Simple Summary
The fusion between sperm and oocyte results in a zygote, which is a single totipotent cell with the ability to develop into a functional organism. Totipotent cells can give rise to different specialized cell types of all lineages. Understanding the interactions between cell signaling pathways, which drive the early embryo to maintain pluripotency, is essential to establishing the optimal embryonic or stem cell culture conditions for biotechnological applications in cattle. Thus, this review summarizes the core of pluripotency genes, strategies for controlling pluripotency, and the potential applications of pluripotency in in vitro production of cattle embryos.
Abstract
Early development in mammals is characterized by the ability of each cell to produce a complete organism plus the extraembryonic, or placental, cells, defined as pluripotency. During subsequent development, pluripotency is lost, and cells begin to differentiate to a particular cell fate. This review summarizes the current knowledge of pluripotency features of bovine embryos cultured in vitro, focusing on the core of pluripotency genes (OCT4, NANOG, SOX2, and CDX2), and main chemical strategies for controlling pluripotent networks during early development. Finally, we discuss the applicability of manipulating pluripotency during the morula to blastocyst transition in cattle species.
1. Introduction
The fusion between sperm and oocyte (two highly differentiated cells) results in a zygote, which is a single totipotent cell with the ability to develop into a functional organism. Totipotent cells can give rise to different specialized cell types of all lineages [1]. Accordingly, by the early blastocyst stage, mammalian embryos are characterized by two morphologically distinct cell populations, outer and inner cells in the morula, which form the surrounding trophectoderm (TE) and the inner cell mass (ICM), respectively, during blastulation. The ICM is formed by totipotent embryonic stem cells (ESCs) that later (in a second wave) differentiate into the pluripotent epiblast (EPI, the nascent embryo proper) and the primitive endoderm (PrE), in nonrodent mammals identified as the hypoblast (HP). The TE, on the other hand, is the precursor to the placenta and the first component of the extraembryonic structures [2,3]. In bovines, many genes show differences in expression in the ICM and TE from those in mouse or human species [4]. The lineage specification in cattle seems to be directed by a different set of regulatory factors [5]. Moreover, the precise molecular interactions governing the ICM/TE specification in this species has not been totally clarified yet [6]. Thus, understanding the interactions between cell signaling pathways, which drive the ICM to maintain pluripotency, is essential to establishing the optimal embryonic or stem cell culture conditions for biotechnological applications [4]. For instance, in the mouse species, the triad of genes OCT4, NANOG and SOX2 are essential factors underlying pluripotency in both ICM and ES cells [7,8,9]. Caudal-type homeodomain transcription factor 2 (CDX2) is essential for segregation of the ICM and TE lineages at the blastocyst stage by ensuring repression of OCT4 and NANOG in the TE [10]. In cattle, the lineages become fully segregated at the late blastocyst stage [5], where OCT4, NANOG, and SOX2 are also critical transcription factors related to pluripotency maintenance in the ICM. Together with CDX2, they are essential for early development and gene expression involved in differentiation of the ICM and TE lineages [11].
Thus, this article focuses on two main topics restricted to bovine species. We review the literature describing pluripotency features of bovine embryos cultured in vitro, focusing on the core of pluripotency genes (OCT4, NANOG, SOX2, and CDX2), and the main chemical strategies for maintaining a pluripotent state during early development. Finally, we discuss the applicability of inducing or maintaining pluripotency in vitro.
2. Pluripotent Core in Early Bovine Development
2.1. OCT4 (POU5F1)
The octamer-binding transcription factor 4 (OCT4) is a transcription factor that belongs to the POU transcription family domain (POU5F1), which is expressed predominantly in pluripotent cells [12].
OCT4 contributes to maintaining cells in an undifferentiated state by modulating expression of different loci involved in pluripotency and cellular differentiation [13]. Furthermore, OCT4 acts as a regulator of cell lineage specification beyond the morula stage and is necessary for pluripotency maintenance and NANOG expression [14]. For instance, silencing of IFNT involves quenching of the transactivation site to inhibit differentiation towards the trophectoderm [15]. The disruption of the OCT4 gene affects blastulation but not the ability of embryos to progress up to the morula stage, suggesting that OCT4 is not required for cell proliferation after EGA [11,16]. Although the absence of OCT4 expression decreases the number of ICM cells and embryonic quality [17], other authors have observed that embryos with different developmental potential, such as those produced by somatic cell nuclear transfer (SCNT), in vivo-derived, and IVF embryos have similar levels of OCT4 expression [18]. In the same line, parthenogenetic embryos having reduced expression of OCT4 showed no reduced cell counts, suggesting that a reduction in OCT4 expression could be not always limitative of the ICM’s viability [19]. Thus, OCT4 transcripts are indicative of pluripotency but would not be considered as a specific marker for embryo quality.
In bovines, OCT4 acts as a regulator of caudal-type homeodomain protein (CDX2) expression and trophectoderm specification [14,16], as well as of the transition of polar and mural trophoblast development [20]. Interestingly, OCT4 is expressed in all cell embryos throughout the morula stage but then becomes restricted to cells of the ICM in the blastocyst stage in mice [1]. Initial studies found that bovine blastocysts expressed OCT4 mRNA only in the ICM and that its presence in the TE would be the cause of high stability of the protein or due to a delay in its clearance [21]. However, other authors have found that its expression is not restricted to pluripotent cells and that it can therefore be found in both ICM and TE cells [22,23,24,25].
During development, the maternal-derived OCT4 transcript is present in the bovine oocyte, but after fertilization, a decrease in its abundance is observed until the time of embryonic genome activation (EGA), followed by a significant increase after the morula stage [16,21,22,26,27]. In OCT4-KO morulae (day 5), ~70% of the nuclei were OCT4 positive, indicating that maternal transcripts could partially maintain OCT4 expression during early development [14]. At the morula stage, OCT4 and CDX2 proteins show global nuclear localization [22,28]. Interestingly, the presence of OCT-4 and homeobox protein NANOG (NANOG) in the TE does not interfere with the expression of trophoblast-specific genes such as CDX2 or interferon tau (IFN-T) [20]. Moreover, it is not possible to increase OCT4 expression by decreasing CDX2, indicating that unlike that in mice, bovine OCT4 is not regulated by CDX2 [14,29]. One author speculated that bovine TE was regulated by different factors and/or that the regulatory region of the OCT4 gene showed variations among species [29]. Additionally, the presence of OCT4 in the TE is related to the maintenance of pluripotency of this tissue to conserve the plasticity of a “non-differentiating trophoblast” [20].
Given that the process of ICM-specific allocation is gradual, OCT4 can be found in both the ICM and in surrounding TE cells of early blastocysts (7 days postfertilization (dpf)). At 8–9 dpf, OCT4-positive blastomeres are predominantly located in the ICM. However, they are still detected within the TE cell population [22,28], where expression levels of OCT4 and CDX2 do not differ between the ICM and the TE [4]. After blastocyst hatching or 9 dpf, OCT4 is located exclusively in ICM cells [26,28,30].
2.2. NANOG (Homeobox Protein NANOG)
NANOG is a member of the homeobox family of DNA-binding transcription factors that is known to maintain the pluripotency of ESCs [8]. Functionally, NANOG is not required for proper segregation of the TE and ICM, but it is required for deriving and maintaining the pluripotent epiblast and for the second lineage commitment [31,32]. Moreover, it seems to be implicated in cell proliferation, probably depending on FGF4 signaling (which is also involved in fate decision and patterning events in the early embryo) from EPI precursor cells [31]. Thus, disruption of the NANOG gene did not affect the blastocyst rate but resulted in a reduced total cell number [31] and an ICM composed mostly of hypoblast cells [32]. In the nascent epiblast, NANOG mediated repression of hypoblast markers, such as SOX17. SOX17 is dependent on MEK signaling, but its FGF4-induced expression depends on NANOG. Therefore, the establishment of the hypoblast lineage depends on epiblast-mediated FGF/MEK signaling [31]. In relation to other markers, the absence of NANOG resulted in lower expression of the epiblast cell marker SOX2 and the hypoblast marker GATA6 without affecting the trophectoderm [32]. Moreover, in bovines, the activation of NANOG might be OCT4 related. Simmet et al. [14] showed that although OCT4-KO bovine blastocysts expressed NANOG at the morula stage (probably remains of maternal origin), it was depleted in later stages, suggesting that NANOG expression is mutually regulated with OCT4 [14] (Figure 1).
