Transport of Acrosomal Enzymes by KIFC1 via the Acroframosomal Cytoskeleton during Spermatogenesis in Macrobrachium rosenbergii (Crustacea, Decapoda, Malacostracea)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Animals and Samples
2.2. Full-Length cDNA Cloning of Mr-kifc1 and Mr-Acrosin
2.3. Multiple Sequence Alignment, Phylogenetic Evolutionary Tree Analysis and Protein Structure Prediction
2.4. Semiquantitative PCR Analysis of Mr-kifc1 and Mr-Acrosin mRNA Expression
2.5. In Situ Hybridization (ISH)
2.5.1. Riboprobe Synthesis
2.5.2. Prehybridization and Hybridization
2.5.3. Detection of the Product Signal
2.6. Antibodies
2.6.1. Prokaryotic Expression
2.6.2. Western Blot Analysis
2.7. Immunofluorescence (IF)
3. Results
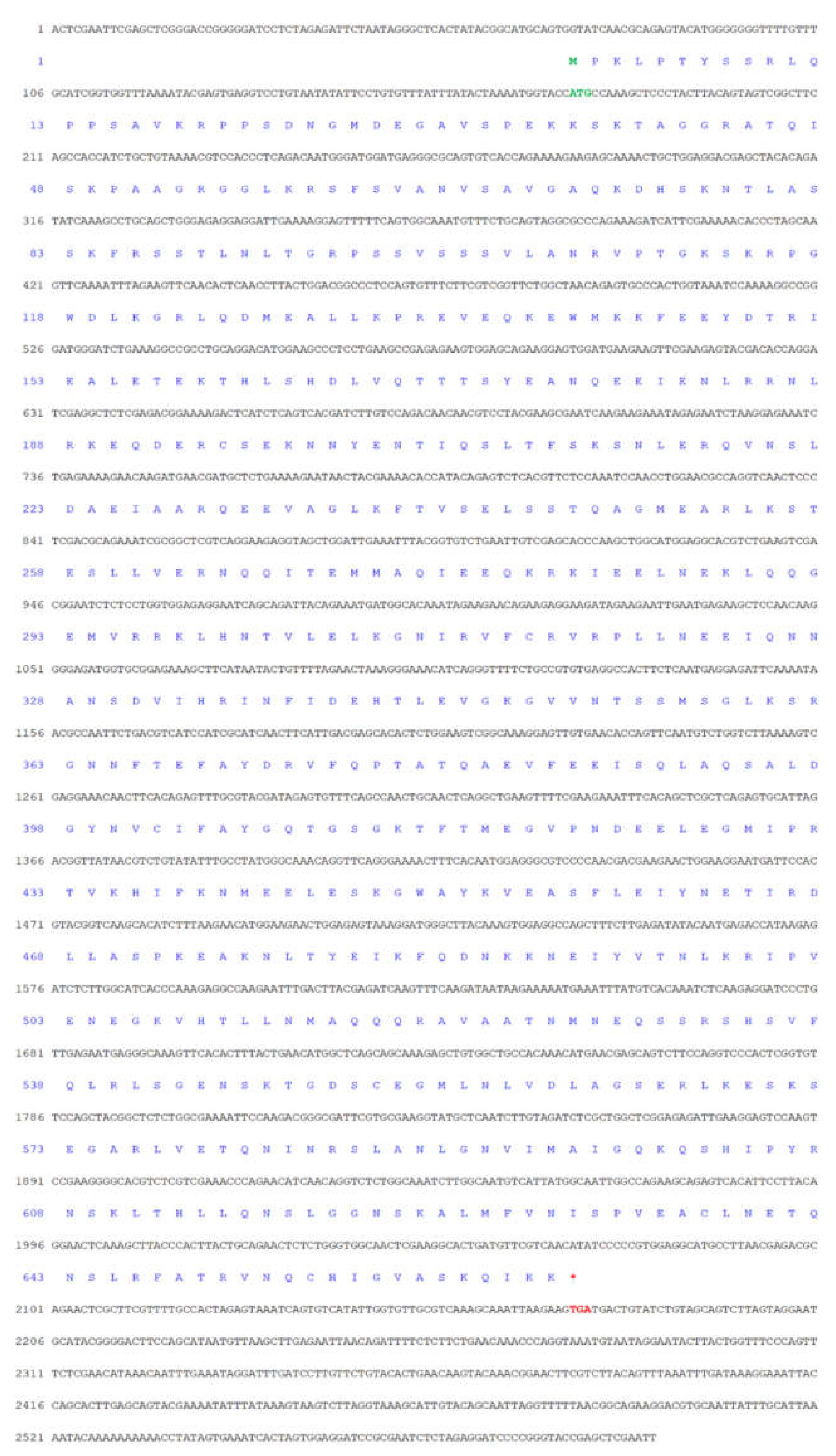
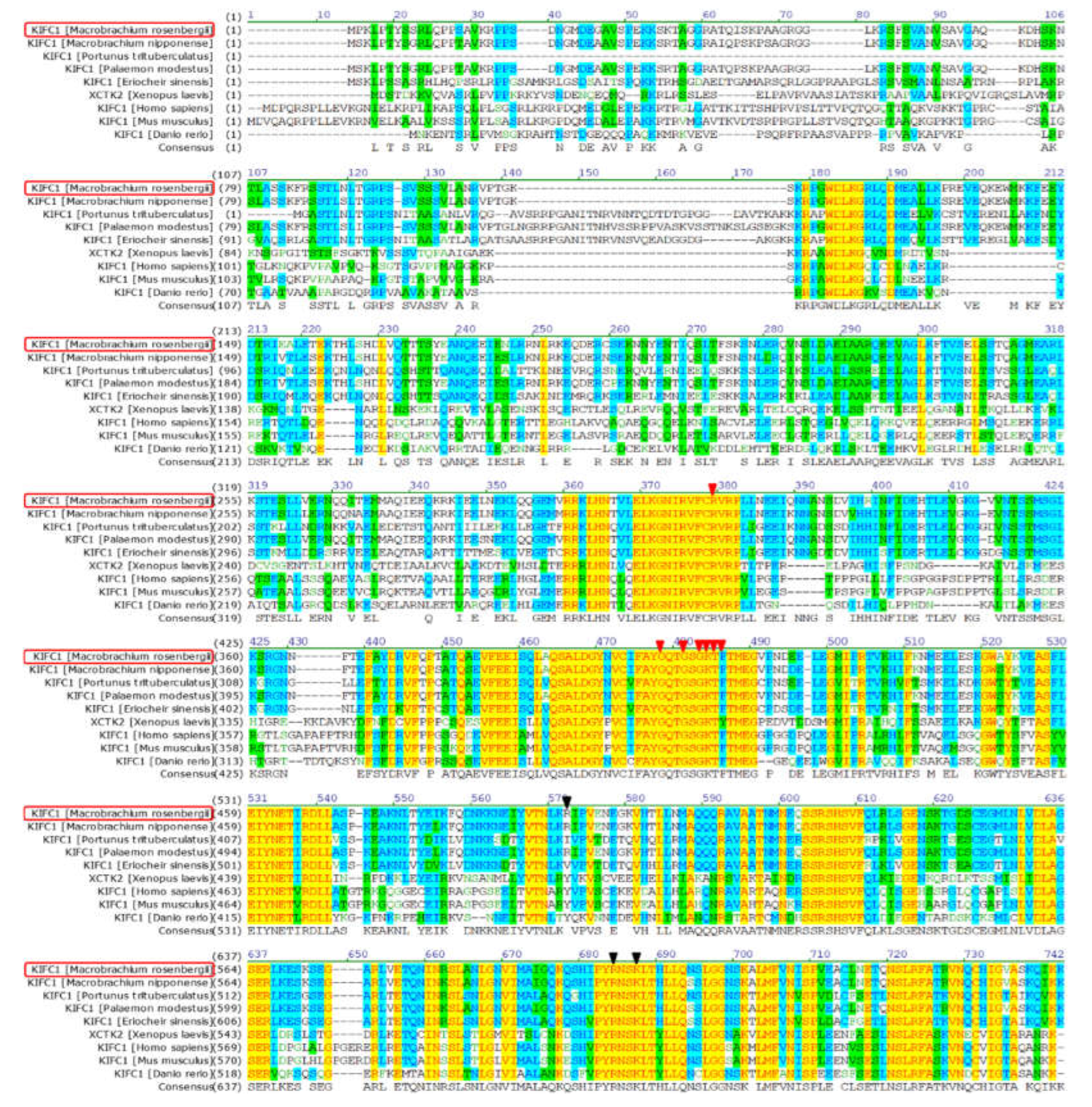
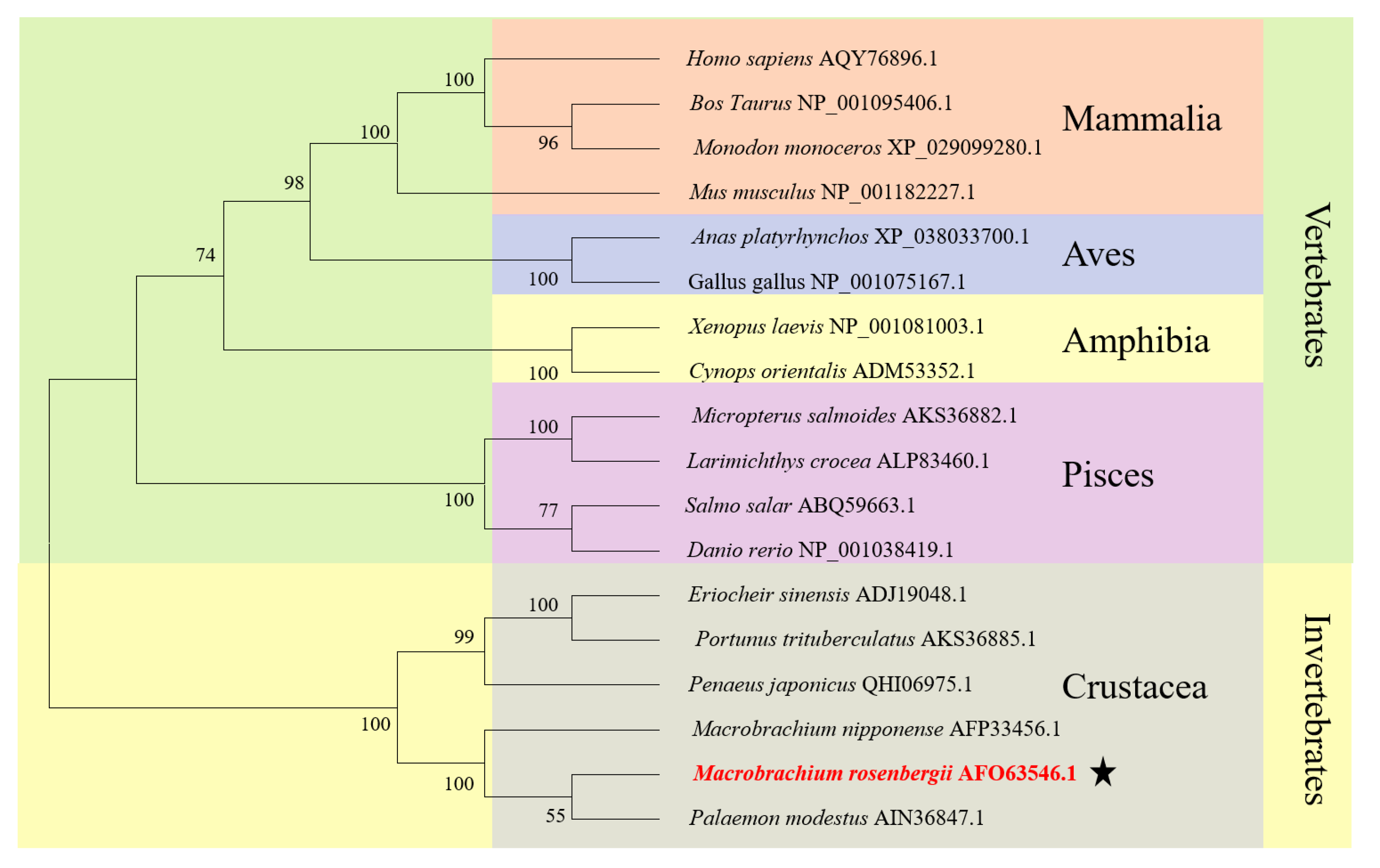
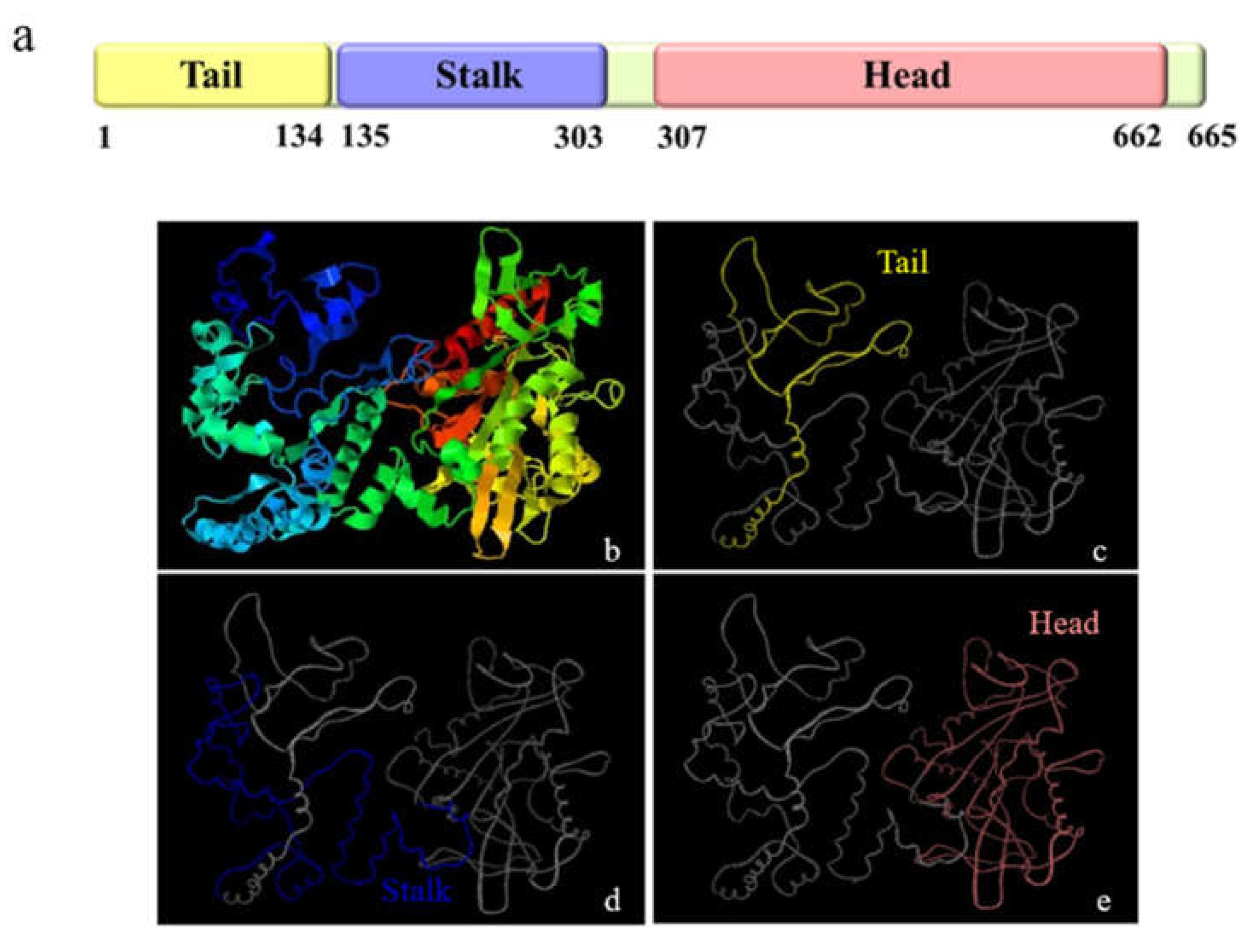
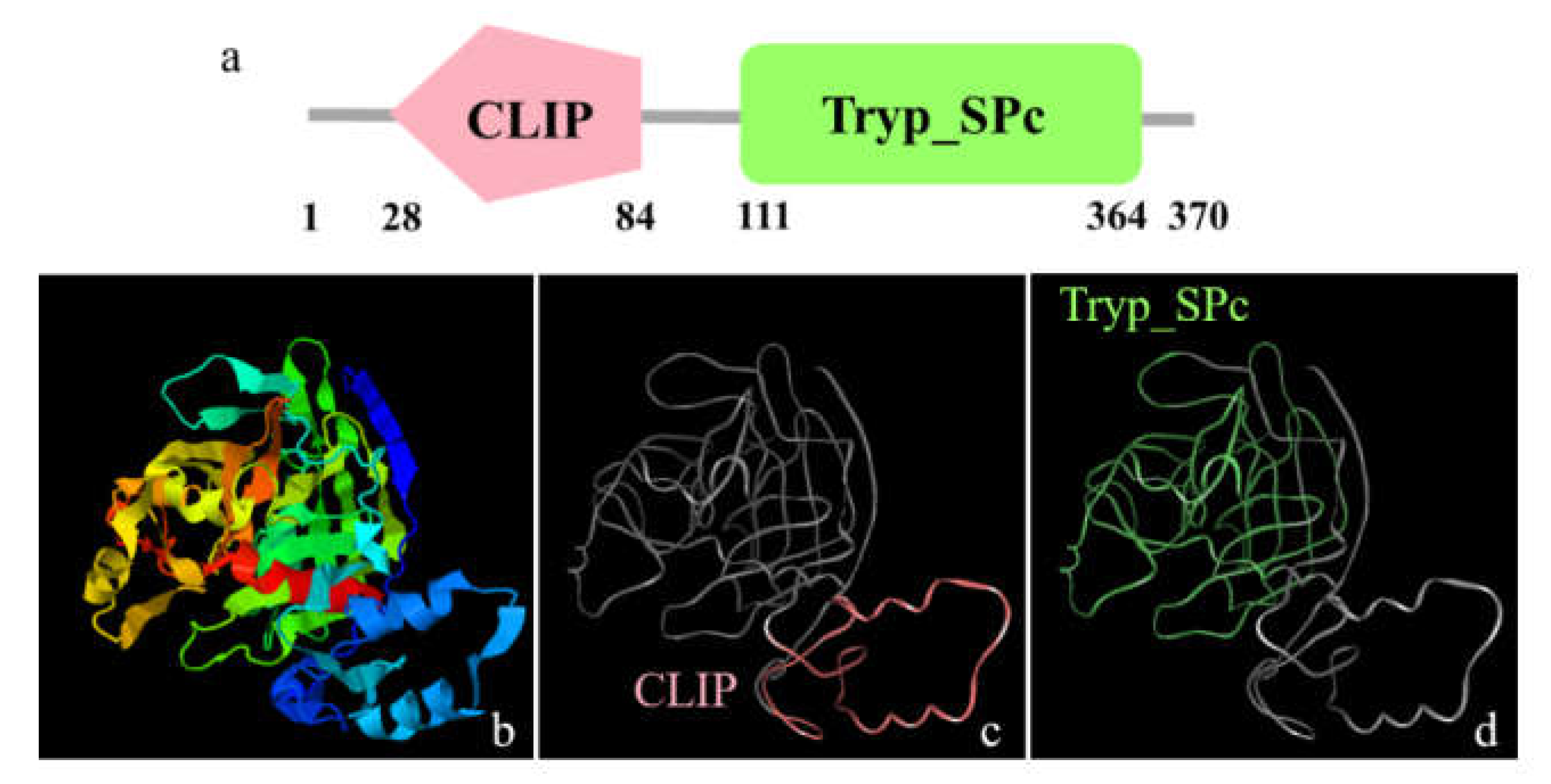
3.1. The Major Features of Mr-kifc1 and Mr-Acrosin
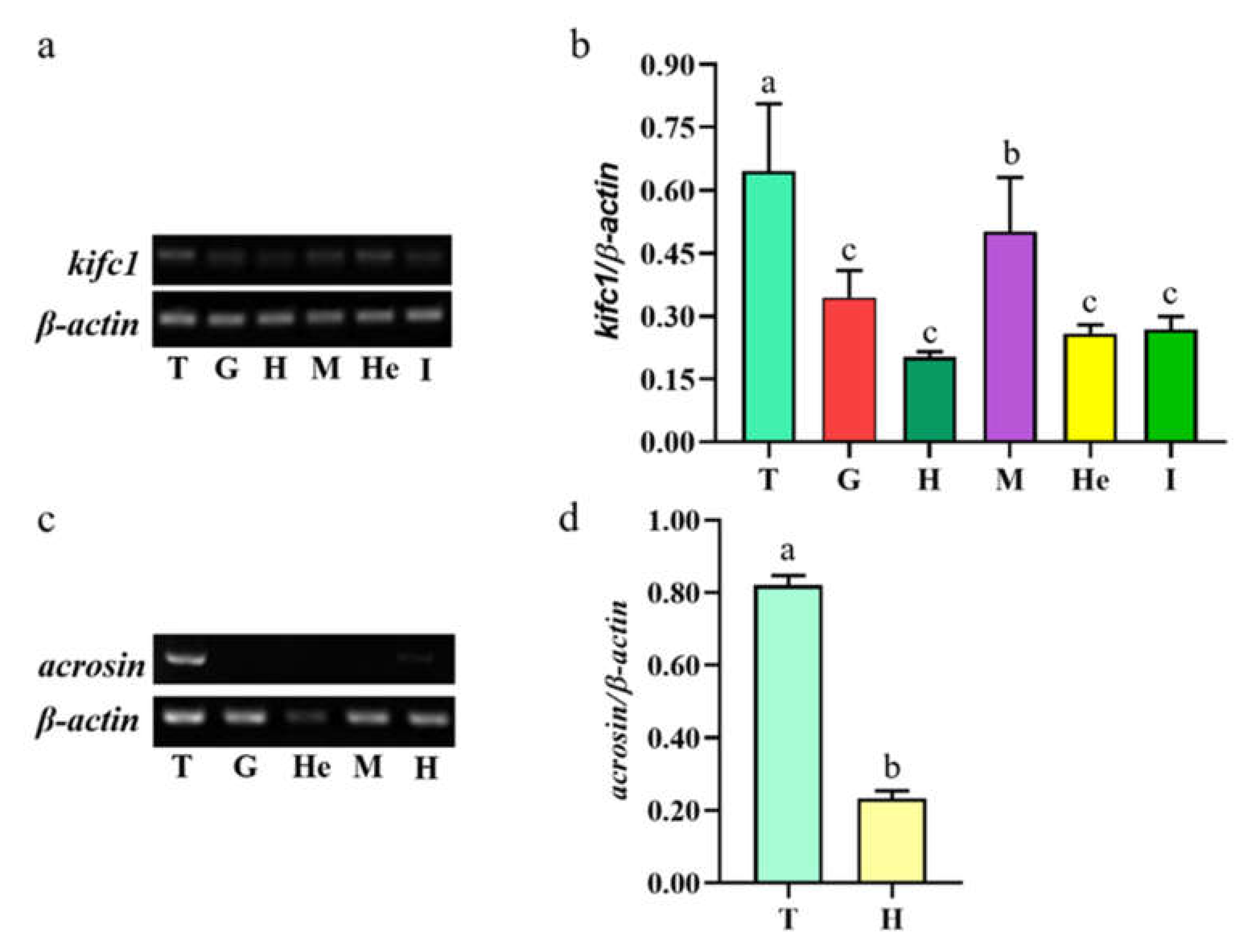
3.2. Mr-kifc1 and Mr-Acrosin mRNA Expression in Different Tissues of M. rosenbergii
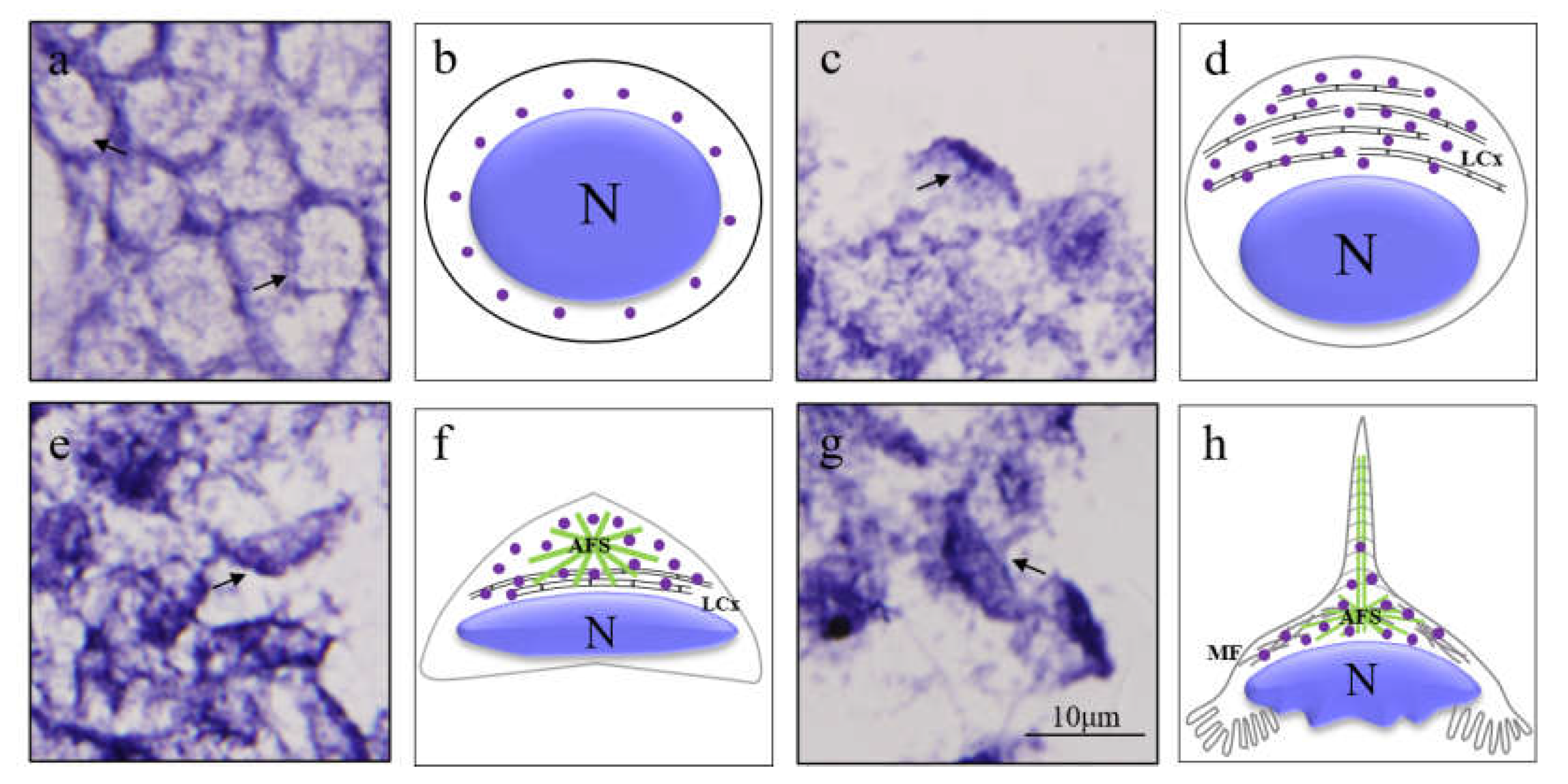
3.3. The Spatial and Temporal Expression Pattern of Mr-kifc1 during Spermatogenesis of M. rosenbergii
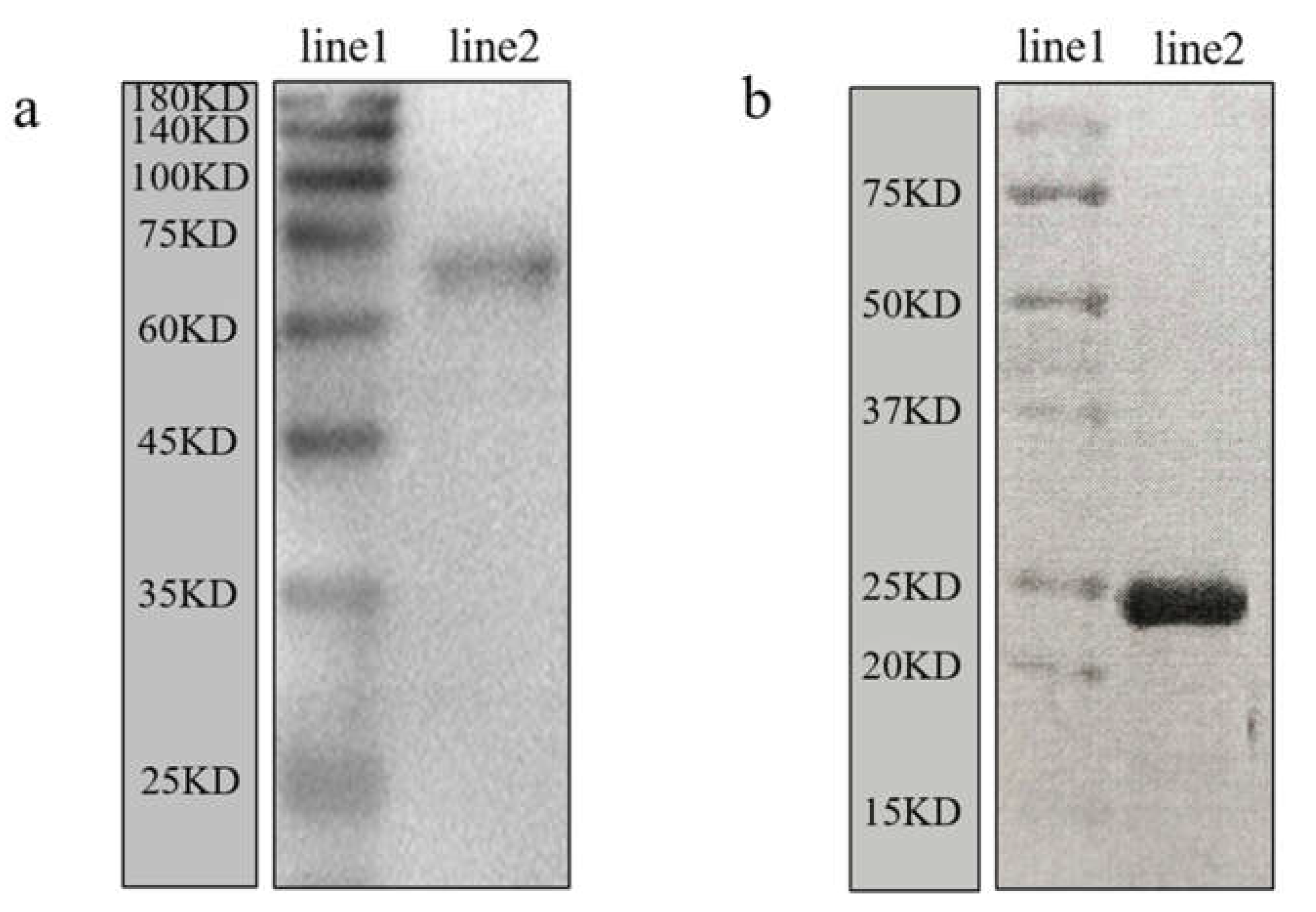
3.4. Validation of the Anti-Mr-KIFC1 Antibody and Anti-Mr-Acrosin Antibody
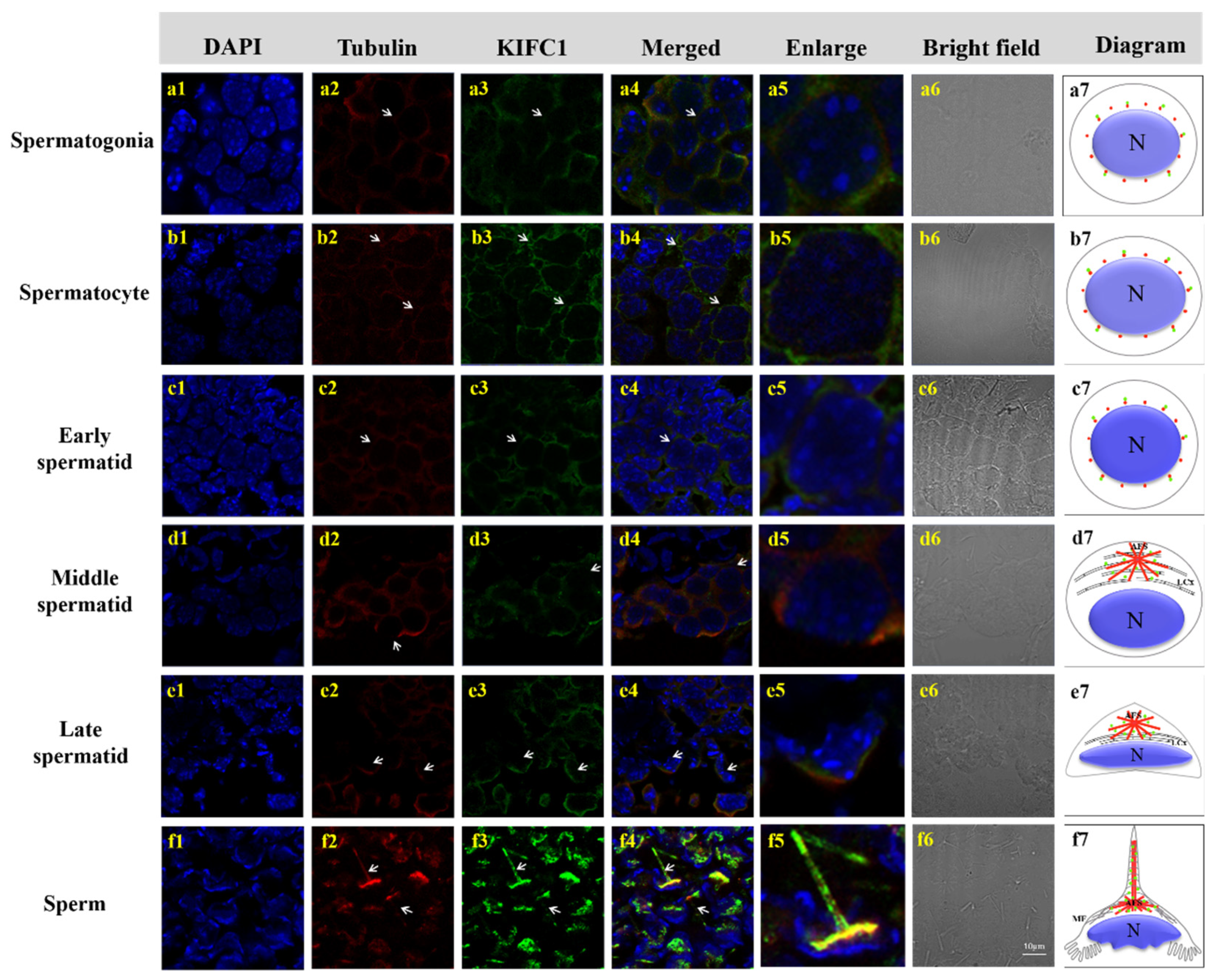
3.5. Colocalization of Mr-KIFC1 and Microtubules during Spermatogenesis of M. rosenbergii
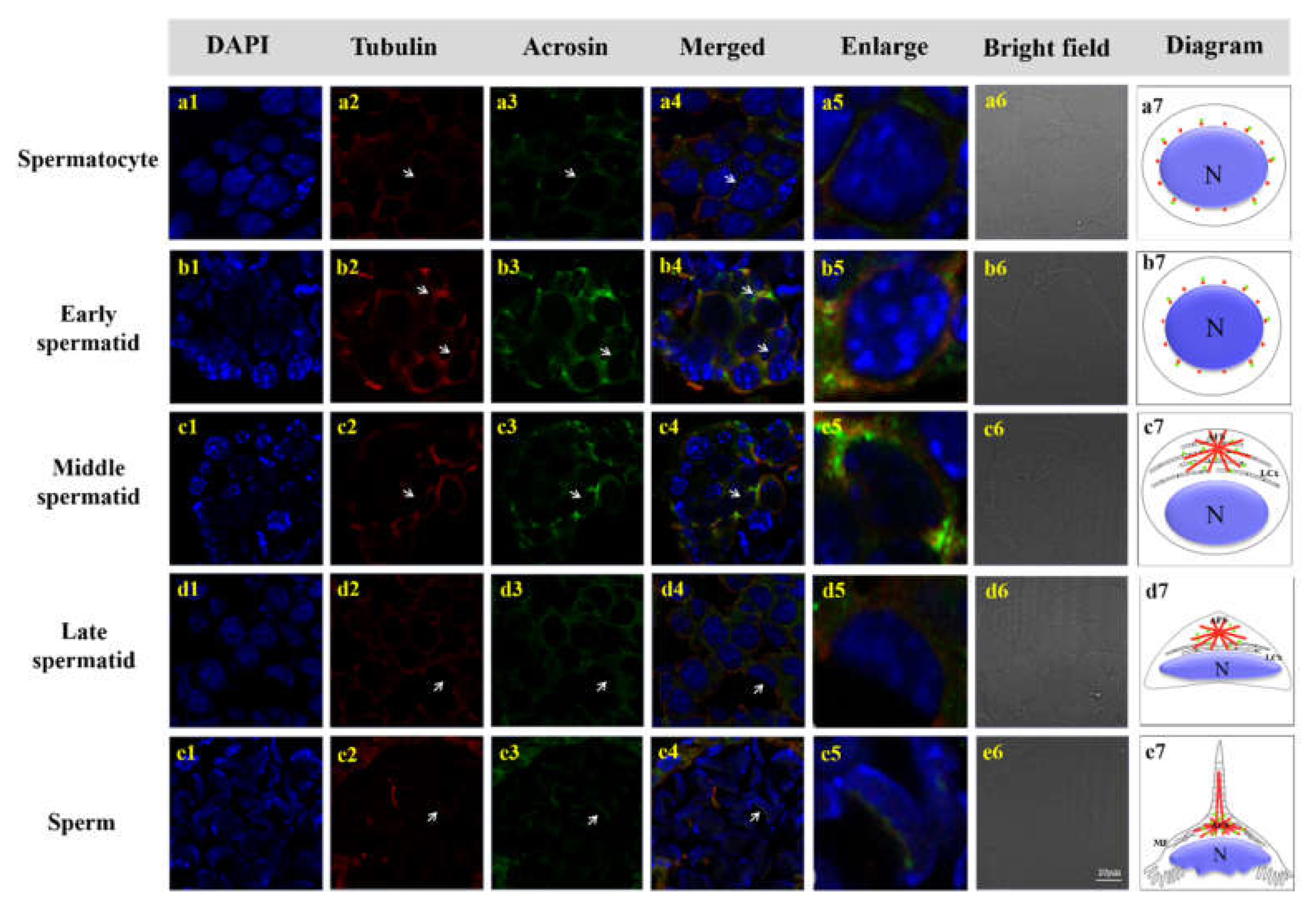
3.6. Colocalization of Mr-Acrosin and Microtubules during Spermatogenesis of M. rosenbergii
4. Discussion
4.1. Structural Features and mRNA Expression Characteristics of Mr-KIFC1 and Mr-Acrosin
4.2. Mr-KIFC1 Participates in Spermiogenesis and Is Closely Related to Nuclear Reshaping and Acrosome Formation
4.3. Mr-KIFC1 Is Involved in Mr-Acrosin Transport in Spermatogenesis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kubota, H.; Avarbock, M.R.; Brinster, R.L. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA 2004, 101, 16489–16494. [Google Scholar] [CrossRef] [PubMed]
