Effects of Dietary Protein Restriction on Colonic Microbiota of Finishing Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Treatment and Experimental Design
2.2. Direction Indicators
2.2.1. Blood Biochemical Parameters
2.2.2. Short-Chain Fatty Acids Analysis
2.2.3. Microbiota Analysis by 16S RNA
2.3. Statistical Analysis
3. Result
3.1. Serum Biochemical Parameters
3.2. SCFAs
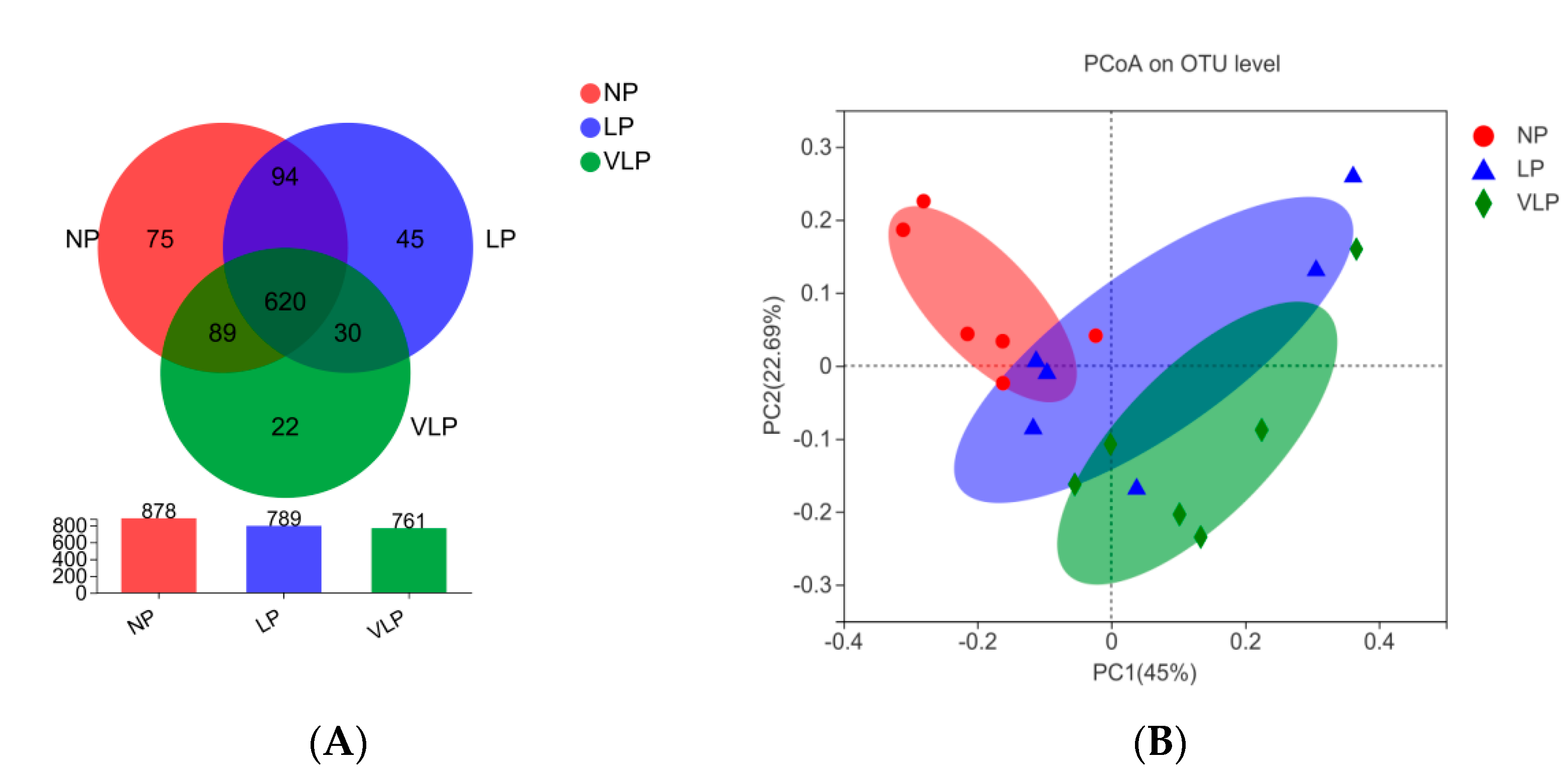
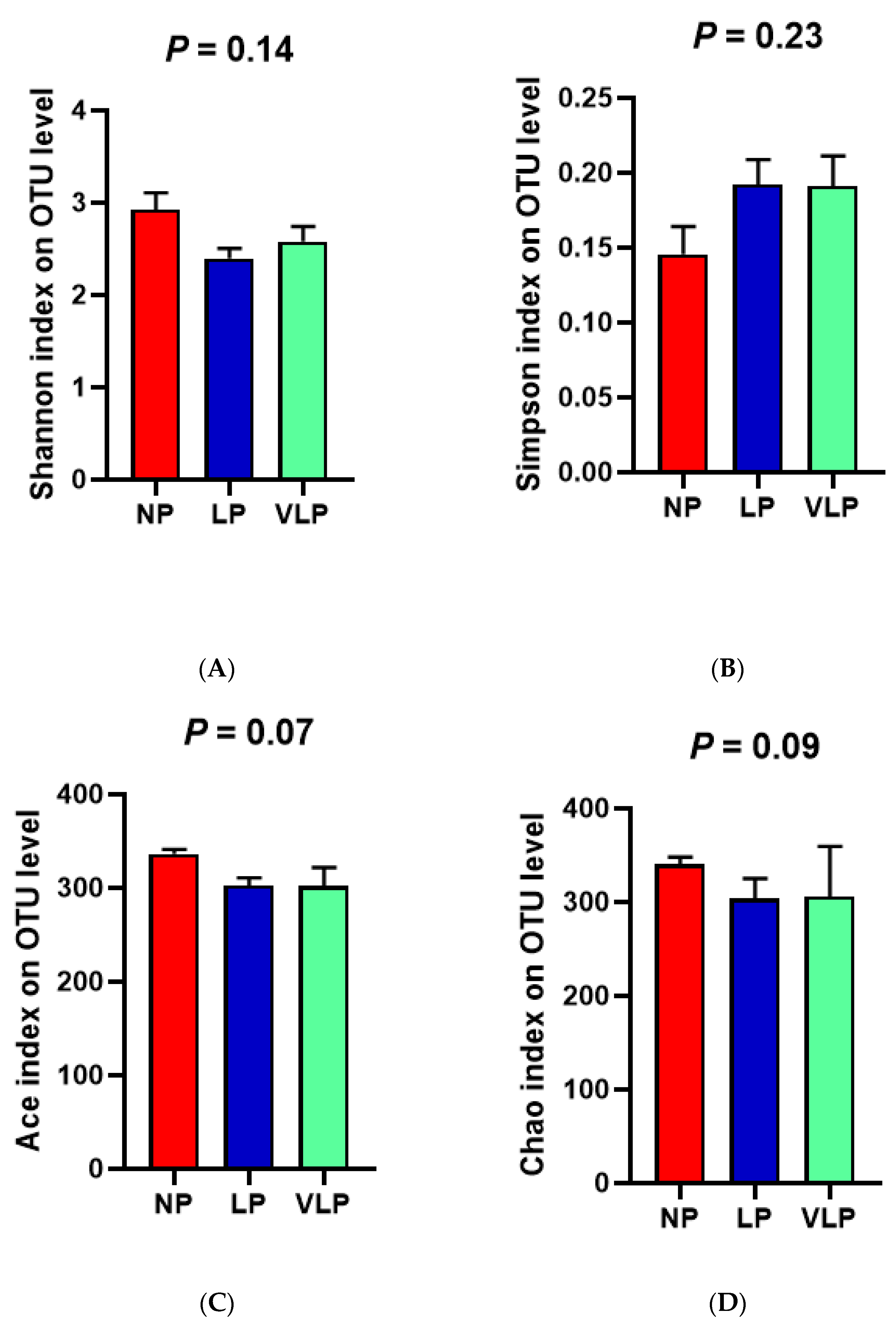
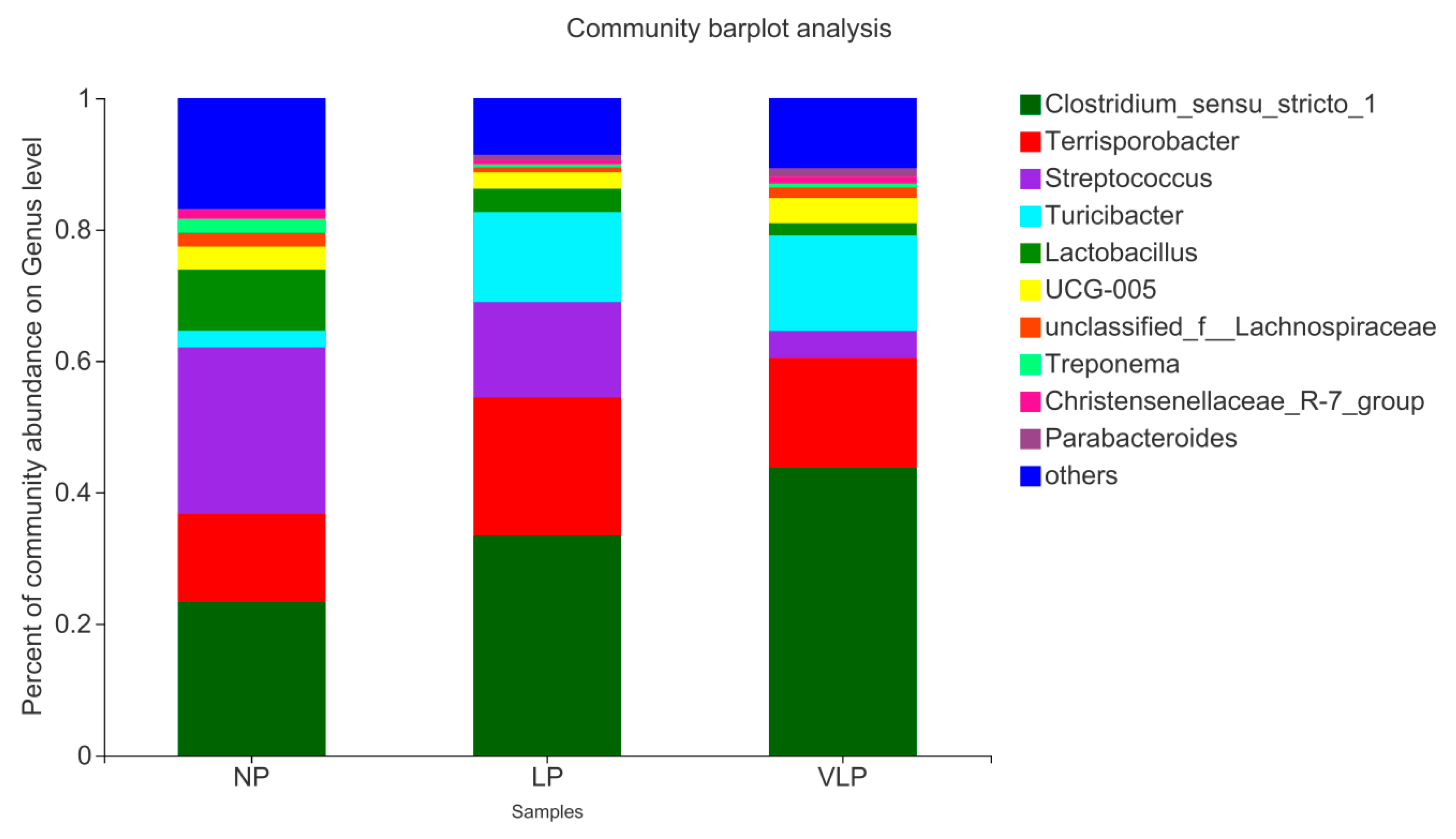
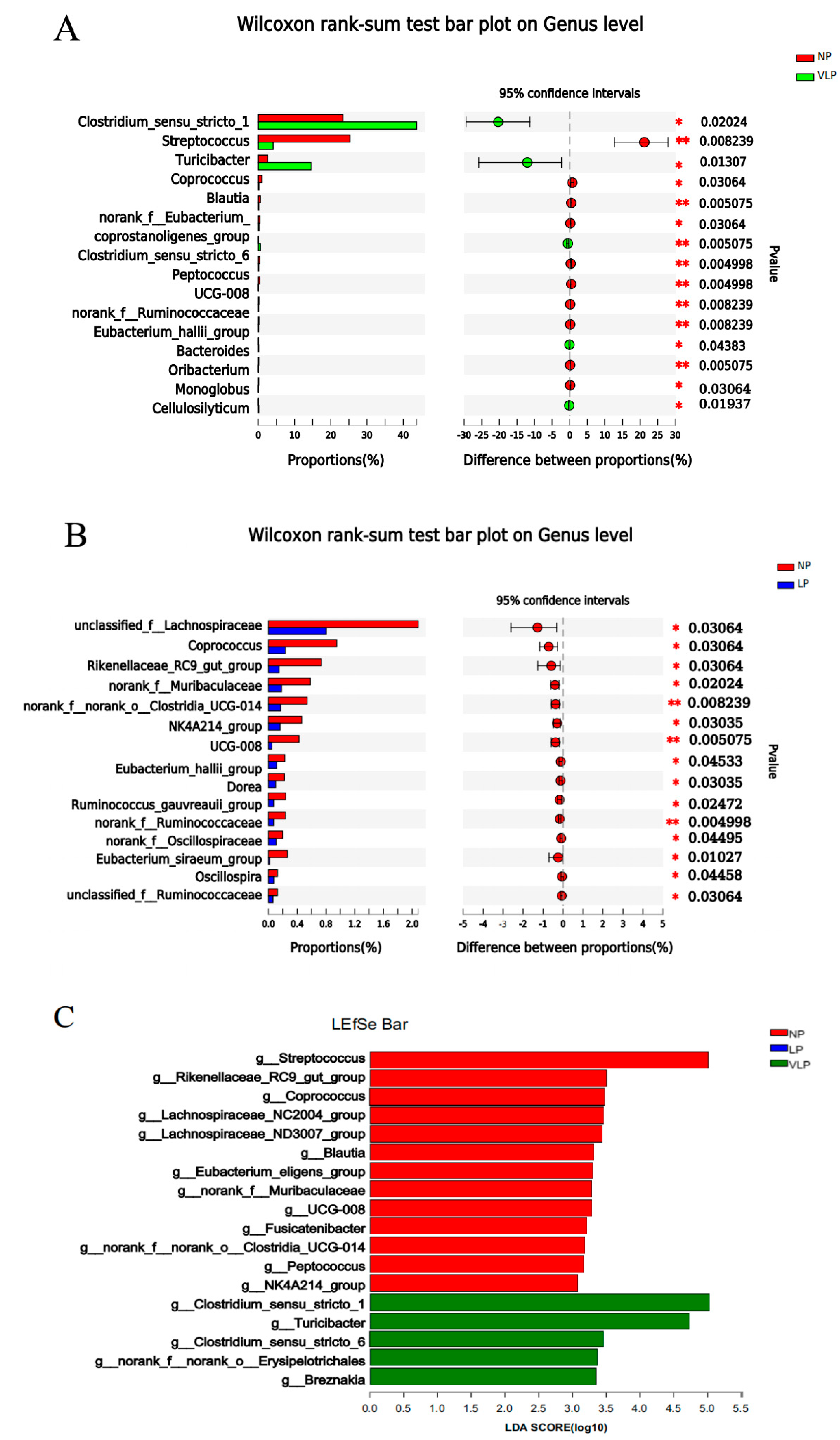
3.3. Structural Changes in The Microbial Community
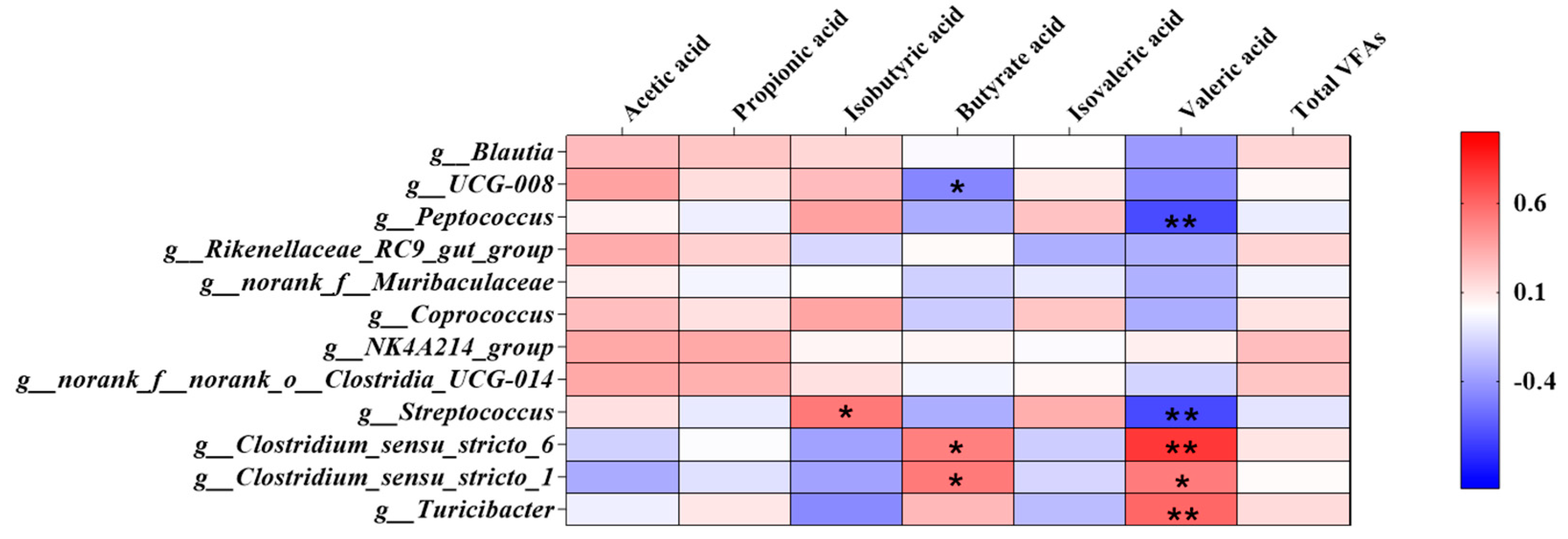
3.4. Correlation of Bacteria and Colonic SCFAs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, G.; Bazer, F.W.; Dai, Z.; Li, D.; Wang, J.; Wu, Z. Amino acid nutrition in animals: Protein synthesis and beyond. Annu. Rev. Anim. Biosci. 2014, 2, 387–417. [Google Scholar] [CrossRef] [PubMed]
- Leek, A.B.; Hayes, E.T.; Curran, T.P.; Callan, J.J.; Beattie, V.E.; Dodd, V.A.; O’Doherty, J.V. The influence of manure composition on emissions of odour and ammonia from finishing pigs fed different concentrations of dietary crude protein. Bioresour. Technol. 2007, 98, 3431–3439. [Google Scholar] [CrossRef] [PubMed]