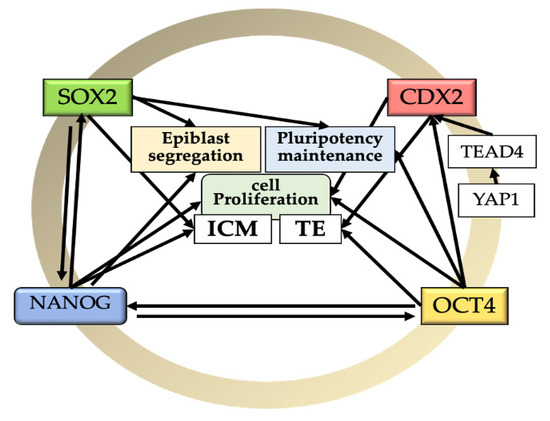
Figure 1.
Relationships among the core of pluripotency factors and epiblast segregation, pluripotency maintenance, and cell proliferation during early bovine development. SOX2: SRY (sex determining region Y)-box 2; OCT4: octamer-binding transcription factor 4 (POU5F1); NANOG: homeobox protein NANOG; TE: trophectoderm; ICM: inner cell mass; TEAD4: TEA Domain Transcription Factor 4; YAP1: Yes Associated Protein 1. Black arrows indicate positive correlations.
In the bovine embryo, NANOG expression begins at the 8–16-cell stage, which is the time of major EGA [22,28]. Although transcripts for NANOG are detected in 5-cell and 8–16-cell stage embryos, the protein is not detectable at these stages [33]. Instead, it appears de novo at the morula stage and in cavitating embryos as a product of the embryonic genome, mainly from the nascent ICM [22,28,33]. In early blastocysts (7 dpi), NANOG is also located in both the ICM and in the surrounding TE cells [34,35]. After hatching, NANOG becomes exclusively ICM-specific [5,26,28,33,36], which has been confirmed by RNA sequencing approaches [4,26,37]. Within the group of cells expressing OCT4 and NANOG, there are cells expressing only NANOG and others expressing both factors, with the NANOG protein being predominantly nuclear and OCT4 nuclear and cytoplasmic [28].
2.3. SOX2
SRY (sex determining region Y)-box 2, also known as SOX2, is a transcription factor essential to maintaining self-renewal, and pluripotency and has been reported as highly expressed in bovine ESCs [38]. SOX2 is necessary for maintaining the undifferentiated state of the bovine ICM [37]. Lacking SOX2 resulted in a blastocyst with a reduced number of blastomeres associated with poor embryonic quality [39,40,41], indicating a role for SOX2 in cell proliferation. Similarly, the knockdown of SOX2 led to the formation of a blastocyst with reduced expression of NANOG; since absence of NANOG results in lower expression of SOX2, this suggests a mutual regulation between SOX2 and NANOG [32,42,43] (Figure 1).
During early development, SOX2 is present in the germinal vesicle and metaphase II (MII) oocyte stages, and it can persist in nuclear and cytoplasmic compartments of four- and eight-cell embryos [22,44,45]. Expression of SOX2 in all nuclei continues in both human and cow embryos up to the formation of an early blastocyst [46]. At the eight-cell stage, it is co-expressed with NANOG, but at the blastocyst stage, it overlaps with NANOG and GATA6 in the ICM [5,26,44]. Although it is restricted to the ICM [4,38], as recently confirmed by RNA sequencing approaches [4,26,37], some embryos can also show weak SOX2 expression in TE cells [5,26,44]. This weak presence of SOX2 in the bovine trophoblast could also imply a delayed commitment of TE cells to differentiation [20,29].
Remarkably, disruption of OCT4 does not affect expression of SOX2, suggesting that initiation of ICM formation is OCT4-independent [16]. In addition, unbalanced overexpression of SOX2 has negative effects on the control of embryonic developmental potential [47]. Dysregulated expression of OCT4 and SOX2 in cloned blastocysts has been related to low developmental competence in cattle [48,49,50]. Thus, SOX2 plays a key role in the formation, maintenance, and plasticity of the ICM compartment and, therefore, on embryonic quality.
2.4. Homeobox Protein CDX2
CDX2 is the master regulator of TE lineage specification [16,29,33,42,51,52]. CDX2 is a key regulator/inducer for formation and functional maintenance of TE. At the genetic level, CDX2 regulates multiple trophoblast genes, such as IFNT, HAND1, ASCL2, SOX15, and ELF5 [26,52], and it is important for maintaining the integrity and proliferation of the trophoblast tissue [11]. It is expressed during the whole period of blastocyst development and localized in the TE and ICM of bovine embryos. The CDX2 transcript is present in oocytes, but it decreases gradually after fertilization [51]. CDX2 protein is found in the cytoplasm of all cells of five-cell embryos, but at the subsequent stages, it is found in the cell nuclei [28]. CDX2 is present at the time of major EGA (8–16-cell stage) and increases afterward from the morula to the blastocyst stage [16,28,29,33,42,52]. At 7–8 dpf, CDX2 segregation to the trophoblast cells can be noted; however, a weak signal is still present within the ICM cells during the time from expanded to hatched blastocysts [28,29,51]. At more advanced developmental stages after 9 dpf, the level of CDX2 is at least three times higher in the TE than in the ICM [28]. In particular, CDX2 transcripts start exceeding those of OCT4 in the TE after hatching around day 9 [29]. Moreover, OCT4 is not required to suppress CDX2 in the bovine ICM [14]. Although it has been reported that CDX2 overexpression downregulates OCT4 [52], others have observed that OCT-4 expression is unaffected by CDX2 downregulation and that the deletion of the OCT4 gene does not affect CDX2 expression in the bovine TE [14,42], ruling out a mutual regulation (Figure 1).
In mice, specification of the TE lineage from the pluripotent early blastomeres involves the Hippo signaling pathway, with activation of CDX2 and TEAD4 (another transcription factor) playing a decisive role [53,54]. Similarly, the TEAD4 transcript is present at the morula stage in the bovine embryo [29], which would activate CDX2 to establish TE lineage [5]. A recent study confirmed that the ICM of bovine possesses the potency to become TE through the YAP1–TEAD4 axis [55]. Thus, although TE cells of the late expanded blastocyst are prone to remaining trophectoderm, they are not yet committed to this fate [29,55].
Interestingly, CDX2-knockdown (CDX2-KD) bovine blastocysts form normal blastocoel cavities and cell numbers and allocations and hatched normally without affecting OCT4, NANOG, or SOX2 [51]. Moreover, the absence of CDX2 promotes the overexpression of TEAD4, probably as a compensatory mechanism. Therefore, expression of TEAD4 may contribute to regulating bovine blastocyst formation along with CDX2 [51].
3. Chemical Modulation of Pluripotency in Early Bovine Development
3.1. WNT (Wingless-Related Mouse Mammary Tumor Virus) Pathway
The WNT signaling pathway is a well-known evolutionary and conserved pathway that regulates crucial aspects of cell fate determination and embryonic development [56]. In cattle, there have been several studies reporting contrasting effects of the activation/inhibition of WNT signaling during the early period of embryonic development (Figure 2, Table 1). One study showed that the activation of WNT signaling by blocking glycogen synthase kinase (GSK3) activity with LiCl2 or CT99021 had inconsistent effects on development to the blastocyst stage. LiCl decreased the proportion of zygotes reaching the blastocyst stage, while CT99021 increased this proportion [57]. Later, a study by Kuijk et al. [33] showed that embryos treated from the zygote to the blastocyst stage in the presence of the GSK3 inhibitor CHIR99021 at 3 µM had higher percentages of NANOG cells in the ICM. In addition, when the GSK3 inhibitor was present from the morula stage onwards, they saw no effects on ICM constitution [33]. Denicol et al. [40] observed that activation of canonical WNT signaling with the agonist AMBMP from day 5, disturbed development until the blastocyst stage and reduced the numbers of TE and ICM cells. This was not surprising, since this molecule also disrupts microtubule organization [58]. Another study observed that blocking GSK3 with CHIR99021 (3 µM) from the morula stage onwards improved blastocyst morphology and epiblast-specific gene expression (NANOG, SOX2) [59]. Similarly, Madeja et al. [12] indicated that WNT activation with CHIR99021 increased the expression of OCT4 and NANOG in the ICM and downregulated CDX2 expression. Meng et al. [36] used forskolin, which activates adenylate cyclase and cAMP/PKA signaling pathway, in turn inactivating GSK3 and thus acting synergistically with WNTs. Forskolin increased NANOG expression threefold [36]. More recently, Warzych et al. [5] also observed that WNT signaling (activated by CHIR99021) increased the levels of NANOG and OCT4 transcripts and NANOG-positive cells within the ICM. Furthermore, the proportion of OCT4-positive cells increased in the TE concomitantly with the downregulation of CDX2 [5]. Likewise, Sidrat et al. [60] used 6 bromoindurbin-3’oxime (6-Bio) as a WNT agonist, observing a higher expression of peroxisome proliferator-activated receptor-delta (PPARδ), which colocalized with Βeta-CATENIN and formed a complex with TCF/LEF transcription factor. In addition, 6-Bio enhanced the expression of Βeta-CATENIN, OCT4, AXIN2, and C-MYC, but CDX2 was downregulated. Moreover, the inhibition of PPARδ with Gsk3787 severely perturbed blastocyst formation and hatching, suggesting an important role for PPARδ as a candidate regulator of the canonical WNT pathway.