- Hess, R.A.; de Franca, L.R. Spermatogenesis and cycle of the seminiferous epithelium. Adv. Exp. Med. Biol. 2008, 636, 1–15. [Google Scholar] [PubMed]
- Hermo, L.; Pelletier, R.M.; Cyr, D.G.; Smith, C.E. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: Background to spermatogenesis, spermatogonia, and spermatocytes. Microsc. Res. Tech. 2010, 73, 241–278. [Google Scholar] [CrossRef]
- Sprando, R.L.; Russell, L.D. Comparative study of cytoplasmic elimination in spermatids of selected mammalian species. Am. J. Anat. 1987, 178, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Sun, W.J.; He, L.; Li, Q.; Wang, Q. Morphological alterations of all stages of spermatogenesis and acrosome reaction in Chinese mitten crab Eriocheir sinensis. Cell Tissue Res. 2015, 360, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Dunleavy, J.E.M.; O’Bryan, M.K.; Stanton, P.G.; O’Donnell, L. The cytoskeleton in spermatogenesis. Reproduction 2019, 157, R53–R72. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.F.; Zhu, J.Q.; Hou, C.C.; Yang, W.X. Identification and expression pattern analysis of Piwi genes during the spermiogenesis of Portunus trituberculatus. Gene 2014, 534, 240–248. [Google Scholar] [CrossRef]
- Wang, Y.T.; Mao, H.; Hou, C.C.; Sun, X.; Wang, D.H.; Zhou, H.; Yang, W.X. Characterization and expression pattern of KIFC1-like kinesin gene in the testis of the Macrobrachium nipponense with discussion of its relationship with structure lamellar complex (LCx) and acroframosome (AFS). Mol. Biol. Rep. 2012, 39, 7591–7598. [Google Scholar] [CrossRef]
- Feng, T.; Paterson, B.; Johnston, S.D. New insights into the spermatogenesis of the black tiger prawn, Penaeus monodon. J. Morphol. 2017, 278, 689–703. [Google Scholar] [CrossRef]
- Ichikawa, Y.; Matsuzaki, M.; Hiyama, G.; Mizushima, S.; Sasanami, T. Sperm-Egg Interaction during Fertilization in Birds. J. Poult. Sci. 2016, 53, 173–180. [Google Scholar] [CrossRef]
- Koch, R.A.; Lambert, C.C. Ultrastructure of sperm, spermiogenesis, and sperm-egg interactions in selected invertebrates and lower vertebrates which use external fertilization. J. Electron. Microsc. Tech. 1990, 16, 115–154. [Google Scholar] [CrossRef] [PubMed]
