RNA-Seq Analysis Reveals Expression Regulatory Divergence of W-Linked Genes between Two Contrasting Chicken Breeds
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. RNA-Seq and Principal Component Analysis
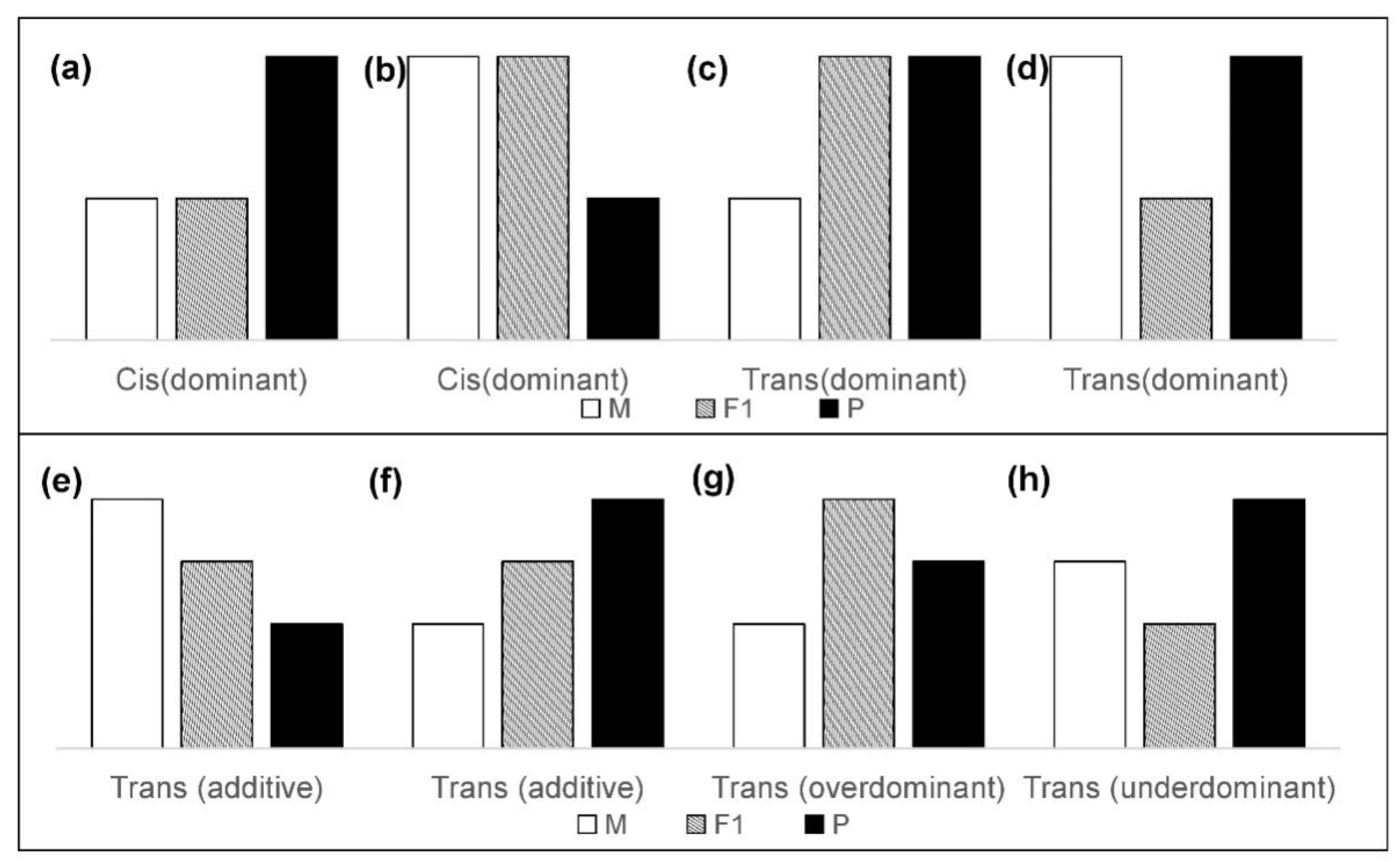
2.3. Classification of Cis- and Trans-Regulatory Categories
- (1)
- Cis: Significant difference between parents (WL and Cor), no significant difference between F1 and their maternal parents (CL and Cor; LC and WL), a significant difference between F1 and their paternal parents (CL and WL; LC and Cor). All “Cis” were considered to have “dominant” inheritance, i.e., expression of the F1 hybrid was only biased towards a single parent (Figure 2a,b).