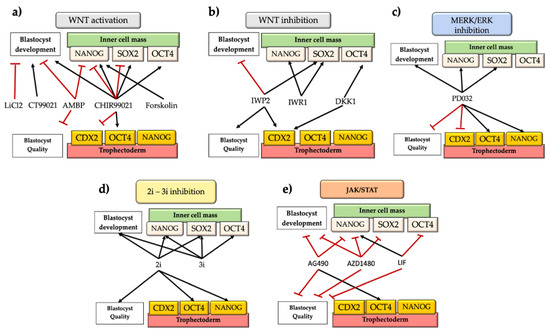
Figure 2.
Effects of small molecules on levels of pluripotency factors (NANOG, SOX2, OCT4, and CDX2), on developmental potential in vitro and embryo quality according to total cell number. (a) Effects of WNT activation on in vitro development and embryo quality; (b) Effects of WNT inhibition on in vitro development and embryo quality; (c) Effects of MERK/ERK inhibition on in vitro development and embryo quality; (d) Effects of 2i-3i inhibition on in vitro development and embryo quality. (e) Effects of JAK/STAT inhibition/activation on in vitro development and embryo quality. Note that some molecules have shown opposites effects across the literature reviewed. SOX2: SRY (sex determining region Y)-box 2; OCT4: POU5F1; NANOG: homeobox protein NANOG; CDX2: homeobox protein CDX2; WNT: wingless-related mouse mammary tumor virus pathway; MEK/ERK: Ras/Raf/Mitogen-activated protein kinase/ERK kinase (MEK)/extracellular-signal-regulated kinase (ERK); 2i-3i inhibition: two-three inhibitors systems (MEK inhibition+WNT activation, and MEK inhibition+WNT activation+FGFR inhibitor, respectively); JAK/STAT: Janus kinase-signal transducer and activator of transcription. Black arrows indicate positive correlations, and red lines indicate negative correlations.

Table 1.
Small-molecule inhibitors and their effects on pluripotency and development of bovine embryos.
On the other hand, other authors have indicated that inhibition of canonical WNT signaling regulates blastocyst development and quality. For instance, Denicol et al. [66] found that the WNT antagonist DKK1 added from the morula to the blastocyst stage promoted differentiation of cells towards trophectoderm and hypoblast lineages [66]. Similarly, exposure to Wnt-C59, which blocks secretion of WNTs, or DKK1, and interferes in the activation of the WNT-FZD-LRP5/6 receptor complex, did not affect development, but Wnt-C59 increased the number of ICM cells, suggesting that regulation of ICM proliferation by endogenous WNTs is independent of the canonical signaling [41]. However, it was recently demonstrated that inhibition of canonical WNT signaling by using an IWR1 inhibitor was crucial for ICM proliferation and derivation of bovine ESCs [38]. Another study indicated that the WNT inhibitor IWP2 increased the total cell number in blastocysts by increasing the number of TE cells and the number of NANOG-positive cells within the ICM but decreasing the percentage of blastocysts [44]. The differences among previous studies could be due to the different specificities and efficacies of the WNT inhibitors used [44]. In fact, recently, Xiao et al. [39] evaluated the effects of different WNT inhibitors on the derivation efficiency of bovine ESCs. They found that canonical WNT signaling was antagonist to pluripotency and that derivation of pluripotent bovine ESCs involved the inhibition of WNT signaling.
However, not all inhibitors showed the same efficacy, with IWR-1 and IWP2 being effective, unlike XAV939 and DKK1. In addition, it was observed that IWR1-inhibition between days 4 and 7.5 after fertilization blocked activation and differentiation into a pSTAT3 positive cell lineage. In the mouse embryo, Stat3 induces differentiation towards the TE lineage when its activation level exceeds certain thresholds [67]. Furthermore, CHIR99021 depressed expression of both NANOG and SOX2 in bovine ESCs and decreased the number and percentage of blastomeres positive for NANOG and SOX2 in the embryo [39]. In this line, other studies have indicated that TE cells highly express transcripts related to WNT signaling [30,37], as observed in the activation of WNT signaling enabling the derivation of the trophoblast stem cell by regulating CDX2 expression through the WNT-YAP/TAZ signaling pathway [68].
Overall, the data indicate that the effects of WNT activation/inhibition depend on the specificity of the inhibitor and time of exposure. In addition, WNT signaling plays a role in TE specification, and the use of specific inhibitors able to interact with JAK and WNT signaling pathways enables the induction of epiblast pluripotency in the blastocyst.
3.2. MEK/ERK Pathway
Molecular interactions of signaling pathways such as MEK/ERK and WNT/β-catenin are critical for cell-to-cell communication and cellular differentiation. Secreted uterine FGF factors induce lineage commitment by activating the mitogen-activated protein kinase (MAPK), comprising MAPK kinase 2 (MAP2K, also known as MAPKK or MEK) and MAPK1/2 (ERK). It has been reported that FGF4 mRNA is present in the trophectoderm of spherical bovine blastocysts [20]. FGF4 can induce the formation of hypoblast and block the formation of epiblast precursors [33], but the role of FGF4 in bovine embryo development differs from that in mice, since FGF4 and MAPK signaling is not essential for bovine hypoblast specification [33].
The suppression of MEK signaling by PD98059 or PD325901 has been performed in numerous studies to detect the importance of MEK/ERK signaling in early development in bovines, although with controversial outcomes. Inhibition of MEK in bovine embryos resulted in ICM with increased epiblast precursors (NANOG+) and decreased hypoblast precursor (GATA6) [33]. Blocking bovine MEK with PD0325901 (0.4 μM) was correlated with improvement in blastocyst morphology and increases in epiblast-specific gene expression (NANOG, SOX2) [59,62]. In addition, trophoblast proliferation, lineage specification, and blastocyst formation were not affected [59,62,69]. This was consistent with studies showing that isolated trophoblast cells did not require active FGF/MEK signaling to survive, proliferate, and maintain CDX2 expression [62,70,71,72]. In agreement with Kuijk et al. [33], under MEK inhibition (PD0325901 0.5 and 2.5 μM), neither embryonic development nor cell numbers were affected, but the proportion of NANOG-positive cells was markedly increased, while the expression of GATA6 was reduced but not completely switched off [31].
The effects of MEK inhibition seem to be dose dependent [5,44]. A study by Canizo et al. [44] indicated that MEK inhibition did not promote epiblast fate but rather prevented hypoblast segregation in cattle. MEK inhibition with PD0325901 at 0.4 μM decreased the numbers of ICM cells, but it had a trophic effect on the TE. Instead, high concentrations of MEK inhibition (between 1 and 2 μM) resulted in abolition of hypoblast segregation, and 10 μM affected both the TE and ICM compartments [44]. Similarly, another study indicated that MEK/ERK downregulation (PD0325901, 1 µM) maintained OCT4 and NANOG within the ICM and prevented their exclusion from the TE, but CDX2 was downregulated [5].
3.3. The Use of Two Inhibitors (2i) and Three Inhibitors (3i) in Early Bovine Embryonic Development
Both the 2i and 3i systems operate within the WNT and the MEK/ERK signaling pathways but use a different set of inhibitors. The 2i system (referring to the combined use of two inhibitors) includes CHIR99021 and MEK inhibition (PD0325901). The 3i (three-inhibitor) system is based on the use of 2i by a MEK/ERK inhibitor (PD184352) and GSK3 inhibitor (CHIR99021) plus an FGF receptor inhibitor (SU5402) [61]. Thus, the 2i system plus the FGF receptor inhibitor (SU5402) involves the suppression of the MAPK/ERK pathway, whereas the inhibition of GSK3 supports WNT activity.