- Vogt, G. Structural specialties, curiosities, and record-breaking features of crustacean reproduction. J. Morphol. 2016, 277, 1399–1422. [Google Scholar] [CrossRef] [PubMed]
- Lynn, J.W. The Reproductive Biology and Gamete Interaction in the Freshwater Prawn Macrobrachium rosenbergii. Ph.D. Thesis, University of California, Los Angeles, CA, USA, 1981. [Google Scholar]
- Hou, C.C.; Yang, W.X. Acroframosome-dependent KIFC1 facilitates acrosome formation during spermatogenesis in the caridean shrimp Exopalaemon modestus. PLoS ONE 2013, 8, e76065. [Google Scholar] [CrossRef] [PubMed]
- Zhe, L.I.; Yang, W.X. Immunocytochemical studies on the acroframosome during spermiogenesis of the caridean shrimp Macrobrachium nipponense (Crustacea, Natantia). Invertebr. Reprod. Dev. 2010, 54, 121–131. [Google Scholar]
- Poljaroen, J.; Vanichviriyakit, R.; Tinikul, Y.; Phoungpetchara, I.; Linthong, V.; Weerachatyanukul, W.; Sobhon, P. Spermatogenesis and distinctive mature sperm in the giant freshwater prawn, Macrobrachium rosenbergii (De Man, 1879). Zool. Anz. 2010, 249, 81–94. [Google Scholar] [CrossRef]
- Lynn, J.W.; Clark, W.H. The Fine Structure of the Mature Sperm of the Freshwater Prawn, Macrobrachium rosenbergii. Biol. Bull. 1983, 164, 459–470. [Google Scholar] [CrossRef]
- Baba, T.; Kashiwabara, S.; Watanabe, K.; Itoh, H.; Michikawa, Y.; Kimura, K.; Takada, M.; Fukamizu, A.; Arai, Y. Activation and maturation mechanisms of boar acrosin zymogen based on the deduced primary structure. J. Biol. Chem. 1989, 264, 11920–11927. [Google Scholar] [CrossRef]
- Mao, H.T.; Yang, W.X. Modes of acrosin functioning during fertilization. Gene 2013, 526, 75–79. [Google Scholar] [CrossRef]
- Adham, I.M.; Nayernia, K.; Engel, W. Spermatozoa lacking acrosin protein show delayed fertilization. Mol. Reprod. Dev. 1997, 46, 370–376. [Google Scholar] [CrossRef]
- Langlois, M.R.; Oorlynck, L.; Vandekerckhove, F.; Criel, A.; Bernard, D.; Blaton, V. Discrepancy between sperm acrosin activity and sperm morphology: Significance for fertilization in vitro. Clin. Chim. Acta 2005, 351, 121–129. [Google Scholar] [CrossRef]
