Genetic Diversity and Trends of Ancestral and New Inbreeding in Deutsch Drahthaar Assessed by Pedigree Data
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Küßner, M. Zur Entwicklung und Bedeutung der Jagd zwischen Thüringer Wald und Harz in der Steinzeit-von den Anfängen bis zum Ende des Neolithikums. Siedl. Archäologie-Gesch.-Geogr. 2015, 32, 161–184. [Google Scholar]
- Deutscher Jagdverband e.V. Natur- und Artenschutz. Available online: https://www.jagdverband.de/rund-um-die-jagd/natur-und-artenschutz (accessed on 7 February 2021).
- Statistic of Born Dogs in VDH. Available online: https://www.vdh.de/ueber-den-vdh/welpenstatistik/ (accessed on 7 February 2021).
- Exhaustive Study of the History of the Deutsch Drahthaar. Available online: https://www.vdd-gna.org/exhaustive-study-of-the-history-of-the-deutsch-drahthaar/ (accessed on 8 February 2021).
- The History of the Wirehaired Pointing Griffon and Eduard Korthals. Available online: https://projectupland.com/hunting-dogs/the-history-of-the-wirehaired-pointing-griffon-and-eduard-korthals-2/ (accessed on 9 March 2021).
- Breeding Standard Deutsch Drahthaar. Available online: http://www.fci.be/de/nomenclature/DEUTSCH-DRAHTHAAR-98.html (accessed on 7 February 2021).
- Der Verein—Die Entstehungsgeschichte. Available online: https://drahthaar.de/pages/der-verein.php (accessed on 8 February 2021).
- Fédération Cynologique Internationale Breeds Nomenclature Deutsch Drahthaar. Available online: http://www.fci.be/en/nomenclature/GERMAN-WIRE-HAIRED-POINTING-DOG-98.html (accessed on 7 February 2021).
- History of the German Wirehaired Pointer. Available online: http://www.vdd-gna.org/history-of-the-german-wirehaired-pointer/ (accessed on 9 February 2021).
- Ubbink, G.; Knol, B.; Bouw, J. The relationship between homozygosity and the occurrence of specific diseases in Bouvier Belge des Flandres dogs in the Netherlands: Inbreeding and disease in the bouvier dog. Vet. Q. 1992, 14, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Leroy, G.; Phocas, F.; Hedan, B.; Verrier, E.; Rognon, X. Inbreeding impact on litter size and survival in selected canine breeds. Vet. J. 2015, 203, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Leroy, G.; Verrier, E.; Meriaux, J.-C.; Rognon, X. Genetic diversity of dog breeds: Within-breed diversity comparing genealogical and molecular data. Anim. Genet. 2009, 40, 323–332. [Google Scholar] [CrossRef]