- PPrandini, A.L.D.O.; Sigolo, S.; Morlacchini, M.; Grilli, E.; Fiorentini, L. Microencapsulated lysine and low-protein diets: Effects on performance, carcass characteristics and nitrogen excretion in heavy growing–finishing pigs. J. Anim. Sci. 2013, 91, 4226–4234. [Google Scholar] [CrossRef] [PubMed]
- Rajilić-Stojanović, M.; De Vos, W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014, 38, 996–1047. [Google Scholar] [CrossRef] [PubMed]
- Willing, B.P.; Van Kessel, A.G. Host pathways for recognition: Establishing gastrointestinal microbiota as relevant in animal health and nutrition. Livest. Sci. 2010, 133, 82–91. [Google Scholar] [CrossRef]
- Gibson, J.A.; Sladen, G.E.; Dawson, A.M. Protein absorption and ammonia production: The effects of dietary protein and removal of the colon. Br. J. Nutr. 1976, 35, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–215. [Google Scholar] [CrossRef]
- Jin, C.; Flavell, R.A. Innate sensors of pathogen and stress: Linking inflammation to obesity. J. Allergy Clin. Immunol. 2013, 132, 287–294. [Google Scholar] [CrossRef]
- Kim, H.B.; Isaacson, R.E. The pig gut microbial diversity: Understanding the pig gut microbial ecology through the next generation high throughput sequencing. Vet. Microbiol. 2015, 177, 242–251. [Google Scholar] [CrossRef]