- Yamagata, K.; Murayama, K.; Okabe, M.; Toshimori, K.; Nakanishi, T.; Kashiwabara, S.; Baba, T. Acrosin accelerates the dispersal of sperm acrosomal proteins during acrosome reaction. J. Biol. Chem. 1998, 273, 10470–10474. [Google Scholar] [CrossRef] [PubMed]
- Howes, L.; Jones, R. Interactions between zona pellucida glycoproteins and sperm proacrosin/acrosin during fertilization. J. Reprod. Immunol. 2002, 53, 181–192. [Google Scholar] [CrossRef]
- Chaudhury, K.; Das, T.; Chakravarty, B.; Bhattacharyya, A.K. Acrosin activity as a potential marker for sperm membrane characteristics in unexplained male infertility. Fertil. Steril. 2005, 83, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Hirokawa, N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 1998, 279, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Hogarth, C.; Itman, C.; Jans, D.A.; Loveland, K.L. Regulated nucleocytoplasmic transport in spermatogenesis: A driver of cellular differentiation? Bioessays 2005, 27, 1011–1025. [Google Scholar] [CrossRef]
- Hirokawa, N.; Noda, Y.; Tanaka, Y.; Niwa, S. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 2009, 10, 682–696. [Google Scholar] [CrossRef]
- Vale, R.D.; Milligan, R.A. The way things move: Looking under the hood of molecular motor proteins. Science 2000, 288, 88–95. [Google Scholar] [CrossRef]
- Sharp, D.J.; Rogers, G.C.; Scholey, J.M. Microtubule motors in mitosis. Nature 2000, 407, 41–47. [Google Scholar] [CrossRef]
- Schliwa, M.; Woehlke, G. Molecular motors. Nature 2003, 422, 759–765. [Google Scholar] [CrossRef]
- Liang, Y.J.; Yang, W.X. Kinesins in MAPK cascade: How kinesin motors are involved in the MAPK pathway? Gene 2019, 684, 1–9. [Google Scholar] [CrossRef]
- Yang, W.X.; Sperry, A.O. C-terminal kinesin motor KIFC1 participates in acrosome biogenesis and vesicle transport. Biol. Reprod. 2003, 69, 1719–1729. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.R.; Liu, M.; Wang, D.H.; Hu, Y.J.; Tan, F.Q.; Yang, W.X. Molecular characterization and expression analysis of a KIFC1-like kinesin gene in the testis of Eumeces chinensis. Mol. Biol. Rep. 2013, 40, 6645–6655. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhu, J.Q.; Yu, H.M.; Tan, F.Q.; Yang, W.X. KIFC1-like motor protein associates with the cephalopod manchette and participates in sperm nuclear morphogenesis in Octopus tankahkeei. PLoS ONE 2010, 5, e15616. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, D.D.; Gao, X.M.; Zhao, Y.Q.; Hou, C.C.; Zhu, J.Q. The C-terminal kinesin motor KIFC1 may participate in nuclear reshaping and flagellum formation during spermiogenesis of Larimichthys crocea. Fish Physiol. Biochem. 2017, 43, 1351–1371. [Google Scholar] [CrossRef]
- Ma, D.D.; Pan, M.Y.; Hou, C.C.; Tan, F.Q.; Yang, W.X. KIFC1 and myosin Va: Two motors for acrosomal biogenesis and nuclear shaping during spermiogenesis of Portunus trituberculatus. Cell Tissue Res. 2017, 369, 625–640. [Google Scholar] [CrossRef]
- Hao, S.L.; Yang, W.X. KIFC1 is essential for normal spermatogenesis and its depletion results in early germ cell apoptosis in the Kuruma shrimp, Penaeus (Marsupenaeus) japonicus. Aging 2019, 11, 12773–12792. [Google Scholar] [CrossRef]
- Gao, X.M.; Mu, D.L.; Hou, C.C.; Zhu, J.Q.; Jin, S.; Wang, C.L. Expression and putative functions of KIFC1 for nuclear reshaping and midpiece formation during spermiogenesis of Phascolosoma esculenta. Gene 2019, 683, 169–183. [Google Scholar] [CrossRef]
- Jiang, H.; Kanost, M.R. The clip-domain family of serine proteinases in arthropods. Insect Biochem. Mol. Biol. 2000, 30, 95–105. [Google Scholar] [CrossRef]
- Verhey, K.J.; Hammond, J.W. Traffic control: Regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 2009, 10, 765–777. [Google Scholar] [CrossRef]
- Zhang, Y.; Sperry, A.O. Comparative analysis of two C-terminal kinesin motor proteins: KIFC1 and KIFC5A. Cell Motil. Cytoskelet. 2004, 58, 213–230. [Google Scholar] [CrossRef]
