Phosphoproteomics Profile of Chicken Cecum in the Response to Salmonella enterica Serovar Enteritidis Inoculation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Inoculation and Sample Collection
2.2. Protein Extraction and Trypsin Digestion
2.3. TMT Labeling and High-Performance Liquid Chromatography (HPLC) Fractionation
2.4. Affinity Enrichment of the Phosphopeptides
2.5. LC-MS/MS Analysis
2.6. Database Search
2.7. Functional Classification of Differentially Phosphorylated Proteins (DPPs) Corresponding to Differentially Phosphorylated Modification Sites
2.8. Functional Enrichment Based on Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway, and Protein Domain of DPPs
2.9. Protein–Protein Interaction (PPI) Network
2.10. Motif Analysis
3. Results
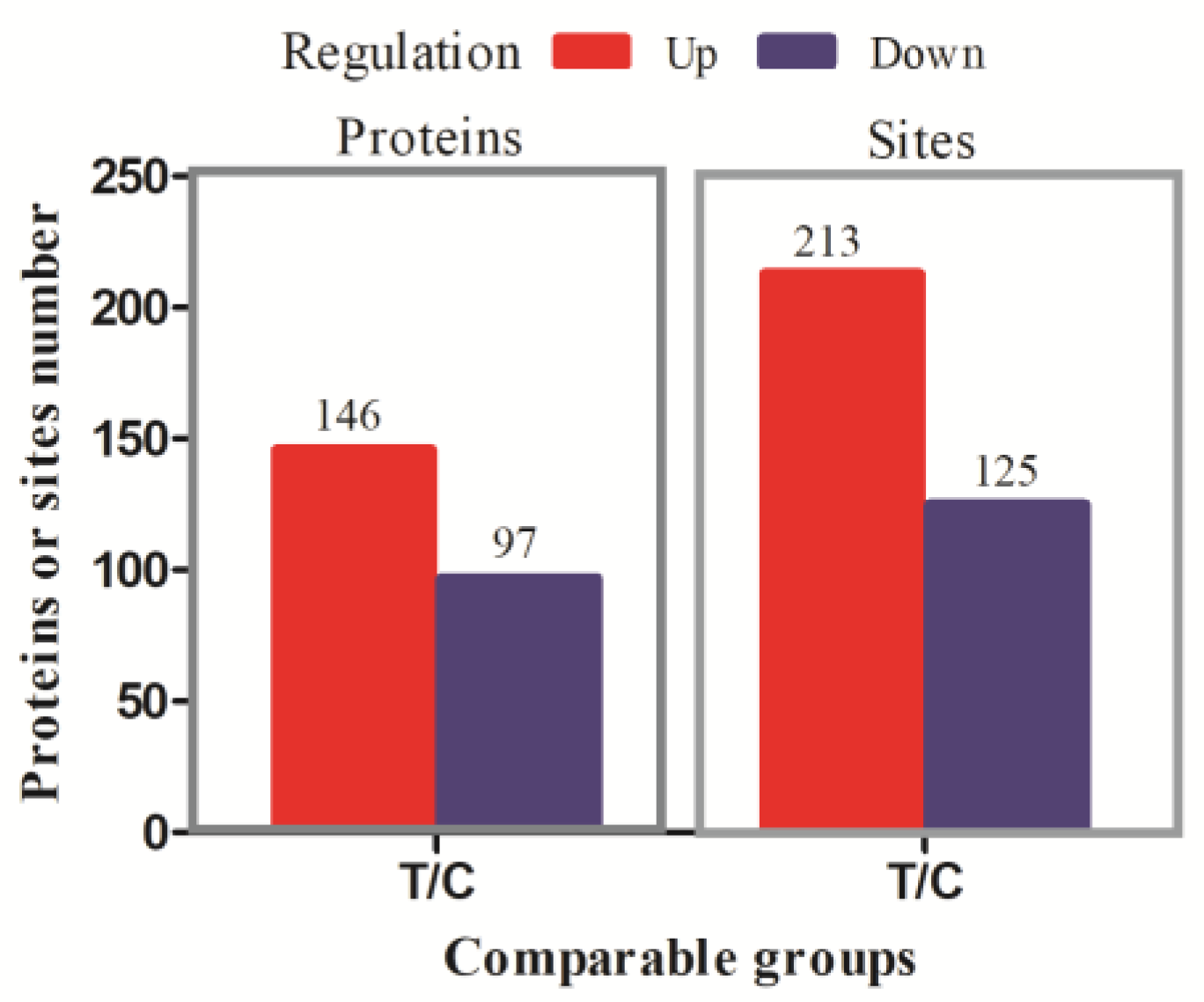
3.1. Identification of Phosphorylated Modification Sites and DPPs of Cecum in Chickens
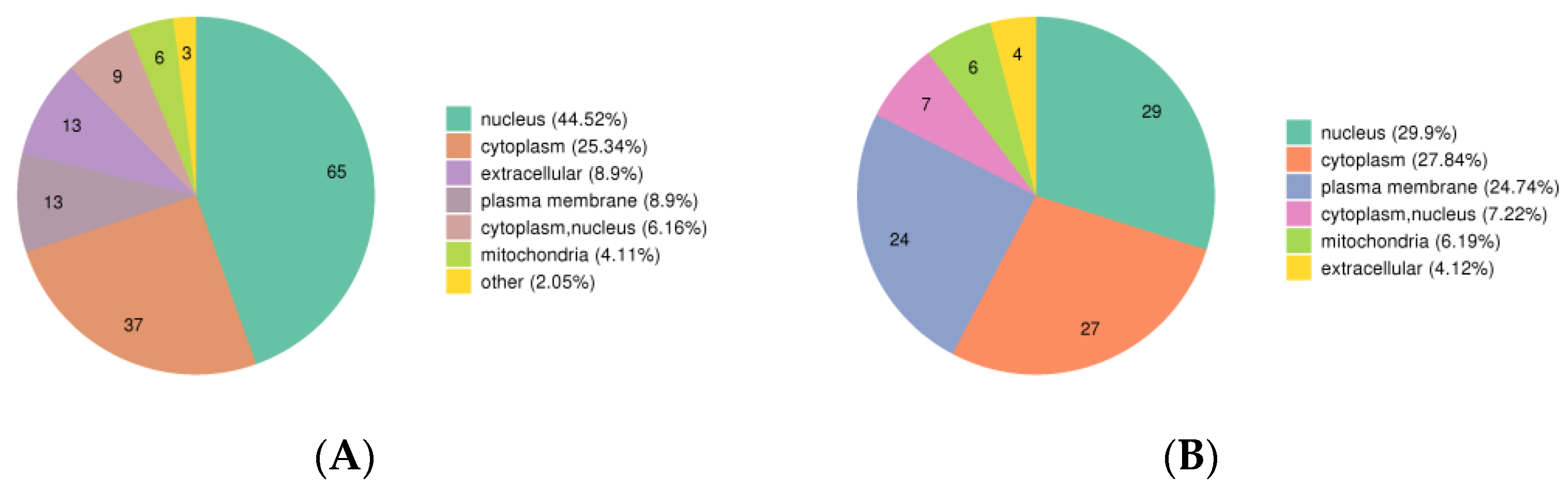
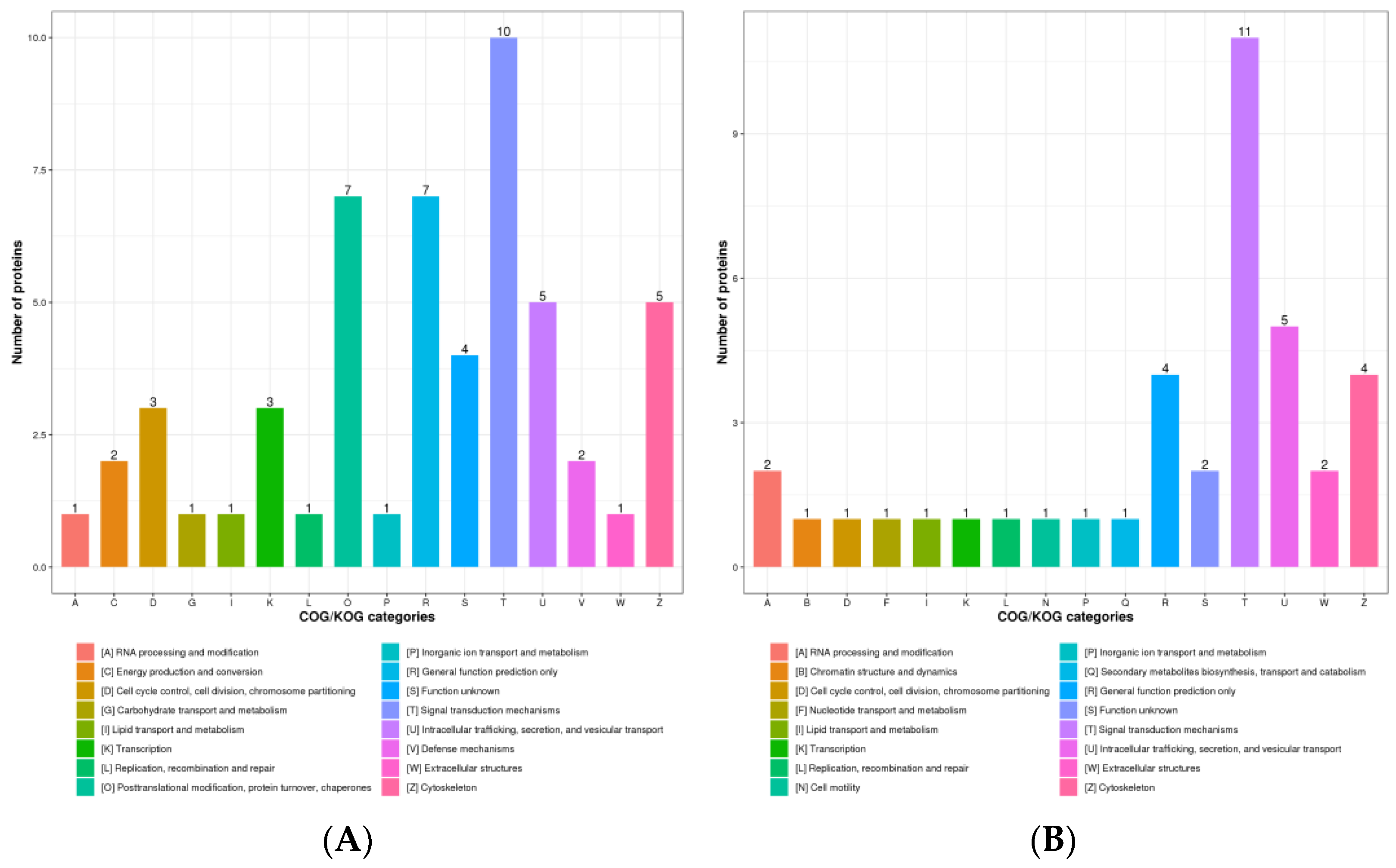
3.2. Subcellular Structure Localization and COG/KOG Functional Classification of DPPs
3.3. Functional Analysis of DPPs in Chicken Cecum
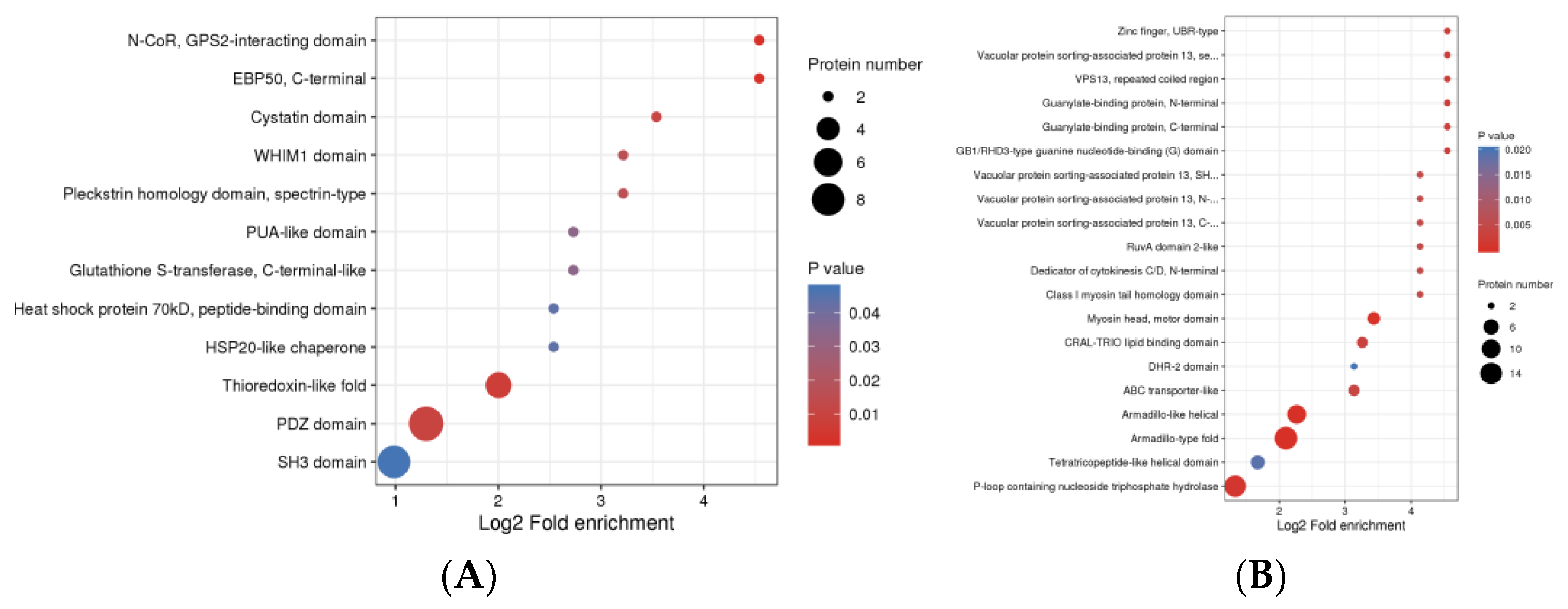
3.4. Protein Domain Analysis
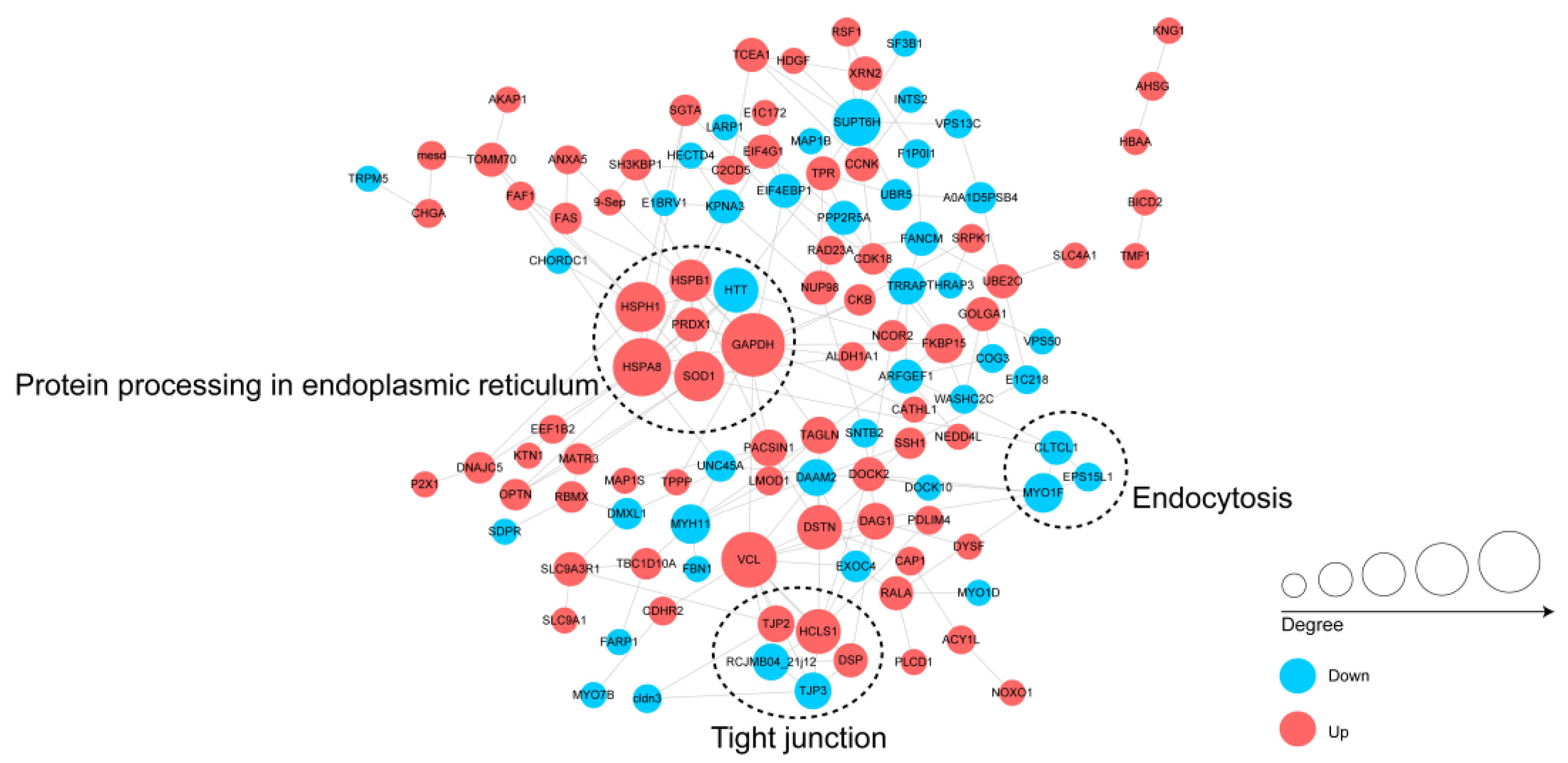
3.5. Protein–Protein Interactions (PPIs) Networks of the DPPs
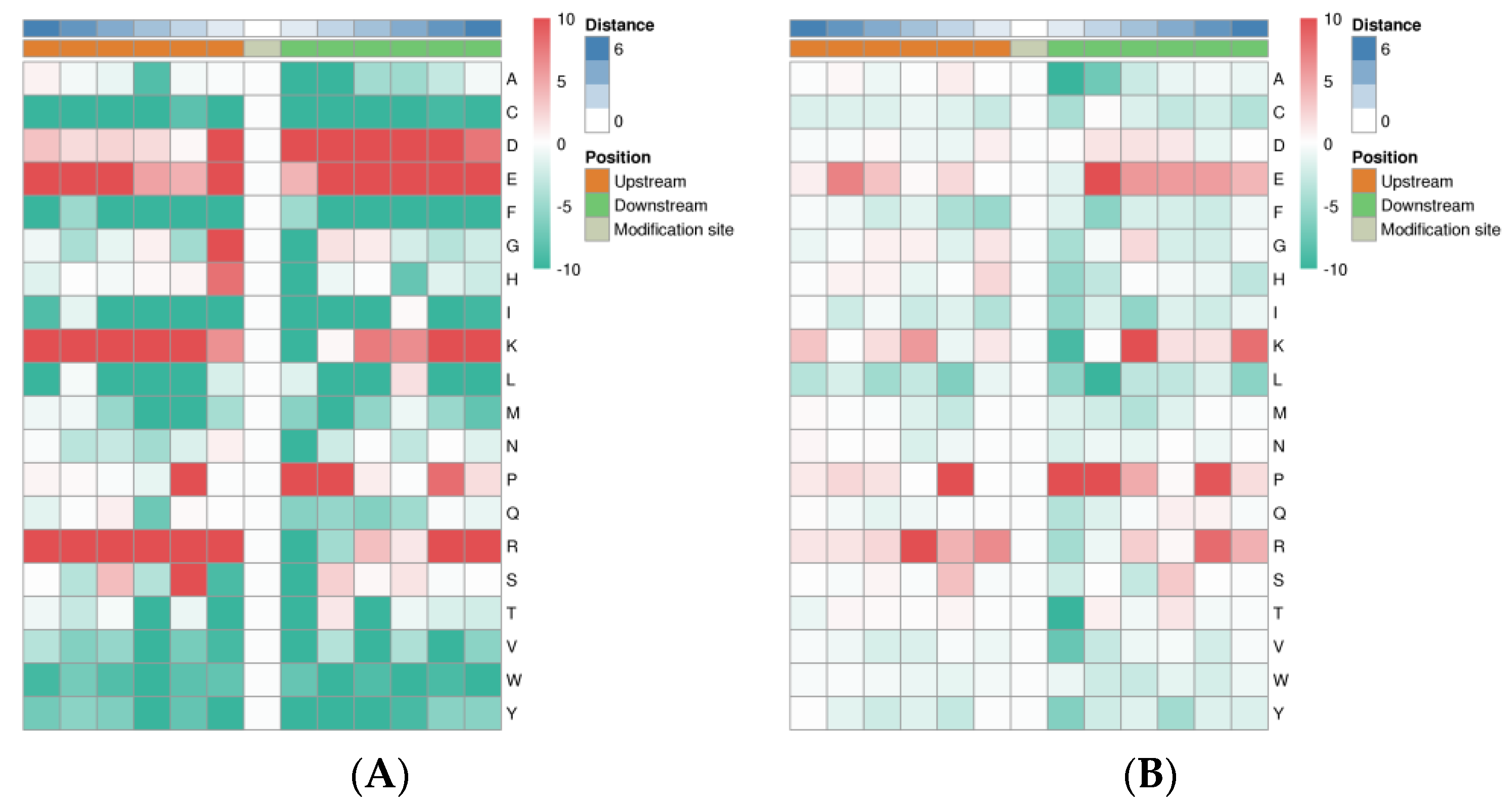
3.6. Motif Analysis of the Phosphorylated Modification Sites
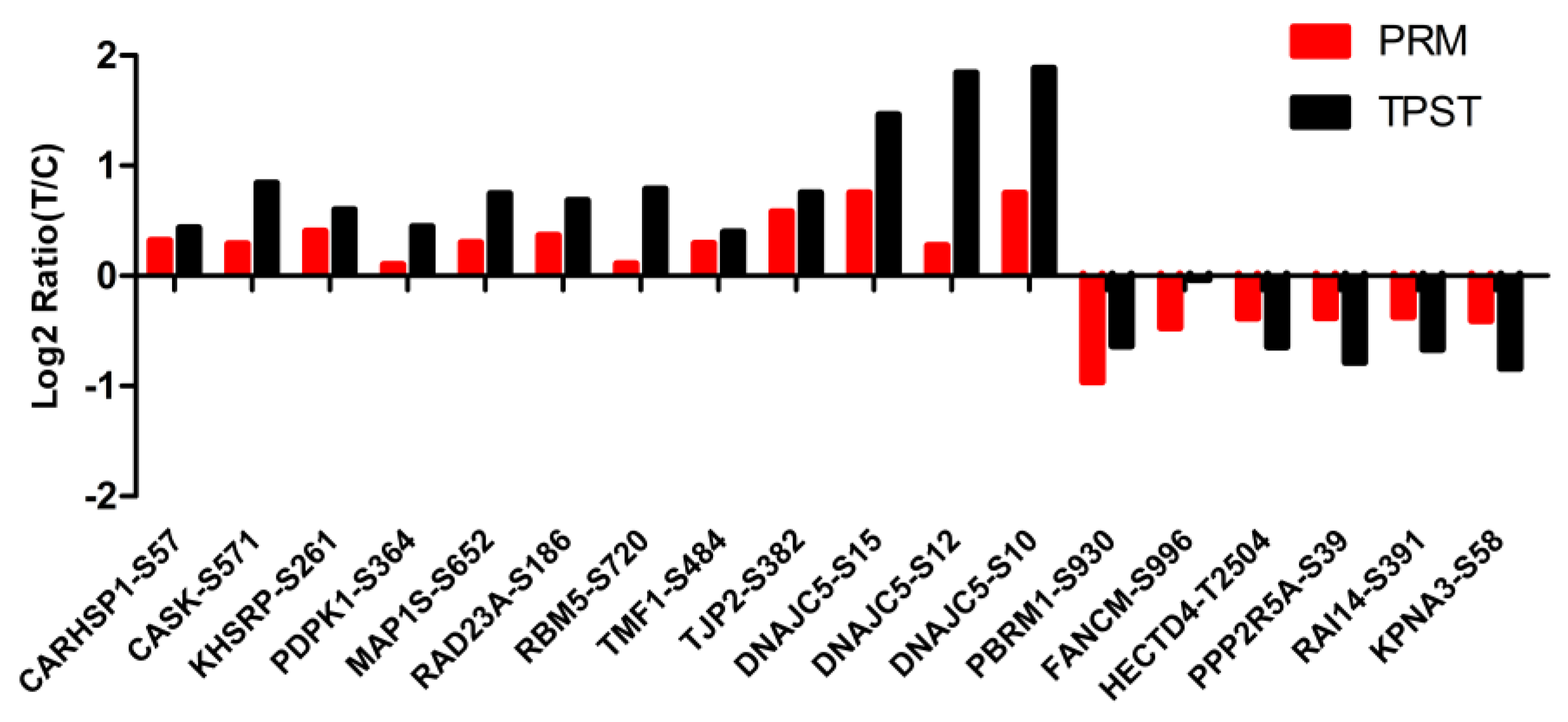
3.7. Validation of the Phosphorylated Modification Sites with Parallel Reaction Monitoring (PRM)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, F.; Geng, Y.; Abbas, W.; Zhen, W.; Wang, S.; Huang, Y.; Guo, Y.; Ma, Q.; Wang, Z. Vitamin D3 nutritional status affects gut health of Salmonella-challenged laying hens. Front. Nutr. 2022, 9, 888580. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, I.; Lingens, J.B.; Chuppava, B.; Wilke, V.; Abd El-Wahab, A.; Buch, J.; Hankel, J.; Ahmed, M.F.E.; Visscher, C. In Vitro evaluation of sodium butyrate on the growth of three Salmonella serovars derived from pigs at a mild acidic pH value. Front. Vet. Sci. 2022, 9, 937671. [Google Scholar] [CrossRef] [PubMed]
- Andino, A.; Hanning, I. Salmonella enterica: Survival, colonization, and virulence differences among serovars. Sci. World J. 2015, 2015, 520179. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Wang, Z.; Tian, X.; Guo, Y.; Zhang, H. Yeast beta-d-glucans induced antimicrobial peptide expressions against Salmonella infection in broiler chickens. Int. J. Biol Macromol. 2016, 85, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Zhen, W.; Shao, Y.; Gong, X.; Wu, Y.; Geng, Y.; Wang, Z.; Guo, Y. Effect of dietary Bacillus coagulans supplementation on growth performance and immune responses of broiler chickens challenged by Salmonella enteritidis. Poult. Sci. 2018, 97, 2654–2666. [Google Scholar] [CrossRef] [PubMed]
- Van Cuyck, H.; Farbos-Granger, A.; Leroy, P.; Yith, V.; Guillard, B.; Sarthou, J.L.; Koeck, J.L.; Kruy, S.L. MLVA polymorphism of Salmonella enterica subspecies isolated from humans, animals, and food in Cambodia. BMC Res. Notes 2011, 4, 306. [Google Scholar] [CrossRef]
- Jajere, S.M. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World 2019, 12, 504–521. [Google Scholar] [CrossRef]
- Wu, G.; Liu, L.; Qi, Y.; Sun, Y.; Yang, N.; Xu, G.; Zhou, H.; Li, X. Splenic gene expression profiling in White Leghorn layer inoculated with the Salmonella enterica serovar Enteritidis. Anim. Genet. 2015, 46, 617–626. [Google Scholar] [CrossRef]