- (2)
- Trans: Significant difference between parents (WL and Cor), a significant difference between F1 and their maternal parents (CL and Cor; LC and WL). The expression relationship between F1 and their paternal parent does not need to be considered. Considering the different inheritance patterns, “Trans” is also subdivided into “dominant” (Figure 2c,d), “additive” (Figure 2e,f), “over-dominant” (Figure 2g), and “under-dominant” (Figure 2h). Literally, “additive” indicates the genes for which expression in the hybrids was between the expression levels of the two parents. Genes for which the expression in hybrids was higher or lower than that in both parents were regarded as “over-dominant” and “under-dominant”.
- (3)
- Conserved: Significant difference between parents (WL and Cor), no significant difference between F1 and their maternal parents (CL and Cor; LC and WL), no significant difference between F1 and their paternal parents (CL and Cor; LC and WL).
3. Results
3.1. Divergence in Gene Expression among Parental Breeds
3.2. Different Expression Clustering Patterns across Tissues and Populations
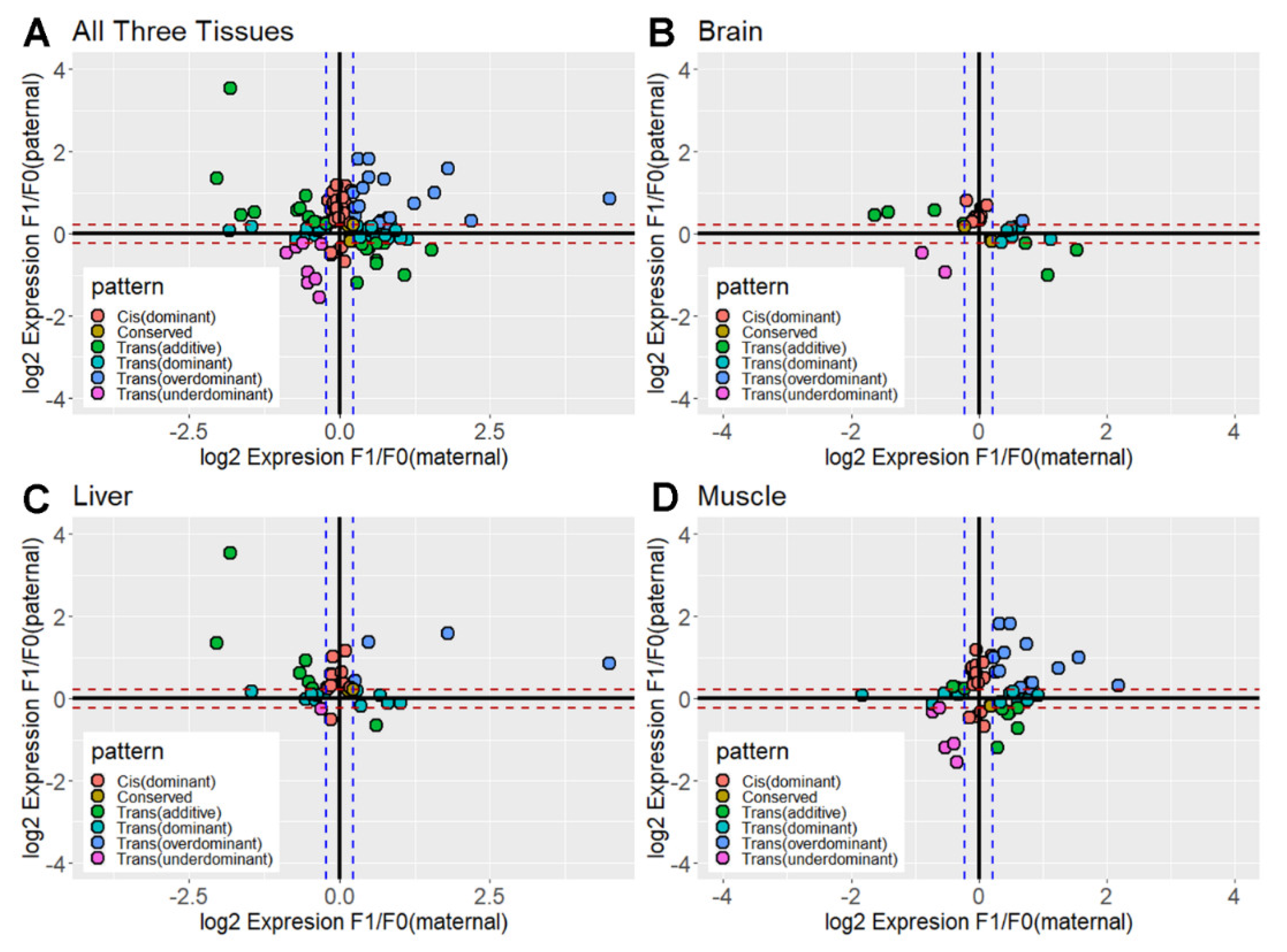
3.3. Contribution of Cis- and Trans-Acting Effects Based on the Classification of Regulatory Divergence
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carroll, S.B. Endless Forms: The Evolution of Gene Regulation and Morphological Diversity. Cell 2000, 101, 577–580. [Google Scholar] [CrossRef]
- Carroll, S.B. Evo-Devo and an Expanding Evolutionary Synthesis: A Genetic Theory of Morphological Evolution. Cell 2008, 134, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.L.; Orgogozo, V. The loci of evolution: How predictable is genetic evolution? Evolution 2008, 62, 2155–2177. [Google Scholar] [CrossRef] [PubMed]
- Cowles, C.R.; Hirschhorn, J.N.; Altshuler, D.; Lander, E.S. Detection of regulatory variation in mouse genes. Nat. Genet. 2002, 32, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Wittkopp, P.J.; Vaccaro, K.; Carroll, S.B. Evolution of yellow Gene Regulation and Pigmentation in Drosophila. Curr. Biol. 2002, 12, 1547–1556. [Google Scholar] [CrossRef]
- Landry, C.R.; Hartl, D.L.; Ranz, J.M. Genome clashes in hybrids: Insights from gene expression. Heredity 2007, 99, 483–493. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Haerum, B.K.; Clark, A. Evolutionary changes in cis and trans gene regulation. Nature 2004, 430, 85–88. [Google Scholar] [CrossRef]
- Wittkopp, P.J. Genomic sources of regulatory variation in cis and in trans. Cell. Mol. Life Sci. 2005, 62, 1779–1783. [Google Scholar] [CrossRef]
- Wray, G.A. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007, 8, 206–216. [Google Scholar] [CrossRef]