- Hou, C.C.; Gao, X.M.; Ni, J.; Mu, D.L.; Yang, H.Y.; Liu, C.; Zhu, J.Q. The expression pattern and potential functions of PHB in the spermiogenesis of Phascolosoma esculenta. Gene 2018, 652, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Flörke-Gerloff, S.; Töpfer-Petersen, E.; Müller-Esterl, W.; Schill, W.B.; Engel, W. Acrosin and the acrosome in human spermatogenesis. Hum. Genet. 1983, 65, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Zahn, A.; Furlong, L.I.; Biancotti, J.C.; Ghiringhelli, P.D.; Marijn-Briggiler, C.I.; Vazquez-Levin, M.H. Evaluation of the proacrosin/acrosin system and its mechanism of activation in human sperm extracts. J. Reprod. Immunol. 2002, 54, 43–63. [Google Scholar] [CrossRef]
- Yang, W.X.; Jefferson, H.; Sperry, A.O. The molecular motor KIFC1 associates with a complex containing nucleoporin NUP62 that is regulated during development and by the small GTPase RAN. Biol. Reprod. 2006, 74, 684–690. [Google Scholar] [CrossRef]
- Finlay, D.R.; Meier, E.; Bradley, P.; Horecka, J.; Forbes, D.J. A complex of nuclear pore proteins required for pore function. J. Cell Biol. 1991, 114, 169–183. [Google Scholar] [CrossRef]
- Jans, D.A.; Xiao, C.Y.; Lam, M.H. Nuclear targeting signal recognition: A key control point in nuclear transport? BioEssays 2000, 22, 532–544. [Google Scholar] [CrossRef]
- Wei, Y.L.; Yang, T.; Kovacs, T.; Yang, W.X. C-terminal kinesin motor es-KIFC1 regulates nuclear formation during spermiogenesis in Chinese mitten crab Eriocheir sinensis. Gene 2019, 719, 144074. [Google Scholar] [CrossRef]
- She, Z.Y.; Pan, M.Y.; Tan, F.Q.; Yang, W.X. Minus end-directed kinesin-14 KIFC1 regulates the positioning and architecture of the Golgi apparatus. Oncotarget 2017, 8, 36469–36483. [Google Scholar] [CrossRef]
- Lee, S.H.; Joo, K.; Jung, E.J.; Hong, H.; Seo, J.; Kim, J. Export of membrane proteins from the Golgi complex to the primary cilium requires the kinesin motor, KIFC1. FASEB J. 2018, 32, 957–968. [Google Scholar] [CrossRef]
- Nath, S.; Bananis, E.; Sarkar, S.; Stockert, R.J.; Sperry, A.O.; Murray, J.W.; Wolkoff, A.W. Kif5B and Kifc1 interact and are required for motility and fission of early endocytic vesicles in mouse liver. Mol. Biol. Cell 2007, 18, 1839–1849. [Google Scholar] [CrossRef]