- Yu, M.; Zhang, C.; Yang, Y.; Mu, C.; Su, Y.; Yu, K.; Zhu, W. Long-term effects of early antibiotic intervention on blood parameters, apparent nutrient digestibility, and fecal microbial fermentation profile in pigs with different dietary protein levels. J. Anim. Sci. Biotechnol. 2017, 8, 60. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tang, L.; Liu, N.; Zhang, F.; Liu, X.; Jiang, Q.; Chen, J.; Ma, X. Comparative Effects of Compound Enzyme and Antibiotics on Growth Performance, Nutrient Digestibility, Blood Biochemical Index, and Intestinal Health in Weaned Pigs. Front. Microbiol. 2021, 12, 768767. [Google Scholar] [CrossRef]
- Figueroa, J.L.; Lewis, A.J.; Miller, P.S.; Fischer, R.L.; Gómez, R.S.; Diedrichsen, R.M. Nitrogen metabolism and growth performance of gilts fed standard corn-soybean meal diets or low-crude protein, amino acid-supplemented diets. J. Anim. Sci. 2002, 80, 2911–2919. [Google Scholar] [CrossRef]
- Zervas, S.; Zijlstra, R.T. Effects of dietary protein and fermentable fiber on nitrogen excretion patterns and plasma urea in grower pigs. J. Anim. Sci. 2002, 80, 3247–3256. [Google Scholar] [CrossRef]
- Kerr, B.J.; McKeith, F.K.; Easter, R.A. Effect on performance and carcass characteristics of nursery to finisher pigs fed reduced crude protein, amino acid-supplemented diets. J. Anim. Sci. 1995, 73, 433–440. [Google Scholar] [CrossRef]