Early studies found that the double inhibition (2i) of MEK and GSK3 offered defined culture conditions for blocking exit from pluripotency. The use of 2i enhanced bovine blastocyst development and expression of epiblast NANOG and SOX2 markers by reducing expression of the hypoblast marker GATA4 [59]. The presence of 2i (0.4–10 μM) from the morula stage (D5) onward increased the numbers of ICM cells, but NANOG and FGF4 were upregulated, and specification towards the hypoblast was reduced, in the ICM after exposure to 3i combinations [62]. From day 2 onward, 3i improved embryonic development-affecting ICM-related genes (OCT4, SOX2, and NANOG) [34]. However, other authors found positive effects of 2i only on blastocyst quality according to total cell and ICM number [61]. Similarly, Warzych et al. [5] observed higher levels of epiblast-related genes (NANOG and OCT4) under the 2i system but no effect on the number of cells in the blastocyst. Likewise, Kuijk et al. [33] did not find any synergetic effects between CHIR99021 and PD032590, as it was recently indicated that modulation of WNT is not sufficient to support enhanced NANOG expression in the epiblast when combined with the ERK inhibitor [44] (Figure 2, Table 1).
Additionally, these pathways seem to be involved in the regulation of apoptosis. A study by Madeja et al. [61] found positive effects of 2i on the ICM constitution; however, the total cell counts in 3i-cultured embryos were reduced. Embryos cultured under 2i or 3i systems also showed higher rates of apoptosis and lower embryonic quality but without changes in BAX, BCL2, or BAK transcripts, suggesting alternative pathways involved in this apoptotic activation.
3.4. JAK/STAT
The Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling pathway mediates cellular responses to growth factors (e.g., EGF) and cytokines (e.g., IL-6). These responses include differentiation, proliferation, apoptosis, migration, and cell survival, depending on the cellular context. Thus, this signaling pathway is essential for numerous homeostatic and developmental processes, including stem cell maintenance [73].
In a study by Meng et al. [36], the authors observed that chemically suppressing JAK/STAT signaling (via JAK2/3) with AG490 and JAK1/2 inhibition with AZD1480 strongly compromised blastocyst development and quality and ICM numbers without affecting the TE. In addition, NANOG was reduced under both AG490 and AZD1480 treatments. The latter also strongly reduced SOX2, KFL4, FGF4, and hypoblast markers (SOX17, PDGFRα) without affecting CDX2. In addition, phosphorylation of STAT3 tyrosine (Y) 705, which is related to JAK1 pluripotency-signal [74], colocalized with NANOG and SOX2 within the ICM in D7 and D8 blastocysts [36], suggesting its role in ICM specification (Figure 2, Table 1).
On the other hand, locally secreted FGF4 can activate both (i) mitogen-activated protein kinase (MAP2K) and (ii) phosphatidylinositol 3-kinase (PI3K)–AKT [5,36,75]. JAK/STAT activation is also triggered by leukemia inhibitory factor (LIF) and related members of the interleukin (IL) family. In bovines, LIF added to the culture medium from four-cell yielded no significant benefit [76]. However, when added to the culture medium from days 5 to 8, it showed adverse effects on in vitro embryonic development based on kinetics, morphology, cell count, and the expression of OCT4 [63]. Similarly, other authors have indicated that LIF did not affect trophoblast or ICM cell numbers [64], nor were any detrimental effects on blastocyst development observed that might be the result of the antimitotic effect of LIF, especially when used in early cleavage stages [65]. Recently, Canizo et al. [44] reported that LIF added to a 2i cocktail boosted the blastocyst yields, and LIF alone promoted expansion of hypoblast in bovine embryos, suggesting that LIF has embryotropic effects in the ICM by increasing NANOG and SOX17 markers. Thus, JAK/STAT signals are required for bovine ICM formation and acquisition of pluripotency markers.
4. The Role of Pluripotency in Biotechnological Applications
4.1. In Vitro Embryo Production
IVP technology has become commercially viable and extensively used for producing embryos in cattle [77]. It is also known that in vitro culture conditions determine embryo quality, expressed as developmental kinetics, blastomere count, EGA efficiency, gene expression, apoptotic rates, etc. [78]. Thus, modifications of the culture system, particularly before the time of EGA, can significantly impact the pluripotency profile and quality of the resulting blastocysts. For example, activation or inhibition of the WNT and silencing of the MEK/ERK signaling pathways alters critical pathways associated with apoptosis, implantation, and maternal recognition of pregnancy [39,61]. However, only few studies have evaluated if control of pluripotency at preimplantation stages can influence postimplantation, delivery, and/or in vivo development in bovine species. For instance, a study by Tribulo et al. [79] found that calves derived from embryos exposed to DKK1 from the morula to the blastocyst stage had lower birth weights than the control group, suggesting that changes in molecular signaling during early developmental stages impact the postnatal phenotype. Recently, Han et al. [34] evaluated the developmental effects of a modified 3i system on bovine and mouse IVF efficiency, and they transferred mouse 3i embryos to surrogate females. They did not find any differences in birth rate, sex ratio, morphology, or body weight compared with the progeny of the control group. In addition, the 3i offspring produced normal pups, indicating that the fertility of mice developed from the inhibited embryos was not affected. In this sense, it would be important to continue studying physiological changes induced by chemical inhibitors to gain greater insight into later impacts on pre- and postimplantation development.
It is well known that most embryos generated by IVF technologies (IVF, SCNT, or ICSI) do not gather the required morphological quality to be transferred [48]. For instance, bovine embryos with low development potential show a precarious balance between pluripotency factors that disturbs later stages of embryonic development [22]. Therefore, the chemical control of cell differentiation pathways and pluripotent profiles raises as a valid strategy for “rescuing” the developmental potential of embryos of lower quality to obtain embryos in vitro efficiently, especially in large animals (Figure 3).
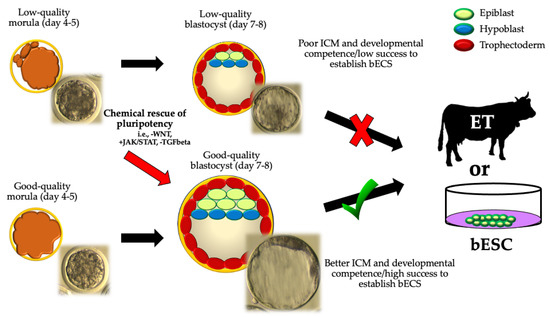
Figure 3.
A theoretical approach to “rescue” in vitro developmental potential from cattle embryos of lower quality (seen as morula with delayed compaction, presence of cell debris, fragmentation and/or slower developmental kinetics). ICM: inner cell mass; ET: embryo transfer; bESCs: bovine embryonic stem cells). Low-quality blastocyst: embryo with delayed blastulation, poor symmetry, and/or cells that are loosely packed for the ICM and trophectoderm. Good-quality blastocyst: embryo with an expanded blastocoel cavity, highly symmetric, absence of cell debris or fragmentation, and highly packed ICM and trophectoderm cells, where a clearly visible ICM can be distinguished during morphological valuation. -WNT: WNT-inhibition; +JAK/STAT: JAK/STAT activation; -TGFbeta: TGFbeta inhibition.
In addition, another approach used to optimize in vitro embryo production efficiency in cattle species has been to supplement culture media with biologically active molecules produced by the reproductive tract or embryo in early pregnancy. Hansen et al. called these “embryokine”, such as CSF2, molecules produced by the female reproductive and embryo tract that control embryonic development and pluripotency [80]. This topic has been reviewed in detail elsewhere [80,81]. Thus, the control of molecular interactions of signaling pathways critical for cellular differentiation and pluripotency leads to strategies seeking to optimize IVP conditions and boost embryonic developmental potential.
4.2. Capturing Pluripotency In Vitro
Currently, because of an improved understanding of pluripotency, stemness from cattle species can be captured in vitro [82] by deriving embryonic stem cells from biparental embryos produced by IVF. Pluripotent stem cells can also be derived from somatic cells, the induced pluripotent stem cells (iPSCs) [83,84]. Bogliotti et al. [38] employed fibroblast growth factor 2 (FGF2) and a canonical WNT signaling pathway inhibitor in their culture conditions and derived stable pluripotent cell lines from bovine blastocysts. Bovine pluripotent cells express the pluripotent markers SOX2 and POU5F1 and are negative for CDX2 and the hypoblast marker GATA6 [82,85,86]. Thus, inhibition of WNT signaling by IWR-1 and stimulation of the FGF2 pathway seem to be essential requirements for deriving bovine ESC lines [86]. Xiao et al. [39] evaluated the effects of different WNT inhibitors on the derivation efficiency of bovine ESCs. They found that canonical WNT signaling was antagonistic to pluripotency and that derivation of pluripotent ESCs involved inhibition of WNT signaling. Nonetheless, not all inhibitors showed the same efficacy, with IWR-1 and IWP2 being effective but not XAV939 and DKK1. Recently, Soto et al. [86] reported a simplified bESC culture system based on a commercially available medium (N2B27) and feeder-free culture conditions based in a chemical substrate (Vitronectin) supplemented with activin A (AA). Activin is a growth factor known to support the expansion of human ESCs [87]. Nonetheless, the effects of AA on the developing bovine blastocyst are adverse for ICM proliferation [88], which would restrict its use only for bESC derivation.