- Shariflou, M.R.; James, J.W.; Nicholas, F.W.; Wade, C.M. A genealogical survey of Australian registered dog breeds. Vet. J. 2011, 189, 203–210. [Google Scholar] [CrossRef]
- Calboli, F.C.; Sampson, J.; Fretwell, N.; Balding, D.J. Population structure and inbreeding from pedigree analysis of purebred dogs. Genet. Soc. Am. 2008, 179, 593–601. [Google Scholar] [CrossRef] [Green Version]
- Machová, K.; Kranjčevičová, A.; Vostrý, L.; Krupa, E. Analysis of Genetic Diversity in the Czech Spotted Dog. Animals 2020, 10, 1416. [Google Scholar] [CrossRef]
- Mäki, K. Population structure and genetic diversity of worldwide Nova Scotia Duck Tolling Retriever and Lancashire Heeler dog populations. J. Anim. Breed. Genet. 2010, 127, 318–326. [Google Scholar] [CrossRef]
- Cole, J.; Franke, D.; Leighton, E. Population structure of a colony of dog guides. J. Anim. Sci. 2004, 82, 2906–2912. [Google Scholar] [CrossRef] [Green Version]
- Michels, P.W.; Distl, O. Genetic Variability in Polish Lowland Sheepdogs Assessed by Pedigree and Genomic Data. Animals 2020, 10, 1520. [Google Scholar] [CrossRef]
- Pfahler, S.; Distl, O. Effective Population Size, Extended Linkage Disequilibrium and Signatures of Selection in the Rare Dog Breed Lundehund. PLoS ONE 2015, 10, e0122680. [Google Scholar] [CrossRef] [PubMed]
- Goleman, M.; Balicki, I.; Radko, A.; Jakubczak, A.; Fornal, A. Genetic diversity of the Polish Hunting Dog population based on pedigree analyses and molecular studies. Livest. Sci. 2019, 229, 114–117. [Google Scholar] [CrossRef]
- Goleman, M.; Balicki, I.; Radko, A.; Rozempolska-Rucińska, I.; Zięba, G. Pedigree and Molecular Analyses in the Assessment of Genetic Variability of the Polish Greyhound. Animals 2021, 11, 353. [Google Scholar] [CrossRef] [PubMed]
- Ács, V.; Bokor, Á.; Nagy, I. Population structure analysis of the border collie dog breed in Hungary. Animals 2019, 9, 250. [Google Scholar] [CrossRef] [Green Version]
- Boccardo, A.; Marelli, S.P.; Pravettoni, D.; Bagnato, A.; Strillacci, M.G. The German Shorthair Pointer Dog Breed (Canis lupus familiaris): Genomic Inbreeding and Variability. Animals 2020, 10, 498. [Google Scholar] [CrossRef] [Green Version]