- Figueroa, J.L.; Lewis, A.J.; Miller, P.S.; Fischer, R.L.; Diedrichsen, R.M. Growth, carcass traits, and plasma amino acid concentrations of gilts fed low-protein diets supplemented with amino acids including histidine, isoleucine, and valine. J. Anim. Sci. 2003, 81, 1529–1537. [Google Scholar] [CrossRef]
- Madeira, M.S.; Rolo, E.A.; Lopes, P.A.; Ramos, D.A.; Alfaia, C.M.; Pires, V.M.; Prates, J.A. Betaine and arginine supplementation of low protein diets improves plasma lipids but does not affect hepatic fatty acid composition and related gene expression profiling in pigs. J. Sci. Food Agric. 2018, 98, 598–608. [Google Scholar] [CrossRef]
- Wang, M.; Sun, H.; Xu, Z. Analysis of Blueberry Plant Rhizosphere Bacterial Diversity and Selection of Plant Growth Promoting Rhizobacteria. Curr. Microbiol. 2022, 79, 331. [Google Scholar] [CrossRef]
- Zhou, L.; Fang, L.; Sun, Y.; Su, Y.; Zhu, W. Effects of the dietary protein level on the microbial composition and metabolomic profile in the hindgut of the pig. Anaerobe 2016, 38, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Liu, P.; Song, P.; Chen, X.; Ma, X. Moderate dietary protein restriction alters the composition of gut microbiota and improves ileal barrier function in adult pig model. Sci. Rep. 2017, 7, 43412. [Google Scholar] [CrossRef] [PubMed]