On the other hand, bovine iPSCs (biPSCs) have been generated from somatic cells using exogenous transcriptional factors combined with small chemical inhibitors supported by current knowledge of pluripotential pathways. Using a combination of seven factors (OCT4, SOX2, NANOG, KLF4, cMYC, LIN28, and KDM4A) and a reprogramming medium containing inhibitors of WNT (IWR1) and H3K79 methyltransferase Dot1L (iDot1L), Su et al. [84] derived primed-like iPSCs from mesenchymal stem cells. OCT4, NANOG, and SOX2 were highly activated in these iPSCs across different passages [84]. Similarly, Pillai et al. [83] enhanced the cellular reprogramming of bovine fibroblasts to biPSCs by forcing expression of OCT4, SOX2, KLF4, and MYC, but they also reported that inhibition of ALK4/5/7 to block TGFβ/activin/nodal signaling together with GSK3β and MEK1/2 supported robust in vitro self-renewal of naive biPSCs. A detailed review of these topics was recently reported [89].
5. Future Perspectives
In general, chemical approaches have been quite successful in modulating and identifying pluripotency pathways involved in early development and cell fate differentiation of mammalian embryos. However, off-target effects complicate small-molecule methods. For example, a study by Xiao et al. [39] found that derivation of bovine ESC can be conducted by IWR-1 and IWP2 but not by XAV939 and DKK1, either because they do not specifically inhibit WNT signaling or because they have additional effects that affect cell function in a canonical, WNT-independent manner. One alternative tool for investigating the functional genomics of bovine pluripotency is gene editing approaches. For instance, Daigneault et al. [16] used CRISPR/Cas9 for targeted disruption of the POU5F1 (OCT4) gene by direct injection into zygotes. Disruption of the bovine POU5F1 locus was highly efficient and was associated with developmental arrest at the morula stage, indicating that POU5F1 is essential for the formation of expanded bovine blastocysts. Similar approaches have been reported [32,90,91]. Likewise, RNA interference has greatly facilitated analysis of loss-of-function phenotypes [92], but correlating these phenotypes with small-molecule inhibition profiles is not always straightforward [93]. Jafarpour et al. [94] reported that downregulation of SUV39H1/H2 (a histone methyltransferase) through siRNA in fibroblasts improved the blastocyst yield of bovine SCNT embryos. However, this effect was not observed after the treatment of fibroblasts with chaetocin, a chemical inhibitor of SUV39H1/H2, possibly because of its off-target effects on other histone methyltransferases. Indeed, gene editing technologies have a wide range of applications in livestock species [95,96], but they also represent valuable tools for investigating functional genomics of the bovine embryo at early stages.
Finally, it is important to highlight the contribution of high-throughput sequencing platforms for dissecting pathways and identifying undefined gene expression sequences associated with pluripotency [83,97,98]. Thus, transcriptomic analysis will continue contributing to the molecular characterization of bovine pluripotency and to the establishment of culture conditions that support the derivation and maintenance of true ESCs.
6. Conclusions
In this article, we review the current knowledge of core pluripotency markers during early development of bovine embryos. We also describe the main pathways involved in pluripotency maintenance and cell differentiation. Embryonic pluripotency depends on the activation of several molecular mechanisms involving different factors. The rigorous balance between maternal clearance and zygotic expression of OCT4, NANOG, SOX2, and CDX2 affects differentiation, proliferation, apoptosis, and embryonic quality. In addition, WNT signaling seems to play a crucial role in driving both cell differentiation and pluripotency maintenance in bovine species. The recent derivation of PSCs (ESCs and iPSCs) is a hallmark of progress concerning pluripotency in cattle species. There is also great potential to optimize in vitro culture conditions by controlling cell differentiation networks, such as by incorporating small molecules and avoiding the need for undefined culture components. In this vein, manipulating pluripotency networks of low-quality embryos produced by IVF technologies can certainly rescue their developmental competence and increase the efficiency of IVP, especially in large species. However, chemical modulation will depend on exposure time, concentrations, and secondary targeting of the small molecule(s). Even the base medium used (e.g., SOF, KSOM, N2B27, etc.) can undoubtedly influence the final outcome. Indeed, the generation of more specific antibodies, genetic engineering, and advanced technologies such as deep sequencing approaches has contributed significantly to understanding how early development in mammals diverges in terms of pluripotent characteristics and to establishing favorable conditions for capturing cattle pluripotency in vitro.
Author Contributions
Conceptualization, L.A., C.O.-S. and F.T.; writing L.A., C.O.-S., F.T. and R.F.—original draft preparation, L.A., C.O.-S., F.T. and R.F.; writing—review and editing, L.A., C.O.-S., F.T. and R.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Universidad de La Frontera, Programa de Formacion de Investigadores Postdoctorales (PDT21-0001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Funded (partially) by the Research Office, Universidad de La Frontera.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rebuzzini, P.; Zuccotti, M.; Garagna, S. Building Pluripotency Identity in the Early Embryo and Derived Stem Cells. Cells 2021, 10, 2049. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.F. Developmental Biology; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Chazaud, C.; Yamanaka, Y.; Pawson, T.; Rossant, J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell 2006, 10, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, M.; Sakatani, M.; Yao, J.; Shanker, S.; Yu, F.; Yamashita, R.; Wakabayashi, S.; Nakai, K.; Dobbs, K.B.; Sudano, M.J.; et al. Global gene expression of the inner cell mass and trophectoderm of the bovine blastocyst. BMC Dev. Biol. 2012, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Warzych, E.; Pawlak, P.; Lechniak, D.; Madeja, Z.E. WNT signalling supported by MEK/ERK inhibition is essential to maintain pluripotency in bovine preimplantation embryo. Dev. Biol. 2020, 463, 63–76. [Google Scholar] [CrossRef]
- Piliszek, A.; Madeja, Z.E. Pre-implantation Development of Domestic Animals. Curr. Top. Dev. Biol. 2018, 128, 267–294. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional expression cloning of NANOG, a pluripotency sustaining factor in embryonic stem cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef]
- Mitsui, K.; Tokuzawa, Y.; Itoh, H.; Segawa, K.; Murakami, M.; Takahashi, K.; Maruyama, M.; Maeda, M.; Yamanaka, S. The homeoprotein NANOG is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003, 113, 631–642. [Google Scholar] [CrossRef]
- Avilion, A.A.; Nicolis, S.K.; Pevny, L.H.; Perez, L.; Vivian, N.; Lovell-Badge, R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003, 17, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Strumpf, D.; Mao, C.A.; Yamanaka, Y.; Ralston, A.; Chawengsaksophak, K.; Beck, F.; Rossant, J. CDX2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005, 132, 2093–2102. [Google Scholar] [CrossRef]