- Ballou, J. Ancestral inbreeding only minimally affects inbreeding depression in mammalian populations. J. Hered. 1997, 88, 169–178. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Hedrick, P.W.; Miller, P.S. Inbreeding depression in the Speke’s gazelle captive breeding program. Conserv. Biol. 2000, 14, 1375–1384. [Google Scholar] [CrossRef]
- Baumung, R.; Farkas, J.; Boichard, D.; Mészáros, G.; Sölkner, J.; Curik, I. GRAIN: A computer program to calculate ancestral and partial inbreeding coefficients using a gene dropping approach. J. Anim. Breed. Genet. 2015, 132, 100–108. [Google Scholar] [CrossRef]
- Boichard, D.; Maignel, L.; Verrier, E. The value of using probabilities of gene origin to measure genetic variability in a population. Genet. Sel. Evol. 1997, 29, 5. [Google Scholar] [CrossRef]
- Lacy, R.C. Analysis of founder representation in pedigrees: Founder equivalents and founder genome equivalents. Zoo Biol. 1989, 8, 111–123. [Google Scholar] [CrossRef]
- Boichard, D. PEDIG: A fortran package for pedigree analysis suited for large populations. In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002; pp. 525–528. [Google Scholar]
- Maignel, L.; Boichard, D.; Verrier, E. Genetic variability of French dairy breeds estimated from pedigree information. Interbull Bull. 1996, 14, 49. [Google Scholar]
- Melka, M.; Stachowicz, K.; Miglior, F.; Schenkel, F. Analyses of genetic diversity in five C anadian dairy breeds using pedigree data. J. Anim. Breed. Genet. 2013, 130, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.; Luo, Z. Computing inbreeding coefficients in large populations. Genet. Sel. Evol. 1992, 24, 305–313. [Google Scholar] [CrossRef]
- Doekes, H.P.; Curik, I.; Nagy, I.; Farkas, J.; Kövér, G.; Windig, J.J. Revised calculation of Kalinowski’s ancestral and new inbreeding coefficients. Diversity 2020, 12, 155. [Google Scholar] [CrossRef] [Green Version]
- Caballero, A.; Toro, M.A. Interrelations between effective population size and other pedigree tools for the management of conserved populations. Genet. Res. 2000, 75, 331–343. [Google Scholar] [CrossRef]