- Hu, G.; Liu, L.; Miao, X.; Zhao, Y.; Li, X. Research Note: IsomiRs of chicken miR-146b-5p are activated upon Salmonella enterica serovar Enteritidis infection. Poult. Sci. 2022, 101, 101977. [Google Scholar] [CrossRef]
- Fan, Y.; Cheng, Y.; Li, Y.; Chen, B.; Wang, Z.; Wei, T.; Zhang, H.; Guo, Y.; Wang, Q.; Wei, Y.; et al. Phosphoproteomic analysis of neonatal regenerative myocardium revealed important roles of checkpoint kinase 1 via activating mammalian target of Rapamycin C1/Ribosomal protein S6 kinase b-1 pathway. Circulation 2020, 141, 1554–1569. [Google Scholar] [CrossRef]
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.C.; Yates, J.R., 3rd. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Min, C.W.; Kim, Y.J.; Kim, S.T. Identification of Msp1-induced signaling components in rice leaves by integrated proteomic and phosphoproteomic analysis. Int. J. Mol Sci. 2019, 20, 4135. [Google Scholar] [CrossRef]
- Huang, J.; Ruan, J.; Tang, X.; Zhang, W.; Ma, H.; Zou, S. Comparative proteomics and phosphoproteomics analyses of DHEA-induced on hepatic lipid metabolism in broiler chickens. Steroids 2011, 76, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Arshad, R.; Meng, Y.; Qiu, N.; Sun, H.; Keast, R.; Rehman, A. Phosphoproteomic analysis of duck egg white and insight into the biological functions of identified phosphoproteins. J. Food Biochem. 2020, 44, e13367. [Google Scholar] [CrossRef] [PubMed]
- Arshad, R.; Meng, Y.; Qiu, N.; Geng, F.; Mine, Y.; Keast, R.; Zhu, C. Phosphoproteomic analysis of duck egg yolk provides novel insights into its characteristics and biofunctions. J. Sci. Food Agric. 2022, 102, 1165–1173. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Sun, H.; Qiu, N.; Keast, R.; Wang, H.; Li, B.; Huang, Q.; Li, S. Comparative quantitative phosphoproteomic analysis of the chicken egg during incubation based on tandem mass tag labeling. J. Agric. Food Chem. 2019, 67, 13353–13361. [Google Scholar] [CrossRef]