- Kujawa-Szewieczek, A.; Adamczak, M.; Kwiecień, K.; Dudzicz, S.; Gazda, M.; Więcek, A. The effect of Lactobacillus plantarum 299v on the incidence of Clostridium difficile infection in high risk patients treated with antibiotics. Nutrients 2015, 7, 10179–10188. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Y.; Lee, Y.; Lu, H.; Chou, C.H.; Wang, C. Analysis of gut microbiota and the effect of lauric acid against necrotic enteritis in Clostridium perfringens and Eimeria side-by-side challenge model. PLoS ONE 2019, 14, e0205784. [Google Scholar] [CrossRef] [PubMed]
- Munyaka, P.M.; Rabbi, M.F.; Khafipour, E.; Ghia, J.E. Acute dextran sulfate sodium (DSS)-induced colitis promotes gut microbial dysbiosis in mice. J. Basic Microbiol. 2016, 56, 986–998. [Google Scholar] [CrossRef]
- Moya, A.; Ferrer, M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016, 24, 402–413. [Google Scholar] [CrossRef]
- La Rosa, S.L.; Leth, M.L.; Michalak, L.; Hansen, M.E.; Pudlo, N.A.; Glowacki, R.; Pereira, G.; Workman, C.T.; Arntzen, M.Ø.; Pope, P.B.; et al. The human gut Firmicute Roseburia intestinalis is a primary degrader of dietary β-mannans. Nat. Commun. 2019, 10, 905. [Google Scholar] [CrossRef]
- Kllingray, L.; Le Gall, G.; Defernez, M.; Beales, I.L.; Franslem-Elumogo, N.; Narbad, A. Microbial taxonomic and metabolic alterations during faecal microbiota transplantation to treat Clostridium difficile infection. J. Infect. 2018, 77, 107–118. [Google Scholar] [CrossRef]