- Sakurai, N.; Takahashi, K.; Emura, N.; Fujii, T.; Hirayama, H.; Kageyama, S.; Hashizume, T.; Sawai, K. The Necessity of OCT-4 and CDX2 for Early Development and Gene Expression Involved in Differentiation of Inner Cell Mass and Trophectoderm Lineages in Bovine Embryos. Cell. Reprogram. 2016, 18, 309–318. [Google Scholar] [CrossRef]
- Madeja, Z.E.; Hryniewicz, K.; Orsztynowicz, M.; Pawlak, P.; Perkowska, A. WNT/β-catenin signaling affects cell lineage and pluripotency-specific gene expression in bovine blastocysts: Prospects for bovine embryonic stem cell derivation. Stem Cells Dev. 2015, 24, 2437–2454. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, V.; Rezende, N.C.; Scotland, K.B.; Shaffer, S.M.; Persson, J.L.; Gudas, L.J.; Mongan, N.P. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009, 18, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Simmet, K.; Zakhartchenko, V.; Philippou-Massier, J.; Blum, H.; Klymiuk, N.; Wolf, E. OCT4/POU5F1 is required for NANOG expression in bovine blastocysts. Proc. Natl. Acad. Sci. USA 2018, 115, 2770–2775. [Google Scholar] [CrossRef] [PubMed]
- Ezashi, T.; Ghosh, D.; Roberts, R.M. Repression of Ets-2-induced transactivation of the tau interferon promoter by Oct-4. Mol. Cell. Biol. 2001, 21, 7883–7891. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Daigneault, B.W.; Rajput, S.; Smith, G.W.; Ross, P.J. Embryonic POU5F1 is Required for Expanded Bovine Blastocyst Formation. Sci. Rep. 2018, 8, 7753. [Google Scholar] [CrossRef] [PubMed]
- Nganvongpanit, K.; Müller, H.; Rings, F.; Hoelker, M.; Jennen, D.; Tholen, E.; Havlicek, V.; Besenfelder, U.; Schellander, K.; Tesfaye, D. Selective degradation of maternal and embryonic transcripts in in vitro produced bovine oocytes and embryos using sequence specific double-stranded RNA. Reproduction 2006, 131, 861–874. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Long, J.E.; Cai, X.; He, L.Q. Gene profiling of cattle blastocysts derived from nuclear transfer, in vitro fertilization and in vivo development based on cDNA library. Anim. Reprod. Sci. 2007, 100, 243–256. [Google Scholar] [CrossRef]
- Gómez, E.; Caamaño, J.N.; Bermejo-Alvarez, P.; Díez, C.; Muñoz, M.; Martín, D.; Carrocera, S.; Gutiérrez-Adán, A. Gene expression in early expanded parthenogenetic and in vitro fertilized bovine blastocysts. J. Reprod. Dev. 2009, 55, 607–614. [Google Scholar] [CrossRef]
- Degrelle, S.A.; Campion, E.; Cabau, C.; Piumi, F.; Reinaud, P.; Richard, C.; Renard, J.P.; Hue, I. Molecular evidence for a critical period in mural trophoblast development in bovine blastocysts. Dev. Biol. 2005, 288, 448–460. [Google Scholar] [CrossRef]
- Kurosaka, S.; Eckardt, S.; McLaughlin, K.J. Pluripotent lineage definition in bovine embryos by Oct4 transcript localization. Biol. Reprod. 2004, 71, 1578–1582. [Google Scholar] [CrossRef]
- Khan, D.R.; Dubé, D.; Gall, L.; Peynot, N.; Ruffini, S.; Laffont, L.; Le Bourhis, D.; Degrelle, S.; Jouneau, A.; Duranthon, V. Expression of pluripotency master regulators during two key developmental transitions: EGA and early lineage specification in the bovine embryo. PLoS ONE 2012, 7, e34110. [Google Scholar] [CrossRef] [PubMed]
- Kirchhof, N.; Carnwath, J.W.; Lemme, E.; Anastassiadis, K.; Scholer, H.; Niemann, H. Expression pattern of Oct-4 in preimplantation embryos of different species. Biol. Reprod. 2000, 63, 1698–1705. [Google Scholar] [CrossRef] [PubMed]
- Van Eijk, M.J.T.; Van Rooijen, M.A.; Modina, S.; Scesi, L.; Folkers, G.; Van Tol, H.T.A.; Bevers, M.M.; Fisher, S.R.; Lewin, H.A.; Rakacolli, D.; et al. Molecular cloning, genetic mapping, and developmental expression of bovine POU5F1. Biol. Reprod. 1999, 60, 1093–1103. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, P.; Dang, Y.; Li, S.; Luo, L.; Hu, B.; Wang, S.; Wang, H.; Zhang, K. Functional roles of the chromatin remodeler SMARCA5 in mouse and bovine preimplantation embryos†. Biol. Reprod. 2021, 105, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Brinkhof, B.; van Tol, H.T.A.; Groot Koerkamp, M.J.A.; Riemers, F.M.; Ijzer, S.G.; Mashayekhi, K.; Haagsman, H.P.; Roelen, B.A.J. A mRNA landscape of bovine embryos after standard and MAPK-inhibited culture conditions: A comparative analysis. BMC Genomics 2015, 16, 277. [Google Scholar] [CrossRef][Green Version]
- Vigneault, C.; McGraw, S.; Massicotte, L.; Sirard, M.A. Transcription factor expression patterns in bovine in vitro-derived embryos prior to maternal-zygotic transition. Biol. Reprod. 2004, 70, 1701–1709. [Google Scholar] [CrossRef]
- Madeja, Z.E.; Sosnowski, J.; Hryniewicz, K.; Warzych, E.; Pawlak, P.; Rozwadowska, N.; Plusa, B.; Lechniak, D. Changes in sub-cellular localisation of trophoblast and inner cell mass specific transcription factors during bovine preimplantation development. BMC Dev. Biol. 2013, 13, 32. [Google Scholar] [CrossRef]
- Berg, D.K.; Smith, C.S.; Pearton, D.J.; Wells, D.N.; Broadhurst, R.; Donnison, M.; Pfeffer, P.L. Trophectoderm lineage determination in cattle. Dev. Cell 2011, 20, 244–255. [Google Scholar] [CrossRef]
- Akizawa, H.; Saito, S.; Kohri, N.; Furukawa, E.; Hayashi, Y.; Bai, H.; Nagano, M.; Yanagawa, Y.; Tsukahara, H.; Takahashi, M.; et al. Deciphering two rounds of cell lineage segregations during bovine preimplantation development. FASEB J. 2021, 35, e21904. [Google Scholar] [CrossRef]
- Springer, C.; Zakhartchenko, V.; Wolf, E.; Simmet, K. Hypoblast Formation in Bovine Embryos Does Not Depend on NANOG. Cells 2021, 10, 2232. [Google Scholar] [CrossRef]
- Ortega, M.S.; Kelleher, A.M.; O’Neil, E.; Benne, J.; Cecil, R.; Spencer, T.E. NANOG is required to form the epiblast and maintain pluripotency in the bovine embryo. Mol. Reprod. Dev. 2020, 87, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Kuijk, E.W.; van Tol, L.T.A.; van de Velde, H.; Wubbolts, R.; Welling, M.; Geijsen, N.; Roelen, B.A.J. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development 2012, 139, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Xiang, J.; Li, C.; Wang, J.; Wang, C.; Zhang, Y.; Li, Z.; Lu, Z.; Yue, Y.; Li, X. MLL1 combined with GSK3 and MAP2K inhibition improves the development of in vitro-fertilized embryos. Theriogenology 2020, 146, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Wang, F.; Chen, Z.; Liu, Z.; Mei, C.; Wu, H.; Huang, J.; Li, C.; Zhou, L.; Lin, L. Isolation and culture of primary bovine embryonic stem cell colonies by a novel method. J. Exp. Zool. A. Ecol. Genet. Physiol. 2009, 311, 368–376. [Google Scholar] [CrossRef]
- Meng, F.; Forrester-Gauntlett, B.; Turner, P.; Henderson, H.; Oback, B. Signal Inhibition Reveals JAK/STAT3 Pathway as Critical for Bovine Inner Cell Mass Development. Biol. Reprod. 2015, 93, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.M.; Cui, L.S.; Hao, H.S.; Wang, H.Y.; Zhao, S.J.; Du, W.H.; Wang, D.; Liu, Y.; Zhu, H.B. Transcriptome analyses of inner cell mass and trophectoderm cells isolated by magnetic-activated cell sorting from bovine blastocysts using single cell RNA-seq. Reprod. Domest. Anim. 2016, 51, 726–735. [Google Scholar] [CrossRef]
- Bogliotti, Y.S.; Wu, J.; Vilarino, M.; Okamura, D.; Soto, D.A.; Zhong, C.; Sakurai, M.; Sampaio, R.V.; Suzuki, K.; Izpisua Belmonte, J.C.; et al. Efficient derivation of stable primed pluripotent embryonic stem cells from bovine blastocysts. Proc. Natl. Acad. Sci. USA 2018, 115, 2090–2095. [Google Scholar] [CrossRef]