- Sun, Y.; Jin, H.; Sun, H.; Sheng, L. A comprehensive identification of chicken egg white phosphoproteomics based on a novel digestion approach. J. Agric. Food Chem. 2020, 68, 9213–9222. [Google Scholar] [CrossRef]
- Hang, J.; Ouyang, H.; Wei, F.; Zhong, Q.; Yuan, W.; Jiang, L.; Liu, Z. Proteomics and phosphoproteomics of chordoma biopsies reveal alterations in multiple pathways and aberrant kinases activities. Front. Oncol. 2022, 12, 941046. [Google Scholar] [CrossRef] [PubMed]
- Ilouz, N.; Harazi, A.; Guttman, M.; Daya, A.; Ruppo, S.; Yakovlev, L.; Mitrani-Rosenbaum, S. In vivo and in vitro genome editing to explore GNE functions. Front. Genome Ed. 2022, 4, 930110. [Google Scholar] [CrossRef]
- Kuffer, S.; Grabowski, J.; Okada, S.; Sojka, N.; Welter, S.; von Hammerstein-Equord, A.; Hinterthaner, M.; Cordes, L.; von Hahn, X.; Muller, D.; et al. Phosphoproteomic analysis identifies TYRO3 as a mediator of sunitinib resistance in metastatic thymomas. Cancers 2022, 14, 4762. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, X.; Zhao, K.; Shi, Y.; Deng, Z.; Yao, W.; Wang, J. Proteomic and phosphoproteomic profiling reveals the oncogenic role of protein kinase D family kinases in cholangiocarcinoma. Cells 2022, 11, 3088. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, T.; Douche, T.; Gianetto, Q.G.; Poncet, S.; El Omrani, N.; Smits, W.K.; Cuenot, E.; Matondo, M.; Martin-Verstraete, I. In-depth characterization of the Clostridioides difficile phosphoproteome to identify Ser/Thr kinase substrates. Mol. Cell. Proteom. 2022, 21, 100428. [Google Scholar] [CrossRef]
- Weng, K.; Huo, W.; Gu, T.; Bao, Q.; Cao, Z.; Zhang, Y.; Zhang, Y.; Xu, Q.; Chen, G. Quantitative phosphoproteomic analysis unveil the effect of marketable ages on meat quality in geese. Food Chem. 2021, 361, 130093. [Google Scholar] [CrossRef]