- Meiklejohn, C.D.; Coolon, J.D.; Hartl, D.L.; Wittkopp, P.J. The roles of cis- and trans-regulation in the evolution of regulatory incompatibilities and sexually dimorphic gene expression. Genome Res. 2014, 24, 84–95. [Google Scholar] [CrossRef]
- Wang, Q.; Jia, Y.X.; Wang, Y.; Jiang, Z.H.; Zhou, X.; Zhang, Z.B.; Nie, C.S.; Li, J.Y.; Yang, N.; Qu, L.J. Evolution of cis- and trans-regulatory divergence in the chicken genome between two contrasting breeds analyzed using three tissue types at one-day-old. BMC Genom. 2019, 20, 933. [Google Scholar] [CrossRef] [PubMed]
- Schaefke, B.; Emerson, J.; Wang, T.-Y.; Lu, M.-Y.J.; Hsieh, L.-C.; Li, W.-H. Inheritance of Gene Expression Level and Selective Constraints on Trans- and Cis-Regulatory Changes in Yeast. Mol. Biol. Evol. 2013, 30, 2121–2133. [Google Scholar] [CrossRef] [PubMed]
- McManus, C.J.; Coolon, J.D.; Duff, M.O.; Eipper-Mains, J.; Graveley, B.R.; Wittkopp, P.J. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 2010, 20, 816–825. [Google Scholar] [CrossRef]
- Fillon, V.; Seguela, A. Chromosomal sexing of birds. Rev. Med. Vet. 1995, 146, 53–58. [Google Scholar]
- Fridolfsson, A.-K.; Cheng, H.; Copeland, N.G.; Jenkins, N.A.; Liu, H.-C.; Raudsepp, T.; Woodage, T.; Chowdhary, B.; Halverson, J.; Ellegren, H. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc. Natl. Acad. Sci. USA 1998, 95, 8147–8152. [Google Scholar] [CrossRef]
- Pigozzi, L.I. Origin and evolution of the sex chromosomes in birds. Biocell 1999, 23, 79–95. [Google Scholar] [PubMed]
- Zhou, Q.; Zhang, J.L.; Bachtrog, D.; An, N.; Huang, Q.F.; Jarvis, E.D.; Gilbert, M.T.P.; Zhang, G.J. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 2014, 346, 1332. [Google Scholar] [CrossRef]
- Mank, J.E. Small but mighty: The evolutionary dynamics of W and Y sex chromosomes. Chromosom. Res. 2012, 20, 21–33. [Google Scholar] [CrossRef]
- Landry, C.R.; Wittkopp, P.J.; Taubes, C.H.; Ranz, J.M.; Clark, A.; Hartl, D.L. Compensatory cis-trans Evolution and the Dysregulation of Gene Expression in Interspecific Hybrids of Drosophila. Genetics 2005, 171, 1813–1822. [Google Scholar] [CrossRef]
- Sung, H.-M.; Wang, T.-Y.; Wang, D.; Huang, Y.-S.; Wu, J.-P.; Tsai, H.-K.; Tzeng, J.-N.; Huang, C.-J.; Lee, Y.-C.; Yang, P.; et al. Roles of Trans and Cis Variation in Yeast Intraspecies Evolution of Gene Expression. Mol. Biol. Evol. 2009, 26, 2533–2538. [Google Scholar] [CrossRef]
- Zhang, X.; O Borevitz, J. Global Analysis of Allele-Specific Expression in Arabidopsis thaliana. Genetics 2009, 182, 943–954. [Google Scholar] [CrossRef]
- He, F.; Zhang, X.; Hu, J.Y.; Turck, F.; Dong, X.; Goebel, U.; Borevitz, J.; De Meaux, J. Genome-wide Analysis of Cis-regulatory Divergence between Species in the Arabidopsis Genus. Mol. Biol. Evol. 2012, 29, 3385–3395. [Google Scholar] [CrossRef] [PubMed]