- Gutiérrez, J.; Cervantes, I.; Goyache, F. Improving the estimation of realized effective population sizes in farm animals. J. Anim. Breed. Genet. 2009, 126, 327–332. [Google Scholar] [CrossRef]
- Cervantes, I.; Goyache, F.; Molina, A.; Valera, M.; Gutiérrez, J. Application of individual increase in inbreeding to estimate realized effective sizes from real pedigrees. J. Anim. Breed. Genet. 2008, 125, 301–310. [Google Scholar] [CrossRef] [Green Version]
- Leroy, G.; Baumung, R. Mating practices and the dissemination of genetic disorders in domestic animals, based on the example of dog breeding. Anim. Genet. 2011, 42, 66–74. [Google Scholar] [CrossRef]
- Cassell, B.; Adamec, V.; Pearson, R. Effect of incomplete pedigrees on estimates of inbreeding and inbreeding depression for days to first service and summit milk yield in Holsteins and Jerseys. J. Dairy Sci. 2003, 86, 2967–2976. [Google Scholar] [CrossRef]
- Lutaaya, B.E.; Misztal, I.; Bertrand, J.; Mabry, J. Inbreeding in populations with incomplete pedigrees. J. Anim. Breed. Genet. 1999, 116, 475–480. [Google Scholar] [CrossRef]
- Jansson, M.; Laikre, L. Recent breeding history of dog breeds in Sweden: Modest rates of inbreeding, extensive loss of genetic diversity and lack of correlation between inbreeding and health. J. Anim. Breed. Genet. 2014, 131, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Vasiliadis, D.; Metzger, J.; Distl, O. Demographic assessment of the Dalmatian dog-effective population size, linkage disequilibrium and inbreeding coefficients. Canine Med. Genet. 2020, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Bijma, P. Long-Term Genetic Contributions: Prediction of Rates of Inbreeding and Genetic Gain in Selected Populations; Wageningen University and Research: Wageningen, The Netherlands, 2000. [Google Scholar]
- Simon, D.L.; Buchenauer, D. Genetic Diversity of European Livestock Breeds; Wageningen Pers: Wageningen, The Netherlands, 1993. [Google Scholar]
- Wijnrocx, K.; François, L.; Stinckens, A.; Janssens, S.; Buys, N. Half of 23 Belgian dog breeds has a compromised genetic diversity, as revealed by genealogical and molecular data analysis. J. Anim. Breed. Genet. 2016, 133, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Velie, B.D.; Wilson, B.J.; Arnott, E.R.; Early, J.B.; McGreevy, P.D.; Wade, C.M. Inbreeding levels in an open-registry pedigreed dog breed: The Australian working kelpie. Vet. J. 2021, 269, 105609. [Google Scholar] [CrossRef]