- Henry, C.; Saadaoui, B.; Bouvier, F.; Cebo, C. Phosphoproteomics of the goat milk fat globule membrane: New insights into lipid droplet secretion from the mammary epithelial cell. Proteomics 2015, 15, 2307–2317. [Google Scholar] [CrossRef]
- Zheng, X.; Su, H.; Wang, L.; Yao, R.; Ma, Y.; Bai, L.; Wang, Y.; Guo, X.; Wang, Z. Phosphoproteomics analysis reveals a pivotal mechanism related to amino acid signals in goat fetal fibroblast. Front. Vet. Sci. 2021, 8, 685548. [Google Scholar] [CrossRef]
- Rodrigues, R.T.; Chizzotti, M.L.; Vital, C.E.; Baracat-Pereira, M.C.; Barros, E.; Busato, K.C.; Gomes, R.A.; Ladeira, M.M.; Martins, T.D. Differences in beef quality between Angus (Bos taurus taurus) and Nellore (Bos taurus indicus) cattle through a proteomic and phosphoproteomic approach. PLoS ONE 2017, 12, e0170294. [Google Scholar] [CrossRef]
- Korf-Klingebiel, M.; Reboll, M.R.; Polten, F.; Weber, N.; Jackle, F.; Wu, X.; Kallikourdis, M.; Kunderfranco, P.; Condorelli, G.; Giannitsis, E.; et al. Myeloid-derived growth factor protects against pressure overload-induced heart failure by preserving sarco/endoplasmic reticulum Ca2+-ATPase expression in cardiomyocytes. Circulation 2021, 144, 1227–1240. [Google Scholar] [CrossRef]
- Kwon, H.K.; Choi, H.; Park, S.G.; Park, W.J.; Kim, D.H.; Park, Z.Y. Integrated quantitative phosphoproteomics and cell-based functional screening reveals specific pathological cardiac hypertrophy-related phosphorylation sites. Mol. Cells 2021, 44, 500–516. [Google Scholar] [CrossRef] [PubMed]