- Farina, F.; Pierobon, P.; Delevoye, C.; Monnet, J.; Dingli, F.; Loew, D.; Quanz, M.; Dutreix, M.; Cappello, G. Kinesin KIFC1 actively transports bare double-stranded DNA. Nucleic Acids Res. 2013, 41, 4926–4937. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, A.K.; Goodpasture, J.C.; Zaneveld, L.J. Acrosin of mouse spermatozoa. Am. J. Physiol. 1979, 237, E40–E44. [Google Scholar] [CrossRef] [PubMed]
- Kashiwabara, S.; Arai, Y.; Kodaira, K.; Baba, T. Acrosin biosynthesis in meiotic and postmeiotic spermatogenic cells. Biochem. Biophys. Res. Commun. 1990, 173, 240–245. [Google Scholar] [CrossRef]
- Klemm, U.; Müller-Esterl, W.; Engel, W. Acrosin, the peculiar sperm-specific serine protease. Hum. Genet. 1991, 87, 635–641. [Google Scholar] [CrossRef]
- Kennedy, W.P.; Kaminski, J.M.; Van der Ven, H.H.; Jeyendran, R.S.; Reid, D.S.; Blackwell, J.; Bielfeld, P.; Zaneveld, L.J. A simple, clinical assay to evaluate the acrosin activity of human spermatozoa. J. Androl. 1989, 10, 221–231. [Google Scholar] [CrossRef]
- Berruti, G.; Paiardi, C. Acrosome biogenesis: Revisiting old questions to yield new insights. Spermatogenesis 2011, 1, 95–98. [Google Scholar] [CrossRef]
- Crosby, J.A.; Barros, C. Effect of recombinant boar beta-acrosin on sperm binding to intact zona pellucida during in vitro fertilization. Biol. Reprod. 1999, 61, 1535–1540. [Google Scholar] [CrossRef]
- Hirokawa, N.; Noda, Y.; Okada, Y. Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr. Opin. Cell Biol. 1998, 10, 60–73. [Google Scholar] [CrossRef]

| Primer Name | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| KIFC1-F | GAGCAGCTTGGGAYYTNAARGG | PCR (kifc1 cloning) |
| KIFC1-R | CCWGTYTGTCCRTANGCRAA | PCR (kifc1 cloning) |
| 5′ KIFC1-R | TCCAGGTTGGATTTGGAGAACGTGAGAC | 5′ RACE (kifc1 cloning) |
| 3′ KIFC1-F | CAAGGGGAGATGGTGCGGAGA | 3′ RACE (kifc1 cloning) |
| KIFC1-BDLF KIFC1-BDLR BDKIFC1-F1 BDKIFC1 R1 | AACGCCAGGTCAACTCCC GAAATGATGGCACAAATAGAAG CGCGGATCCATGCCAAAGCTCCCTACTT CCGGAATTC CTGCTGATTCCTCTCCACC | qPCR qPCR Antibody Antibody |
| ACR-F1 | GGGGAGCACAACTTGATCACAGAGAGAGA | PCR (acr cloning) |
| ACR-F2 | TGCTCATTGCTTCAAGAAACAACTGGGA | PCR (acr cloning) |
| ACR-R1 | GGGGAACCACAGCCAGGGAT | PCR (acr cloning) |