- Xiao, Y.; Amaral, T.F.; Ross, P.J.; Soto, D.A.; Diffenderfer, K.E.; Pankonin, A.R.; Jeensuk, S.; Tríbulo, P.; Hansen, P.J. Importance of WNT-dependent signaling for derivation and maintenance of primed pluripotent bovine embryonic stem cells†. Biol. Reprod. 2021, 105, 52–63. [Google Scholar] [CrossRef]
- Denicol, A.C.; Dobbs, K.B.; McLean, K.M.; Carambula, S.F.; Loureiro, B.; Hansen, P.J. Canonical WNT signaling regulates development of bovine embryos to the blastocyst stage. Sci. Rep. 2013, 3, 1266. [Google Scholar] [CrossRef]
- Tribulo, P.; da Silva Leão, B.C.; Lehloenya, K.C.; Mingoti, G.Z.; Hansen, P.J. Consequences of endogenous and exogenous WNT signaling for development of the preimplantation bovine embryo. Biol. Reprod. 2017, 96, 1129–1141. [Google Scholar] [CrossRef]
- Goissis, M.D.; Cibelli, J.B. Functional characterization of CDX2 during bovine preimplantation development in vitro. Mol. Reprod. Dev. 2014, 81, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Mistri, T.K.; Arindrarto, W.; Ng, W.P.; Wang, C.; Lim, L.H.; Sun, L.; Chambers, I.; Wohland, T.; Robson, P. Dynamic changes in SOX2 spatio-temporal expression promote the second cell fate decision through Fgf4/ Fgfr2 signaling in preimplantation mouse embryos. Biochem. J. 2018, 475, 1075–1089. [Google Scholar] [CrossRef] [PubMed]
- Canizo, J.R.; Ynsaurralde Rivolta, A.E.; Vazquez Echegaray, C.; Suvá, M.; Alberio, V.; Aller, J.F.; Guberman, A.S.; Salamone, D.F.; Alberio, R.H.; Alberio, R. A dose-dependent response to MEK inhibition determines hypoblast fate in bovine embryos. BMC Dev. Biol. 2019, 19, 13. [Google Scholar] [CrossRef] [PubMed]
- Kuijk, E.W.; Du Puy, L.; Van Tol, H.T.A.; Oei, C.H.Y.; Haagsman, H.P.; Colenbrander, B.; Roelen, B.A.J. Differences in early lineage segregation between mammals. Dev. Dyn. 2008, 237, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Gerri, C.; McCarthy, A.; Alanis-Lobato, G.; Demtschenko, A.; Bruneau, A.; Loubersac, S.; Fogarty, N.M.E.; Hampshire, D.; Elder, K.; Snell, P.; et al. Initiation of a conserved trophectoderm program in human, cow and mouse embryos. Nature 2020, 587, 443–447. [Google Scholar] [CrossRef]
- Velásquez, A.E.; Veraguas, D.; Cabezas, J.; Manríquez, J.; Castro, F.O.; Rodríguez-Alvarez, L.L. The expression level of SOX2 at the blastocyst stage regulates the developmental capacity of bovine embryos up to day-13 of in vitro culture. Zygote 2019, 27, 398–404. [Google Scholar] [CrossRef]
- Hall, V.J.; Ruddock, N.T.; French, A.J. Expression profiling of genes crucial for placental and preimplantation development in bovine in vivo, in vitro, and nuclear transfer blastocysts. Mol. Reprod. Dev. 2005, 72, 16–24. [Google Scholar] [CrossRef]
- Rodríguez-Alvarez, L.; Manriquez, J.; Velasquez, A.; Castro, F.O. Constitutive expression of the embryonic stem cell marker OCT4 in bovine somatic donor cells influences blastocysts rate and quality after nucleus transfer. In Vitro Cell. Dev. Biol. Anim. 2013, 49, 657–667. [Google Scholar] [CrossRef]
- Rodŕguez-Alvarez, L.; Cox, J.; Tovar, H.; Einspanier, R.; Castro, F.O. Changes in the expression of pluripotency-associated genes during preimplantation and peri-implantation stages in bovine cloned and in vitro produced embryos. Zygote 2010, 18, 269–279. [Google Scholar] [CrossRef]
- Wu, X.; Song, M.; Yang, X.; Liu, X.; Liu, K.; Jiao, C.; Wang, J.; Bai, C.; Su, G.; Liu, X.; et al. Establishment of bovine embryonic stem cells after knockdown of CDX2. Sci. Rep. 2016, 6, 28343. [Google Scholar] [CrossRef]
- Schiffmacher, A.T.; Keefer, C.L. CDX2 regulates multiple trophoblast genes in bovine trophectoderm CT-1 cells. Mol. Reprod. Dev. 2013, 80, 826–839. [Google Scholar] [CrossRef]
- Nishioka, N.; Inoue, K.-I.; Adachi, K.; Kiyonari, H.; Ota, M.; Ralston, A.; Yabuta, N.; Hirahara, S.; Stephenson, R.O.; Ogonuki, N.; et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 2009, 16, 398–410. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Kohn, M.J.; Karavanova, I.; Kaneko, K.J.; Vullhorst, D.; DePamphilis, M.L.; Buonanno, A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 2007, 134, 3827–3836. [Google Scholar] [CrossRef] [PubMed]
- Kohri, N.; Akizawa, H.; Iisaka, S.; Bai, H.; Yanagawa, Y.; Takahashi, M.; Komatsu, M.; Kawai, M.; Nagano, M.; Kawahara, M. Trophectoderm regeneration to support full-term development in the inner cell mass isolated from bovine blastocyst. J. Biol. Chem. 2019, 294, 19209–19223. [Google Scholar] [CrossRef] [PubMed]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, I.M.; Garcia-Herreros, M.; Fair, T.; Lonergan, P. Identification and regulation of glycogen synthase kinase-3 during bovine embryo development. Reproduction 2010, 140, 83–92. [Google Scholar] [CrossRef]
- Werner, M.; del Castillo, U.; Ventrella, R.; Brotslaw, E.; Mitchell, B. The small molecule AMBMP disrupts microtubule growth, ciliogenesis, cell polarity, and cell migration. Cytoskeleton 2018, 75, 450–457. [Google Scholar] [CrossRef]
- Harris, D.; Huang, B.; Oback, B. Inhibition of MAP2K and GSK3 signaling promotes bovine blastocyst development and epiblast-associated expression of pluripotency factors. Biol. Reprod. 2013, 88, 74. [Google Scholar] [CrossRef]
- Sidrat, T.; Khan, A.A.; Idrees, M.; Joo, M.D.; Xu, L.; Lee, K.L.; Kong, I.K. Role of Wnt Signaling During In-Vitro Bovine Blastocyst Development and Maturation in Synergism with PPARδ Signaling. Cells 2020, 9, 923. [Google Scholar] [CrossRef]
- Madeja, Z.E.; Warzych, E.; Pawlak, P.; Lechniak, D. Inhibitor mediated WNT and MEK/ERK signalling affects apoptosis and the expression of quality related genes in bovine in vitro obtained blastocysts. Biochem. Biophys. Res. Commun. 2019, 510, 403–408. [Google Scholar] [CrossRef]
- McLean, Z.; Meng, F.; Henderson, H.; Turner, P.; Oback, B. Increased MAP kinase inhibition enhances epiblast-specific gene expression in bovine blastocysts. Biol. Reprod. 2014, 91, 49. [Google Scholar] [CrossRef][Green Version]
- Vejlsted, M.; Avery, B.; Gjorret, J.O.; Maddox-Hyttel, P. Effect of leukemia inhibitory factor (LIF) on in vitro produced bovine embryos and their outgrowth colonies. Mol. Reprod. Dev. 2005, 70, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; De Frutos, C.; Díez, C.; Caamaño, J.N.; Facal, N.; Duque, P.; García-Ochoa, C.; Gómez, E. Effects of human versus mouse leukemia inhibitory factor on the in vitro development of bovine embryos. Theriogenology 2007, 67, 1092–1095. [Google Scholar] [CrossRef] [PubMed]
- Kocyigit, A.; Cevik, M. Leucemia inhibitory factor; investigating the time-dependent effect on viability of vitrified bovine embryos. Reprod. Domest. Anim. 2017, 52, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Denicol, A.C.; Block, J.; Kelley, D.E.; Pohler, K.G.; Dobbs, K.B.; Mortensen, C.J.; Ortega, M.S.; Hansen, P.J. The WNT signaling antagonist Dickkopf-1 directs lineage commitment and promotes survival of the preimplantation embryo. FASEB J. 2014, 28, 3975–3986. [Google Scholar] [CrossRef] [PubMed]
- Tai, C.I.; Schulze, E.N.; Ying, Q.L. Stat3 signaling regulates embryonic stem cell fate in a dose-dependent manner. Biol. Open 2014, 3, 958–965. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, C.; Han, X.; Zhou, Z.; Uyunbilig, B.; Huang, X.; Li, R.; Li, X. Wnt3a Activates the WNT-YAP/TAZ Pathway to Sustain CDX2 Expression in Bovine Trophoblast Stem Cells. DNA Cell Biol. 2019, 38, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Huang, B.; Kallingappa, P.K.; Oback, B. Dual kinase inhibition promotes pluripotency in finite bovine embryonic cell lines. Stem Cells Dev. 2013, 22, 1728–1742. [Google Scholar] [CrossRef]