- Leo, K.T.; Chou, C.L.; Yang, C.R.; Park, E.; Raghuram, V.; Knepper, M.A. Bayesian analysis of dynamic phosphoproteomic data identifies protein kinases mediating GPCR responses. Cell Commun. Signal. 2022, 20, 80. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Cai, D.; Huang, H.; Zhang, H.; Bai, R.; Zhao, X.; Sun, H.; Qin, P. Phosphoproteomic profiling of the hippocampus of offspring rats exposed to prenatal stress. Brain Behav. 2021, 11, e2233. [Google Scholar] [CrossRef]
- Huang, S.; Zheng, F.; Liu, L.; Meng, S.; Cai, W.; Zhang, C.; Dai, W.; Liu, D.; Hong, X.; Tang, D.; et al. Integrated proteome and phosphoproteome analyses of peripheral blood mononuclear cells in primary Sjogren syndrome patients. Aging 2020, 13, 1071–1095. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, J.; Liu, F.; Hu, X.; Lu, Y.; Yan, S.; Dai, D.; Yang, X.; Zhu, Z.; Guo, Q. Proteome and phosphoproteome profiling reveals the regulation mechanism of hibernation in a freshwater leech (Whitmania pigra). J. Proteom. 2020, 229, 103866. [Google Scholar] [CrossRef]
- Huang, Z.; Liu, Y.; Qi, G.; Brand, D.; Zheng, S.G. Role of vitamin A in the immune system. J. Clin. Med. 2018, 7, 258. [Google Scholar] [CrossRef]
- Tanumihardjo, S.A.; Russell, R.M.; Stephensen, C.B.; Gannon, B.M.; Craft, N.E.; Haskell, M.J.; Lietz, G.; Schulze, K.; Raiten, D.J. Biomarkers of nutrition for development (BOND)-vitamin A review. J. Nutr. 2016, 146, 1816S–1848S. [Google Scholar] [CrossRef]
- Sun, C.; Zhu, T.; Zhu, Y.; Li, B.; Zhang, J.; Liu, Y.; Juan, C.; Yang, S.; Zhao, Z.; Wan, R.; et al. Hepatotoxic mechanism of diclofenac sodium on broiler chicken revealed by iTRAQ-based proteomics analysis. J. Vet. Sci. 2022, 23, e56. [Google Scholar] [CrossRef]
- Li, D.; Li, F.; Jiang, K.; Zhang, M.; Han, R.; Jiang, R.; Li, Z.; Tian, Y.; Yan, F.; Kang, X.; et al. Integrative analysis of long noncoding RNA and mRNA reveals candidate lncRNAs responsible for meat quality at different physiological stages in Gushi chicken. PLoS ONE 2019, 14, e0215006. [Google Scholar] [CrossRef]