- Voges, S.; Distl, O. Inbreeding trends and pedigree analysis of Bavarian mountain hounds, Hanoverian hounds and Tyrolean hounds. J. Anim. Breed. Genet. 2009, 126, 357–365. [Google Scholar] [CrossRef]
- Kumpulainen, M.; Anderson, H.; Svevar, T.; Kangasvuo, I.; Donner, J.; Pohjoismäki, J. Founder representation and effective population size in old versus young breeds—Genetic diversity of Finnish and Nordic Spitz. J. Anim. Breed. Genet. 2017, 134, 422–433. [Google Scholar] [CrossRef] [Green Version]
- Doekes, H.P.; Veerkamp, R.F.; Bijma, P.; de Jong, G.; Hiemstra, S.J.; Windig, J.J. Inbreeding depression due to recent and ancient inbreeding in Dutch Holstein-Friesian dairy cattle. Genet. Sel. Evol. 2019, 51, 54. [Google Scholar] [CrossRef] [Green Version]
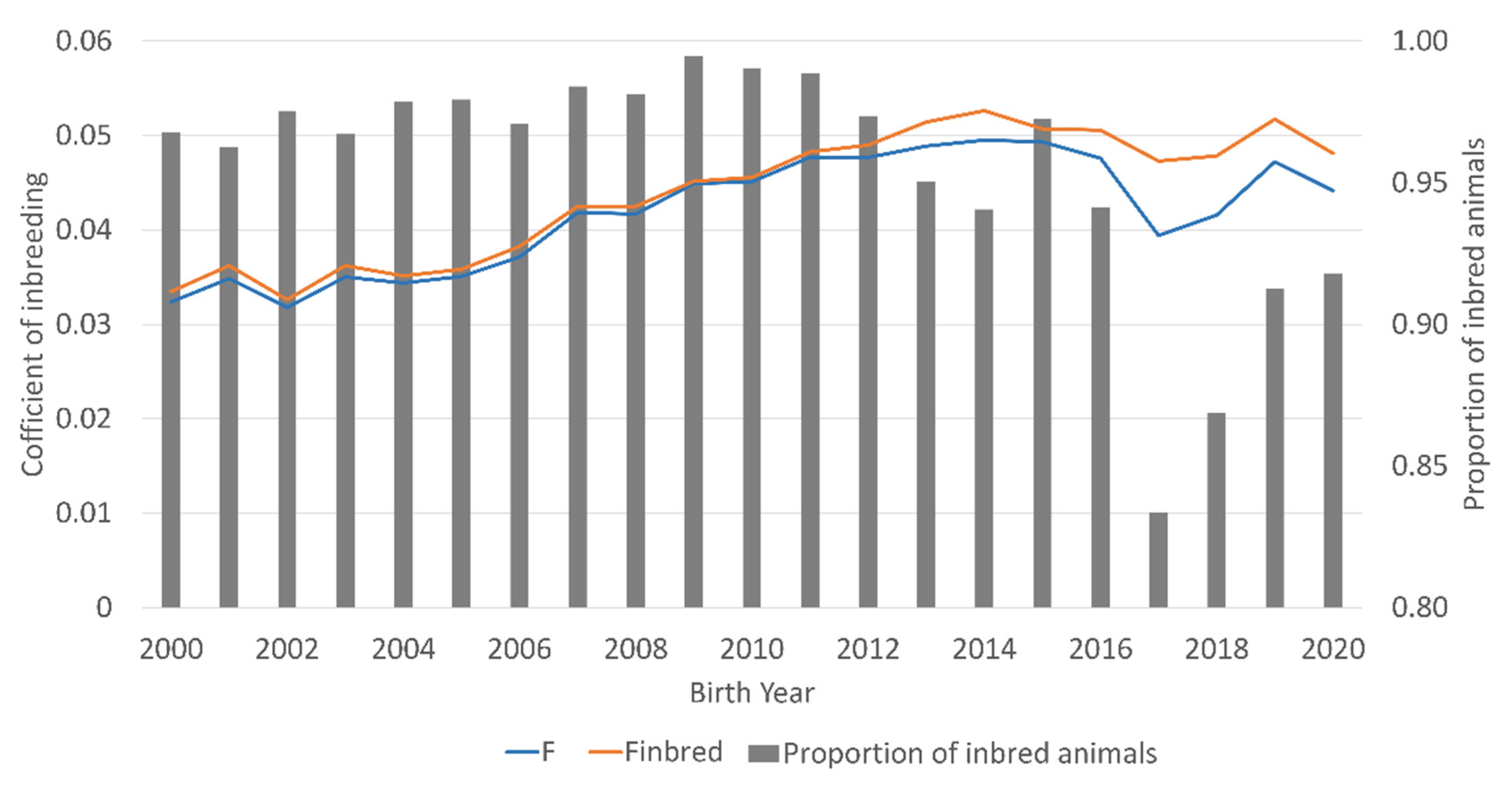

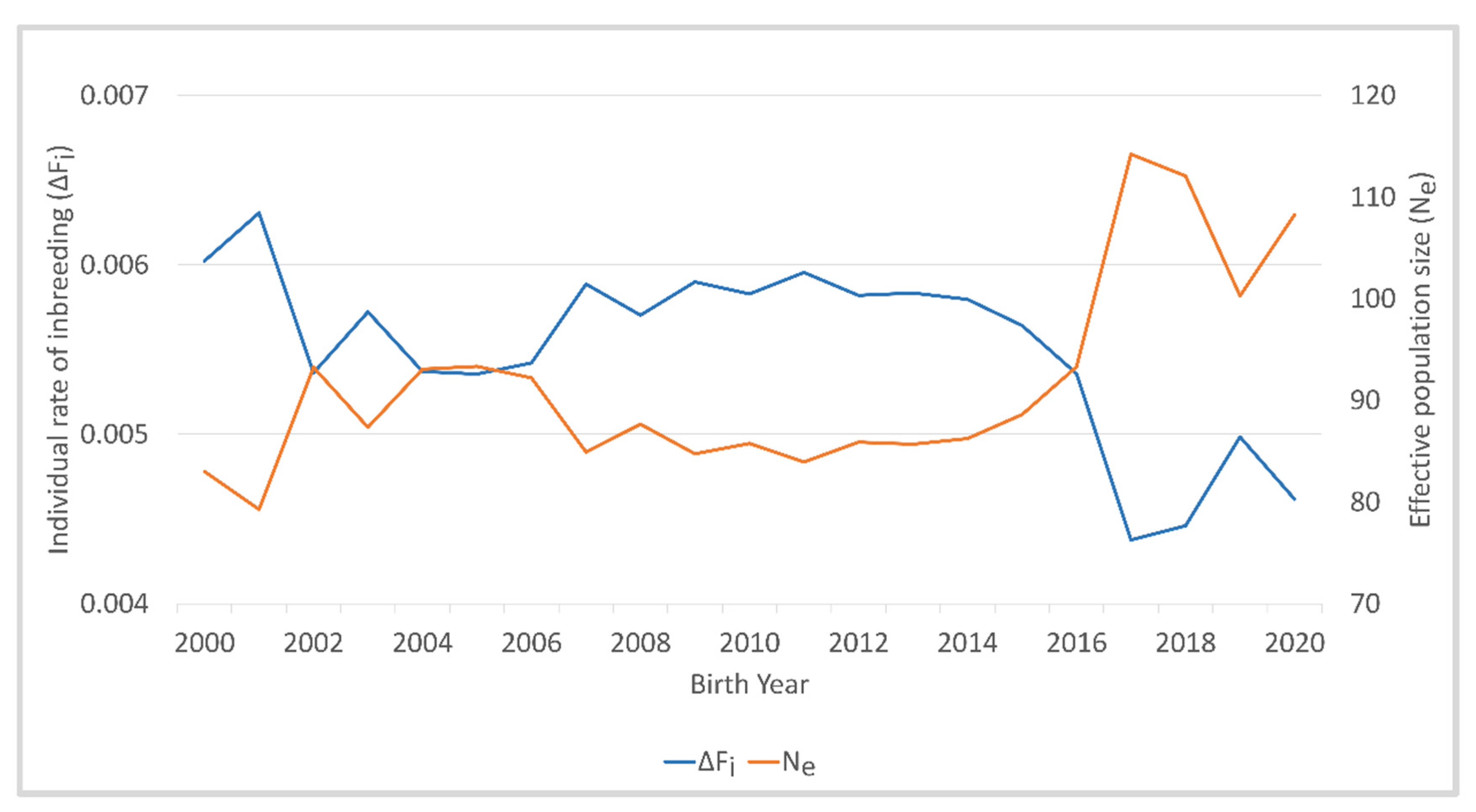
| Parameter | Reference Population from 2000 to 2020 |
|---|---|
| Reference population | 65,927 |
| Inbred animals in reference population | 62,984 |
| Mean equivalent generations (GE) | 8.62 |
| Mean generation interval in years | 4.42 |
| Number of founders (f) | 814 |
| Effective number of founders (fe) | 65.5 |
| Effective number of ancestors (fa) | 37.8 |
| Effective number of founder genomes (fg) | 16.2 |
| fe/f | 0.08 |
| fa/fe | 0.58 |
| fg/fe | 0.25 |
| Ancestors explaining 30% of the gene pool | 5 |
| Ancestors explaining 40% of the gene pool | 8 |
| Ancestors explaining 50% of the gene pool | 13 |
| Ancestors explaining 60% of the gene pool | 19 |
| Ancestors explaining 70% of the gene pool | 29 |
| Ancestors explaining 80% of the gene pool | 49 |
| Ancestors explaining 90% of the gene pool | 110 |
| Ancestors explaining 95% of the gene pool | 177 |
| Effective population size (Ne) | 91.6 |
| ΔFi | 0.00551 |
| ID | Birth Year | Sex | Marginal Contribution | Cumulated Marginal Contributions | Number of Progeny |
|---|---|---|---|---|---|
| 112374 | 1980 | male | 0.0846 | 0.0846 | 117 |
| 144135 | 1989 | male | 0.0577 | 0.1422 | 218 |
| 70425 | 1969 | female | 0.0573 | 0.1995 | 16 |
| 87288 | 1973 | female | 0.0407 | 0.2402 | 28 |