- Bell, G.D.M.; Kane, N.C.; Rieseberg, L.H.; Adams, K.L. RNA-Seq Analysis of Allele-Specific Expression, Hybrid Effects, and Regulatory Divergence in Hybrids Compared with Their Parents from Natural Populations. Genome Biol. Evol. 2013, 5, 1309–1323. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.K.S.; Liu, B.; Wang, J.; Zhang, Y.; Yang, X.; Zhang, Z.J.; Meng, Q.S.; Zhou, J.; Li, D.W.; Zhang, J.J.; et al. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature 2004, 432, 717–722. [Google Scholar] [CrossRef]
- Rubin, C.-J.; Zody, M.C.; Eriksson, J.; Meadows, J.R.S.; Sherwood, E.; Webster, M.T.; Jiang, L.; Ingman, M.; Sharpe, T.; Ka, S.; et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 2010, 464, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Y.; Li, Y.; Li, M.; Che, T.D.; Tian, S.L.; Chen, B.L.; Zhou, X.M.; Zhang, G.L.; Gaur, U.; Luo, M.J.; et al. Population genomics identifies patterns of genetic diversity and selection in chicken. BMC Genom. 2019, 20, 263. [Google Scholar] [CrossRef] [PubMed]
- Moghadam, H.K.; Pointer, M.A.; Wright, A.E.; Berlin, S.; Mank, J.E. W chromosome expression responds to female-specific selection. Proc. Natl. Acad. Sci. USA 2012, 109, 8207–8211. [Google Scholar] [CrossRef]
- Peichel, C.L. Convergence and divergence in sex-chromosome evolution. Nat. Genet. 2017, 49, 321–322. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: AnRPackage for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Gibson, G.; Riley-Berger, R.; Harshman, L.; Kopp, A.; Vacha, S.; Nuzhdin, S.; Wayne, M. Extensive Sex-Specific Nonadditivity of Gene Expression in Drosophila melanogaster. Genetics 2004, 167, 1791–1799. [Google Scholar] [CrossRef] [PubMed]
- Lippman, Z.; Zamir, D. Heterosis: Revisiting the magic. Trends Genet. 2007, 23, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013, 14, 471–482. [Google Scholar] [CrossRef]
- Yang, X.; Schadt, E.E.; Wang, S.; Wang, H.; Arnold, A.P.; Ingram-Drake, L.; Drake, T.A.; Lusis, A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006, 16, 995–1004. [Google Scholar] [CrossRef]
- Brawand, D.; Soumillon, M.; Necsulea, A.; Julien, P.; Csardi, G.; Harrigan, P.; Weier, M.; Liechti, A.; Aximu-Petri, A.; Kircher, M.; et al. The evolution of gene expression levels in mammalian organs. Nature 2011, 478, 343–348. [Google Scholar] [CrossRef]
- Catalán, A.; Hutter, S.; Parsch, J. Population and sex differences in Drosophila melanogaster brain gene expression. BMC Genom. 2012, 13, 654. [Google Scholar] [CrossRef]