- Qing, X.; Zeng, D.; Wang, H.; Ni, X.; Lai, J.; Liu, L.; Khalique, A.; Pan, K.; Jing, B. Analysis of hepatic transcriptome demonstrates altered lipid metabolism following Lactobacillus johnsonii BS15 prevention in chickens with subclinical necrotic enteritis. Lipids Health Dis. 2018, 17, 93. [Google Scholar] [CrossRef]
- Dar, M.A.; Ahmad, S.M.; Bhat, B.A.; Dar, T.A.; Haq, Z.U.; Wani, B.A.; Shabir, N.; Kashoo, Z.A.; Shah, R.A.; Ganai, N.A.; et al. Comparative RNA-Seq analysis reveals insights in Salmonella disease resistance of chicken; and database development as resource for gene expression in poultry. Genomics 2022, 114, 110475. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Fan, W.; Everaert, N.; Liu, R.; Li, Q.; Zheng, M.; Cui, H.; Zhao, G.; Wen, J. Messenger RNA sequencing and pathway analysis provide novel insights into the susceptibility to Salmonella enteritidis infection in chickens. Front. Genet. 2018, 9, 256. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Tampe, R. Structural and mechanistic principles of ABC transporters. Annu. Rev. Biochem. 2020, 89, 605–636. [Google Scholar] [CrossRef] [PubMed]
- Modi, A.; Roy, D.; Sharma, S.; Vishnoi, J.R.; Pareek, P.; Elhence, P.; Sharma, P.; Purohit, P. ABC transporters in breast cancer: Their roles in multidrug resistance and beyond. J. Drug Target. 2022, 30, 927–947. [Google Scholar] [CrossRef] [PubMed]
- Qiao, D.; He, Q.; Cheng, X.; Yao, Y.; Nair, V.; Shao, H.; Qin, A.; Qian, K. Regulation of avian leukosis virus subgroup J replication by Wnt/beta-catenin signaling pathway. Viruses 2021, 13, 1968. [Google Scholar] [CrossRef]