| 145543 | 1989 | female | 0.0362 | 0.2765 | 34 |
| 106677 | 1978 | female | 0.0358 | 0.3123 | 8 |
| 134384 | 1986 | male | 0.0350 | 0.3473 | 155 |
| 129824 | 1984 | female | 0.0286 | 0.3759 | 46 |
| 90647 | 1974 | female | 0.0272 | 0.4031 | 7 |
| 79470 | 1971 | male | 0.0266 | 0.4296 | 10 |
| 110712 | 1979 | male | 0.0243 | 0.4539 | 52 |
| 90556 | 1974 | male | 0.0237 | 0.4776 | 11 |
| 109594 | 1979 | male | 0.0213 | 0.4989 | 7 |
| 105465 | 1978 | male | 0.0206 | 0.5195 | 21 |
| 94096 | 1975 | male | 0.0172 | 0.5367 | 8 |
| Method | Inbreeding Coefficients |
|---|---|
| F | 0.042 |
| Finbred | 0.044 |
| Fgd | 0.042 |
| Fa_Bal | 0.145 |
| Fa_Kal | 0.016 |
| FNew | 0.026 |
| AHC | 0.168 |
| Inbreeding Coefficients | F | Fgd | Fa_Bal | Fa_Kal | FNew | AHC |
|---|---|---|---|---|---|---|
| F | 1.00 | 0.41 | 0.87 | 0.95 | 0.40 | |
| Fgd | 0.41 | 0.87 | 0.95 | 0.40 | ||
| Fa_Bal | 0.68 | 0.18 | 1.00 | |||
| Fa_Kal | 0.67 | 0.68 | ||||
| FNew | 0.16 |
| Population | Average Coancestry within Parents (Φ) | F | Deviation from Random Mating (α) | Ne | Nec |
|---|---|---|---|---|---|
| Reference | 0.027 | 0.042 | 0.01528 | 92 | 139 |
| 2000–2005 | 0.021 | 0.034 | 0.01328 | 88 | 143 |
| 2006–2010 | 0.029 | 0.042 | 0.01339 | 87 | 127 |
| 2011–2015 | 0.036 | 0.049 | 0.01345 | 86 | 115 |
| 2016–2020 | 0.038 | 0.044 | 0.00624 | 105 | 116 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michels, P.W.; Distl, O. Genetic Diversity and Trends of Ancestral and New Inbreeding in Deutsch Drahthaar Assessed by Pedigree Data. Animals 2022, 12, 929. https://doi.org/10.3390/ani12070929
Michels PW, Distl O. Genetic Diversity and Trends of Ancestral and New Inbreeding in Deutsch Drahthaar Assessed by Pedigree Data. Animals. 2022; 12(7):929. https://doi.org/10.3390/ani12070929
Chicago/Turabian StyleMichels, Paula Wiebke, and Ottmar Distl. 2022. "Genetic Diversity and Trends of Ancestral and New Inbreeding in Deutsch Drahthaar Assessed by Pedigree Data" Animals 12, no. 7: 929. https://doi.org/10.3390/ani12070929
APA StyleMichels, P. W., & Distl, O. (2022). Genetic Diversity and Trends of Ancestral and New Inbreeding in Deutsch Drahthaar Assessed by Pedigree Data. Animals, 12(7), 929. https://doi.org/10.3390/ani12070929






