain. Biology 2022, 11, 1796. [Google Scholar] [CrossRef]
- Galván, A.L.; Magnet, A.; Izquierdo, F.; Fenoy, S.; Rueda, C.; Fernández-Vadillo, C.; Henriques-Gil, N.; del Aguila, C. Molecular characterization of human-pathogenic microsporidia and Cyclospora cayetanensis isolated from various water sources in Spain: A year-long longitudinal study. Appl. Environ. Microbiol. 2013, 79, 449–459. [Google Scholar] [CrossRef]
- Nogales, M.; Rodríguez-Luengo, J.L.; Marrero, P. Ecological effects and distribution of invasive non-native mammals on the Canary Islands. Mammal Rev. 2006, 36, 49–65. [Google Scholar] [CrossRef]
- Pan, G.; Bao, J.; Ma, Z.; Song, Y.; Han, B.; Ran, M.; Li, C.; Zhou, Z. Invertebrate host responses to microsporidia infections. Dev. Comp. Immunol. 2018, 83, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Hinney, B.; Sak, B.; Joachim, A.; Kváč, M. More than a rabbit’s tale—Encephalitozoon spp. in wild mammals and birds. Int. J. Parasitol. Parasit. Wildl. 2016, 5, 76–87. [Google Scholar] [CrossRef]
- Medina, F.M. First record of Algerian hedgehog Atelerix algirus (Lereboullet, 1842) in La Palma Island biosphere reserve. Galemys 2016, 28, 61–62. [Google Scholar] [CrossRef]
- Lobo, M.L.; Xiao, L.; Antunes, F.; Matos, O. Microsporidia as emerging pathogens and the implication for public health: A 10 year study on HIV-positive and –negative patients. Int. J. Parasitol. 2012, 42, 197–205. [Google Scholar] [CrossRef] [PubMed]
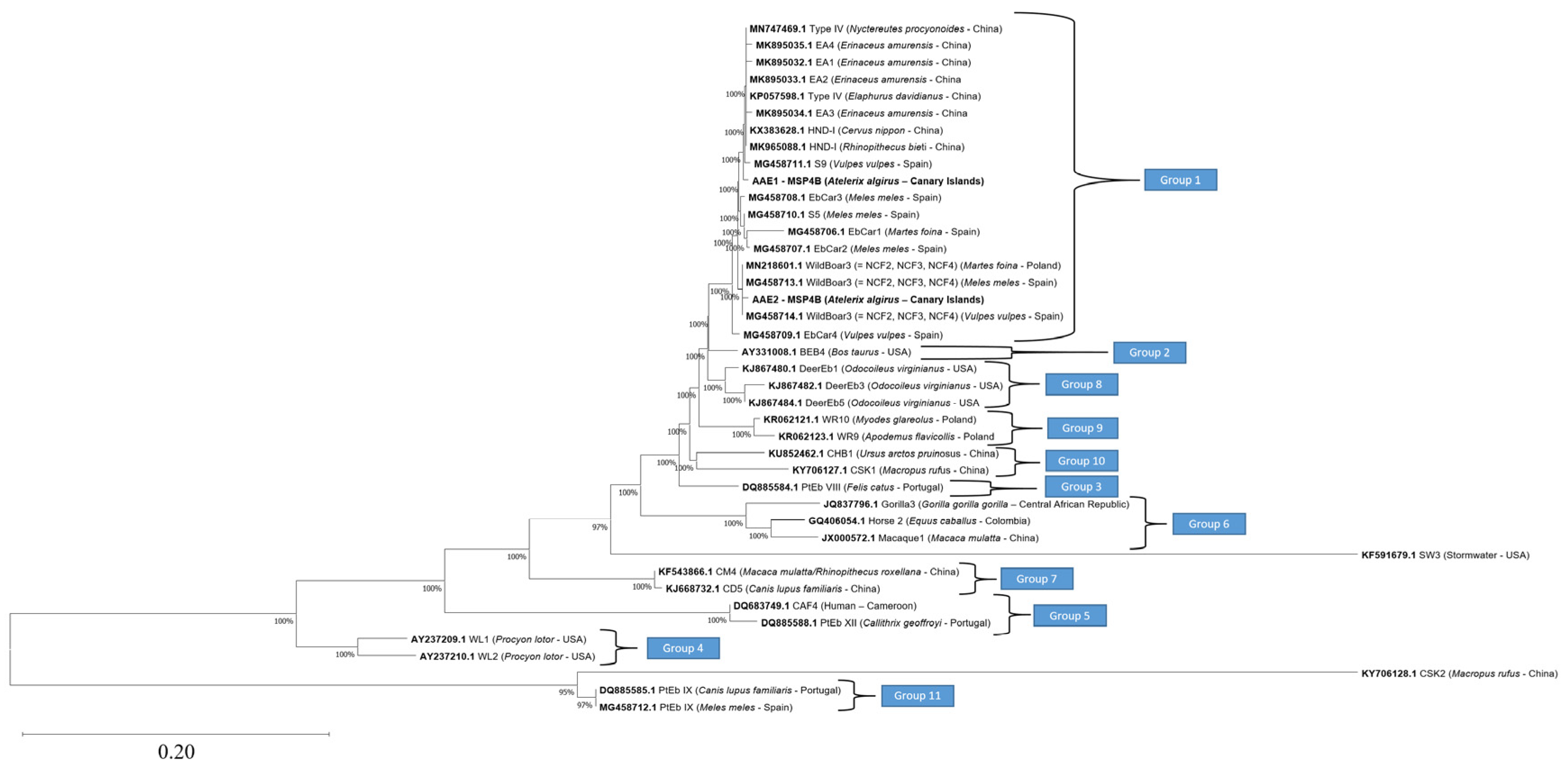
| Genotype (Host) | Nucleotide Position (5′→ 3′) 1 | |||||||
|---|---|---|---|---|---|---|---|---|
| 31 | 32 | 51 | 52 | 53 | 86 | 131 | 155 | |
| AAE1 (Atelerix algirus) | A | T | G | T | - | G | G | A |
| HND-I (Rhinopithecus bieti) | G | T | G | T | A | G | G | A |
| EA1 (Erinaceus amurensis) | G | C | G | T | A | G | G | A |
| EA2 (Erinaceus amurensis) | G | T | - | - | A | G | G | A |
| EA3 (Erinaceus amurensis) | G | T | G | T | A | A | G | A |