- Navarro, D.M.; Abelilla, J.J.; Stein, H.H. Structures and characteristics of carbohydrates in diets fed to pigs: A review. J. Anim. Sci. Biotechnol. 2019, 10, 39. [Google Scholar] [CrossRef]
- Morita, T.; Kasaoka, S.; Kiriyama, S. Physiological functions of resistant proteins: Proteins and peptides regulating large bowel fermentation of indigestible polysaccharide. J. AOAC Int. 2004, 87, 792–796. [Google Scholar] [CrossRef]
- Han, P.; Ma, X.; Yin, J. The effects of lipoic acid on soybean β-conglycinin-induced anaphylactic reactions in a rat model. Arch. Anim. Nutr. 2010, 64, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Cummings, J.H.; Allison, C. Protein degradation by human intestinal bacteria. Microbiology 1986, 132, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Meerveld, G.V.; Johnson, A.C.; Grundy, D. Gastrointestinal physiology and function. In Gastrointestinal Pharmacology; Springer: Cham, Switzerland, 2017; pp. 1–16. [Google Scholar]
- Zhao, Y.; Tian, G.; Chen, D.; Zheng, P.; Yu, J.; He, J.; Yu, B. Dietary protein levels and amino acid supplementation patterns alter the composition and functions of colonic microbiota in pigs. Anim. Nutr. 2020, 6, 143–151. [Google Scholar] [CrossRef] [PubMed]
| Item | Dietary Treatments a | ||
|---|---|---|---|
| NP | LP | VLP | |
| Corn | 78.17 | 87.38 | 83.86 |