- Shimada, A.; Nakano, H.; Takahashi, T.; Imai, K.; Hashizume, K. Isolation and characterization of a bovine blastocyst-derived trophoblastic cell line, BT-1: Development of a culture system in the absence of feeder cell. Placenta 2001, 22, 652–662. [Google Scholar] [CrossRef]
- Talbot, N.C.; Caperna, T.J.; Edwards, J.L.; Garrett, W.; Wells, K.D.; Ealy, A.D. Bovine blastocyst-derived trophectoderm and endoderm cell cultures: Interferon tau and transferrin expression as respective in vitro markers. Biol. Reprod. 2000, 62, 235–247. [Google Scholar] [CrossRef]
- Yang, Q.E.; Fields, S.D.; Zhang, K.; Ozawa, M.; Johnson, S.E.; Ealy, A.D. Fibroblast growth factor 2 promotes primitive endoderm development in bovine blastocyst outgrowths. Biol. Reprod. 2011, 85, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.A. The JAK/STAT Pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011205. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Han, X.; Uyunbilig, B.; Zhang, M.; Duo, S.; Zuo, Y.; Zhao, Y.; Yun, T.; Tai, D.; Wang, C.; et al. Establishment of bovine trophoblast stem-like cells from in vitro-produced blastocyst-stage embryos using two inhibitors. Stem Cells Dev. 2014, 23, 1501–1514. [Google Scholar] [CrossRef] [PubMed]
- Madsen, R.R. PI3K in stemness regulation: From development to cancer. Biochem. Soc. Trans. 2020, 48, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Sirisathien, S.; Hernandez-Fonseca, H.J.; Bosch, P.; Hollet, B.R.; Lott, J.D.; Brackett, B.G. Effect of leukemia inhibitory factor on bovine embryos produced in vitro under chemically defined conditions. Theriogenology 2003, 59, 1751–1763. [Google Scholar] [CrossRef]
- Van Wagtendonk-De Leeuw, A.M. Ovum pick up and in vitro production in the bovine after use in several generations: A 2005 status. Theriogenology 2006, 65, 914–925. [Google Scholar] [CrossRef] [PubMed]
- Lonergan, P.; Fair, T. In vitro-produced bovine embryos: Dealing with the warts. Theriogenology 2008, 69, 17–22. [Google Scholar] [CrossRef]
- Tríbulo, P.; Bernal Ballesteros, B.H.; Ruiz, A.; Tríbulo, A.; Tríbulo, R.J.; Tríbulo, H.E.; Bo, G.A.; Hansen, P.J. Consequences of exposure of embryos produced in vitro in a serum-containing medium to dickkopf-related protein 1 and colony stimulating factor 2 on blastocyst yield, pregnancy rate, and birth weight. J. Anim. Sci. 2017, 95, 4407–4412. [Google Scholar] [CrossRef]
- Hansen, P.J.; Dobbs, K.B.; Denicol, A.C. Programming of the preimplantation embryo by the embryokine colony stimulating factor 2. Anim. Reprod. Sci. 2014, 149, 59–66. [Google Scholar] [CrossRef]
- Ealy, A.D.; Speckhart, S.L.; Wooldridge, L.K. Cytokines That Serve as Embryokines in Cattle. Animals 2021, 11, 2313. [Google Scholar] [CrossRef]
- Navarro, M.; Soto, D.A.; Pinzon, C.A.; Wu, J.; Ross, P.J. Livestock pluripotency is finally captured in vitro. Reprod. Fertil. Dev. 2019, 32, 11–39. [Google Scholar] [CrossRef]
- Pillai, V.V.; Koganti, P.P.; Kei, T.G.; Gurung, S.; Butler, W.R.; Selvaraj, V. Efficient induction and sustenance of pluripotent stem cells from bovine somatic cells. Biol. Open 2021, 10, bio058756. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Wang, L.; Fan, Z.; Liu, Y.; Zhu, J.; Kaback, D.; Oudiz, J.; Patrick, T.; Yee, S.P.; Tian, X.; et al. Establishment of Bovine-Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2021, 22, 10489. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Kobayashi, T.; Planells, B.; Klisch, D.; Spindlow, D.; Masaki, H.; Bornelöv, S.; Stirparo, G.G.; Matsunari, H.; Uchikura, A.; et al. Pluripotent stem cells related to embryonic disc exhibit common self-renewal requirements in diverse livestock species. Development 2021, 148, dev199901. [Google Scholar] [CrossRef] [PubMed]
- Soto, D.A.; Navarro, M.; Zheng, C.; Halstead, M.M.; Zhou, C.; Guiltinan, C.; Wu, J.; Ross, P.J. Simplification of culture conditions and feeder-free expansion of bovine embryonic stem cells. Sci. Rep. 2021, 11, 11045. [Google Scholar] [CrossRef] [PubMed]
- Vallier, L.; Mendjan, S.; Brown, S.; Ching, Z.; Teo, A.; Smithers, L.E.; Trotter, M.W.B.; Cho, C.H.H.; Martinez, A.; Rugg-Gunn, P.; et al. Activin/Nodal signalling maintains pluripotency by controlling NANOG expression. Development 2009, 136, 1339–1349. [Google Scholar] [CrossRef]
- Xiao, Y.; Sosa, F.; Ross, P.J.; Diffenderfer, K.E.; Hansen, P.J. Regulation of NANOG and SOX2 expression by activin A and a canonical WNT agonist in bovine embryonic stem cells and blastocysts. Biol. Open 2021, 10, bio058669. [Google Scholar] [CrossRef]
- Kim, D.; Roh, S. Strategy to Establish Embryo-Derived Pluripotent Stem Cells in Cattle. Int. J. Mol. Sci. 2021, 22, 5011. [Google Scholar] [CrossRef]
- Owen, J.R.; Hennig, S.L.; McNabb, B.R.; Mansour, T.A.; Smith, J.M.; Lin, J.C.; Young, A.E.; Trott, J.F.; Murray, J.D.; Delany, M.E.; et al. One-step generation of a targeted knock-in calf using the CRISPR-Cas9 system in bovine zygotes. BMC Genomics 2021, 22, 118. [Google Scholar] [CrossRef]
- Savy, V.; Alberio, V.; Canel, N.G.; Ratner, L.D.; Gismondi, M.I.; Ferraris, S.F.; Fernandez-Martín, R.; Knott, J.G.; Bevacqua, R.J.; Salamone, D.F. CRISPR-on for activation of endogenous SMARCA4 and TFAP2C expression in bovine embryos. Reproduction 2020, 159, 767–778. [Google Scholar] [CrossRef]
- Takahashi, K.; Ross, P.J.; Sawai, K. The necessity of ZSCAN4 for preimplantation development and gene expression of bovine embryos. J. Reprod. Dev. 2019, 65, 319–326. [Google Scholar] [CrossRef]
- Weiss, W.A.; Taylor, S.S.; Shokat, K.M. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat. Chem. Biol. 2007, 3, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Jafarpour, F.; Zadegan, F.G.; Ostadhosseini, S.; Hajian, M.; Kiani-Esfahani, A.; Nasr-Esfahani, M.H. siRNA inhibition and not chemical inhibition of Suv39h1/2 enhances pre-implantation embryonic development of bovine somatic cell nuclear transfer embryos. PLoS ONE 2020, 15, e0233880. [Google Scholar] [CrossRef] [PubMed]
- Jabbar, A.; Zulfiqar, F.; Mahnoor, M.; Mushtaq, N.; Zaman, M.H.; Din, A.S.U.; Khan, M.A.; Ahmad, H.I. Advances and Perspectives in the Application of CRISPR-Cas9 in Livestock. Mol. Biotechnol. 2021, 63, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Serna, S.; Vilarino, M.; Park, I.; Gadea, J.; Ross, P.J. Livestock Gene Editing by One-step Embryo Manipulation. J. Equine Vet. Sci. 2020, 89, 103025. [Google Scholar] [CrossRef]
- Xiang, J.; Wang, H.; Zhang, Y.; Wang, J.; Liu, F.; Han, X.; Lu, Z.; Li, C.; Li, Z.; Gao, Y.; et al. LCDM medium supports the derivation of bovine extended pluripotent stem cells with embryonic and extraembryonic potency in bovine-mouse chimeras from iPSCs and bovine fetal fibroblasts. FEBS J. 2021, 288, 4394–4411. [Google Scholar] [CrossRef]
- Sagi, I.; De Pinho, J.C.; Zuccaro, M.V.; Atzmon, C.; Golan-Lev, T.; Yanuka, O.; Prosser, R.; Sadowy, A.; Perez, G.; Cabral, T.; et al. Distinct Imprinting Signatures and Biased Differentiation of Human Androgenetic and Parthenogenetic Embryonic Stem Cells. Cell Stem Cell 2019, 25, 419–432.e9. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).