- Kogut, M.H.; Arsenault, R.J. A Role for the non-canonical Wnt-beta-catenin and TGF-beta signaling pathways in the induction of tolerance during the establishment of a Salmonella enterica serovar Enteritidis persistent cecal infection in chickens. Front. Vet. Sci. 2015, 2, 33. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.; Nusse, R. Wnt/beta-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Dixit, V.M. Mechanisms and functions of inflammasomes. Cell 2014, 157, 1013–1022. [Google Scholar] [CrossRef]
- Hao, E.Y.; Wang, D.H.; Chang, L.Y.; Huang, C.X.; Chen, H.; Yue, Q.X.; Zhou, R.Y.; Huang, R.L. Melatonin regulates chicken granulosa cell proliferation and apoptosis by activating the mTOR signaling pathway via its receptors. Poult. Sci. 2020, 99, 6147–6162. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, X.; Zhao, Y.; Li, H.; Ren, Y.; Hu, G.; Yang, J.; Liu, L.; Li, X. Phosphoproteomics Profile of Chicken Cecum in the Response to Salmonella enterica Serovar Enteritidis Inoculation. Animals 2023, 13, 78. https://doi.org/10.3390/ani13010078
Miao X, Zhao Y, Li H, Ren Y, Hu G, Yang J, Liu L, Li X. Phosphoproteomics Profile of Chicken Cecum in the Response to Salmonella enterica Serovar Enteritidis Inoculation. Animals. 2023; 13(1):78. https://doi.org/10.3390/ani13010078
Chicago/Turabian StyleMiao, Xiuxiu, Ya’nan Zhao, Huilong Li, Yanru Ren, Geng Hu, Jingchao Yang, Liying Liu, and Xianyao Li. 2023. "Phosphoproteomics Profile of Chicken Cecum in the Response to Salmonella enterica Serovar Enteritidis Inoculation" Animals 13, no. 1: 78. https://doi.org/10.3390/ani13010078
APA StyleMiao, X., Zhao, Y., Li, H., Ren, Y., Hu, G., Yang, J., Liu, L., & Li, X. (2023). Phosphoproteomics Profile of Chicken Cecum in the Response to Salmonella enterica Serovar Enteritidis Inoculation. Animals, 13(1), 78. https://doi.org/10.3390/ani13010078