- Huylmans, A.K.; Parsch, J. Population- and Sex-Biased Gene Expression in the Excretion Organs of Drosophila melanogaster. G3 Genes Genomes Genet. 2014, 4, 2307–2315. [Google Scholar] [CrossRef] [PubMed]
- Mank, J.; Hultin-Rosenberg, L.; Webster, M.T.; Ellegren, H. The unique genomic properties of sex-biased genes: Insights from avian microarray data. BMC Genom. 2008, 9, 148. [Google Scholar] [CrossRef] [PubMed]
- Pointer, M.A.; Harrison, P.W.; Wright, A.E.; Mank, J.E. Masculinization of Gene Expression Is Associated with Exaggeration of Male Sexual Dimorphism. PLoS Genet. 2013, 9, e1003697. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Wright, A.E.; Zimmer, F.; Dean, R.; Montgomery, S.H.; Pointer, M.A.; Mank, J.E. Sexual selection drives evolution and rapid turnover of male gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, 4393–4398. [Google Scholar] [CrossRef]
- Mank, J.E.; Nam, K.; Brunström, B.; Ellegren, H. Ontogenetic Complexity of Sexual Dimorphism and Sex-Specific Selection. Mol. Biol. Evol. 2010, 27, 1570–1578. [Google Scholar] [CrossRef]
- Perry, J.C.; Harrison, P.W.; Mank, J.E. The Ontogeny and Evolution of Sex-Biased Gene Expression in Drosophila melanogaster. Mol. Biol. Evol. 2014, 31, 1206–1219. [Google Scholar] [CrossRef]
- Ayers, K.L.; Davidson, N.M.; Demiyah, D.; Roeszler, K.N.; Grützner, F.; Sinclair, A.H.; Oshlack, A.; Smith, C.A. RNA sequencing reveals sexually dimorphic gene expression before gonadal differentiation in chicken and allows comprehensive annotation of the W-chromosome. Genome Biol. 2013, 14, R26. [Google Scholar] [CrossRef]
- Jackson, S.; Diamond, J. Metabolic and Digestive Responses to Artificial Selection in Chickens. Evolution 1996, 50, 1638–1650. [Google Scholar] [CrossRef]
- Xue, Q.; Zhang, G.X.; Li, T.T.; Ling, J.J.; Zhang, X.Q.; Wang, J.Y. Transcriptomic profile of leg muscle during early growth in chicken. PLoS ONE 2017, 12, e0173824. [Google Scholar] [CrossRef]
- Metzger, B.P.H.; Wittkopp, P.J.; Coolon, J.D. Evolutionary Dynamics of Regulatory Changes Underlying Gene Expression Divergence among Saccharomyces Species. Genome Biol. Evol. 2017, 9, 843–854. [Google Scholar] [CrossRef]
- Shi, X.L.; Ng, D.W.-K.; Zhang, C.Q.; Comai, L.; Ye, W.X.; Chen, Z.J. Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat. Commun. 2012, 3, 950. [Google Scholar] [CrossRef] [PubMed]
- Combes, M.-C.; Hueber, Y.; Dereeper, A.; Rialle, S.; Herrera, J.-C.; Lashermes, P. Regulatory divergence between parental alleles determines gene expression patterns in hybrids. Genome Biol. Evol. 2015, 7, 1110–1121. [Google Scholar] [CrossRef] [PubMed]
- Crow, J.F. Dominance and overdominance. In Proceedings of the International Symposium on the Genetics and Exploitation of Heterosis in Crops, Mexico City, Mexico, 17–22 August 1999; pp. 49–58. [Google Scholar]