| ACR-R2 ACR-R3 | TCCAATCCACGAGACGCCAT GCCCTGAAAGGAAGTTACCCA | PCR (acr cloning) PCR (acr cloning) |
| 5′ACR-R1 3′ACR-F1 UPM-long UPM-short NUP 3′Outer 3′Inner BD-Acr-F1 BD-Acr-R1Acrosin-BDLFAcrosin-BDLR β-actin-F β-actin-R | ACCCTTGAGTTTGCCAGAAT GCAGGAGGCGAGGGTAAAGA CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT CTAATACGACTCACTATAGGGC AAGCAGTGGTATCAACGCAGAGT TATTGGGCTGATTCTTGATGACA CGCGGATCCTCCACTAGTGATTTCACTATAGG CCGGAATTCAGCTTCTGGTTCTGTGGAG CCGCTCGAGCTTGTTTGGGTAACTTCCT GAGCTTCTGGTTCTGTGGAG CCTTGTTTGGGTAACTTCCT CAGGAATCGCTGACAGAATG GAAGGTAGAAAGAGAAGCCAAGA | 5′ RACE (acr cloning) 3′ RACE (acr cloning) 5′ RACE (cloning) 5′ RACE (cloning) 5′ RACE (cloning) 3′ RACE (cloning) 3′ RACE (cloning) Antibody Antibody qPCR qPCR Positive control of qPCR Positive control of qPCR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, L.; Xiang, Q.-M.; Zhu, J.-Q.; Chen, Y.-E.; Tang, D.-J.; Zhang, C.-D.; Hou, C.-C. Transport of Acrosomal Enzymes by KIFC1 via the Acroframosomal Cytoskeleton during Spermatogenesis in Macrobrachium rosenbergii (Crustacea, Decapoda, Malacostracea). Animals 2022, 12, 991. https://doi.org/10.3390/ani12080991
Chang L, Xiang Q-M, Zhu J-Q, Chen Y-E, Tang D-J, Zhang C-D, Hou C-C. Transport of Acrosomal Enzymes by KIFC1 via the Acroframosomal Cytoskeleton during Spermatogenesis in Macrobrachium rosenbergii (Crustacea, Decapoda, Malacostracea). Animals. 2022; 12(8):991. https://doi.org/10.3390/ani12080991
Chicago/Turabian StyleChang, Le, Qiu-Meng Xiang, Jun-Quan Zhu, Yin-Er Chen, Dao-Jun Tang, Chun-Dan Zhang, and Cong-Cong Hou. 2022. "Transport of Acrosomal Enzymes by KIFC1 via the Acroframosomal Cytoskeleton during Spermatogenesis in Macrobrachium rosenbergii (Crustacea, Decapoda, Malacostracea)" Animals 12, no. 8: 991. https://doi.org/10.3390/ani12080991
APA StyleChang, L., Xiang, Q.-M., Zhu, J.-Q., Chen, Y.-E., Tang, D.-J., Zhang, C.-D., & Hou, C.-C. (2022). Transport of Acrosomal Enzymes by KIFC1 via the Acroframosomal Cytoskeleton during Spermatogenesis in Macrobrachium rosenbergii (Crustacea, Decapoda, Malacostracea). Animals, 12(8), 991. https://doi.org/10.3390/ani12080991