| Soybean meal | 16.56 | 7.40 | - |
| Soybean oil | 1.08 | 0.05 | 2.71 |
| Stone powder | 1.01 | 1.08 | 1.10 |
| Premix b | 2.00 | 2.00 | 2.00 |
| Mould inhibitor | 0.05 | 0.05 | 0.05 |
| L-Lysine HCL | 0.22 | 0.48 | 0.72 |
| Potassium chloride | - | 0.25 | 0.51 |
| Sodium chloride | 0.50 | 0.50 | 0.50 |
| Calcium hydrogen phosphate | 0.37 | 0.41 | 0.50 |
| DL-Methionine | 0.03 | 0.10 | 0.19 |
| Tryptophan | 0.01 | 0.05 | 0.09 |
| Threonine | - | 0.11 | 0.24 |
| Valine | - | 0.07 | 0.23 |
| Isoleucine | - | 0.06 | 0.21 |
| Histidine | - | 0.01 | 0.10 |
| Phenylalanine | - | - | 0.15 |
| Filler | - | - | 6.84 |
| Total | 100.00 | 100.00 | 100.00 |
| Calculated nutrient levels | |||
| Net energy, kcal/kg | 2475 | 2475 | 2475 |
| Crude protein | 13.50 | 10.76 | 8.02 |
| Potassium | 0.52 | 0.52 | 0.52 |
| Calcium | 0.52 | 0.52 | 0.52 |
| Dispensable amino acids | |||
| SID Lysine | 0.73 | 0.73 | 0.73 |
| SID Methionine | 0.23 | 0.27 | 0.31 |
| SID Cystine | 0.20 | 0.16 | 0.11 |
| SID Methionine + cystine | 0.42 | 0.42 | 0.42 |
| SID threonine | 0.46 | 0.46 | 0.46 |
| SID tryptophan | 0.13 | 0.13 | 0.13 |
| Items 1 | NP | LP | VLP | SEM 2 | p-Value |
|---|---|---|---|---|---|
| TBA | 23.452 | 20.983 | 41.785 | 5.051 | 0.06 |
| GLU | 2.538 | 5.112 | 3.393 | 1.117 | 0.30 |
| TC | 1.593 | 1.65 | 1.81 | 0.135 | 0.52 |