| EA4 (Erinaceus amurensis) | G | T | G | T | A | G | G | G |
| S9 (Vulpes vulpes) | G | T | G | T | A | G | A | A |
| Location | Sample Size (n) | Nested-PCR Confirmed Samples (n) | E. bieneusi Genotypes (n) | E. cuniculi Genotypes (n) | Undetermined Species |
|---|---|---|---|---|---|
| Adeje | 1 | 1 | - | - | 1 |
| Arafo | 1 | - | - | - | - |
| Arona | 7 | 6 | AAE1 (5) Undetermined genotype (1) * | - | - |
| El Rosario | 3 | 2 | AAE1 (1) | I (1) | - |
| El Sauzal | 1 | 1 | AAE2 (1) | - | - |
| Granadilla de Abona | 3 | 2 | AAE1 (2) | - | - |
| Güímar | 1 | - | - | - | - |
| Icod de Los Vinos | 1 | - | - | - | - |
| Las Palmas de Gran Canaria | 3 | 3 | AAE1 (3) | - | - |
| San Cristóbal de La Laguna | 7 | 2 | - | - | 2 |
| San Miguel de Abona | 1 | - | - | - | - |
| Santa Cruz de Tenerife | 5 | 3 | AAE1 (2) | I (1) | - |
| Tacoronte | 2 | 2 | AAE2 (2) | - | - |
| TOTAL | 36 | 22 | 17 | 2 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baz-González, E.; Abreu-Acosta, N.; Foronda, P. High Prevalence of Microsporidia in the North African Hedgehog (Atelerix algirus) in the Canary Islands, Spain. Animals 2023, 13, 1756. https://doi.org/10.3390/ani13111756
Baz-González E, Abreu-Acosta N, Foronda P. High Prevalence of Microsporidia in the North African Hedgehog (Atelerix algirus) in the Canary Islands, Spain. Animals. 2023; 13(11):1756. https://doi.org/10.3390/ani13111756
Chicago/Turabian StyleBaz-González, Edgar, Néstor Abreu-Acosta, and Pilar Foronda. 2023. "High Prevalence of Microsporidia in the North African Hedgehog (Atelerix algirus) in the Canary Islands, Spain" Animals 13, no. 11: 1756. https://doi.org/10.3390/ani13111756
APA StyleBaz-González, E., Abreu-Acosta, N., & Foronda, P. (2023). High Prevalence of Microsporidia in the North African Hedgehog (Atelerix algirus) in the Canary Islands, Spain. Animals, 13(11), 1756. https://doi.org/10.3390/ani13111756