- Stupar, R.M.; Hermanson, P.J.; Springer, N.M. Nonadditive Expression and Parent-of-Origin Effects Identified by Microarray and Allele-Specific Expression Profiling of Maize Endosperm. Plant. Physiol. 2007, 145, 411–425. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant. Sci. 2010, 15, 57–71. [Google Scholar] [CrossRef]
- Gu, H.C.; Qi, X.; Jia, Y.; Zhang, Z.; Nie, C.; Li, X.; Li, J.; Jiang, Z.; Wang, Q.; Qu, L. Inheritance patterns of the transcriptome in hybrid chickens and their parents revealed by expression analysis. Sci. Rep. 2019, 9, 10. [Google Scholar] [CrossRef]
- Mai, C.; Wen, C.; Xu, Z.; Xu, G.; Chen, S.; Zheng, J.; Sun, C.; Yang, N. Genetic basis of negative heterosis for growth traits in chickens revealed by genome-wide gene expression pattern analysis. J. Anim. Sci. Biotechnol. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Denver, D.R.; Morris, K.; Streelman, J.T.; Kim, S.K.; Lynch, M.; Thomas, W.K. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat. Genet. 2005, 37, 544–548. [Google Scholar] [CrossRef]



| Tissues | Groups | Number of Genes | |||||
|---|---|---|---|---|---|---|---|
| Cis | Trans | Conserved | |||||
| Cis (Dominant) | Trans (Dominant) | Trans (Additive) | Trans (Overdominant) | Trans (Underdominant) | |||
| Brain | 1 | 11 | 0 | 4 | 0 | 1 | 1 |
| 2 | 0 | 10 | 3 | 1 | 1 | 2 | |
| Liver | 1 | 8 | 6 | 6 | 1 | 0 | 0 |
| 2 | 7 | 6 | 1 | 3 | 1 | 3 | |
| Muscle | 1 | 11 | 6 | 2 | 5 | 8 | 0 |
| 2 | 7 | 10 | 7 | 4 | 2 | 2 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, H.; Wang, L.; Lv, X.; Yang, W.; Chen, Y.; Li, K.; Zhang, J.; Jia, Y.; Ning, Z.; Qu, L. RNA-Seq Analysis Reveals Expression Regulatory Divergence of W-Linked Genes between Two Contrasting Chicken Breeds. Animals 2022, 12, 1218. https://doi.org/10.3390/ani12091218
Gu H, Wang L, Lv X, Yang W, Chen Y, Li K, Zhang J, Jia Y, Ning Z, Qu L. RNA-Seq Analysis Reveals Expression Regulatory Divergence of W-Linked Genes between Two Contrasting Chicken Breeds. Animals. 2022; 12(9):1218. https://doi.org/10.3390/ani12091218
Chicago/Turabian StyleGu, Hongchang, Liang Wang, Xueze Lv, Weifang Yang, Yu Chen, Kaiyang Li, Jianwei Zhang, Yaxiong Jia, Zhonghua Ning, and Lujiang Qu. 2022. "RNA-Seq Analysis Reveals Expression Regulatory Divergence of W-Linked Genes between Two Contrasting Chicken Breeds" Animals 12, no. 9: 1218. https://doi.org/10.3390/ani12091218
APA StyleGu, H., Wang, L., Lv, X., Yang, W., Chen, Y., Li, K., Zhang, J., Jia, Y., Ning, Z., & Qu, L. (2022). RNA-Seq Analysis Reveals Expression Regulatory Divergence of W-Linked Genes between Two Contrasting Chicken Breeds. Animals, 12(9), 1218. https://doi.org/10.3390/ani12091218