| TG | 0.347 | 0.367 | 0.467 | 0.04 | 0.13 |
| HDL-C | 0.653 | 0.667 | 0.797 | 0.073 | 0.35 |
| LDL-C | 0.735 | 0.677 | 0.563 | 0.075 | 0.31 |
| Items 1 | NP | LP | VLP | SEM 2 | p-Value |
|---|---|---|---|---|---|
| Acetic acid | 416.07 | 382.35 | 390.74 | 21.41 | 0.53 |
| Propionic acid | 165.42 | 160.43 | 163.01 | 10.51 | 0.95 |
| Isobutyric acid | 16.03 | 14.78 | 13.05 | 1.23 | 0.27 |
| Butyric acid | 99.43 | 116.50 | 129.12 | 13.76 | 0.23 |
| Isovaleric acid | 25.24 | 24.55 | 24.54 | 2.51 | 0.97 |
| Valeric acid | 18.47 b | 20.42 b | 30.40 a | 2.78 | 0.03 |
| Total SCFAs | 734.66 | 719.02 | 750.86 | 42.83 | 0.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Fan, Z. Effects of Dietary Protein Restriction on Colonic Microbiota of Finishing Pigs. Animals 2023, 13, 9. https://doi.org/10.3390/ani13010009
Liu S, Fan Z. Effects of Dietary Protein Restriction on Colonic Microbiota of Finishing Pigs. Animals. 2023; 13(1):9. https://doi.org/10.3390/ani13010009
Chicago/Turabian StyleLiu, Shanghang, and Zhiyong Fan. 2023. "Effects of Dietary Protein Restriction on Colonic Microbiota of Finishing Pigs" Animals 13, no. 1: 9. https://doi.org/10.3390/ani13010009
APA StyleLiu, S., & Fan, Z. (2023). Effects of Dietary Protein Restriction on Colonic Microbiota of Finishing Pigs. Animals, 13(1), 9. https://doi.org/10.3390/ani13010009




