Molecular Detection and Genetic Characterization of Porcine Circovirus 2 (PCV-2) in Black-Backed Jackal (Lupulella mesomelas) in Namibia
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
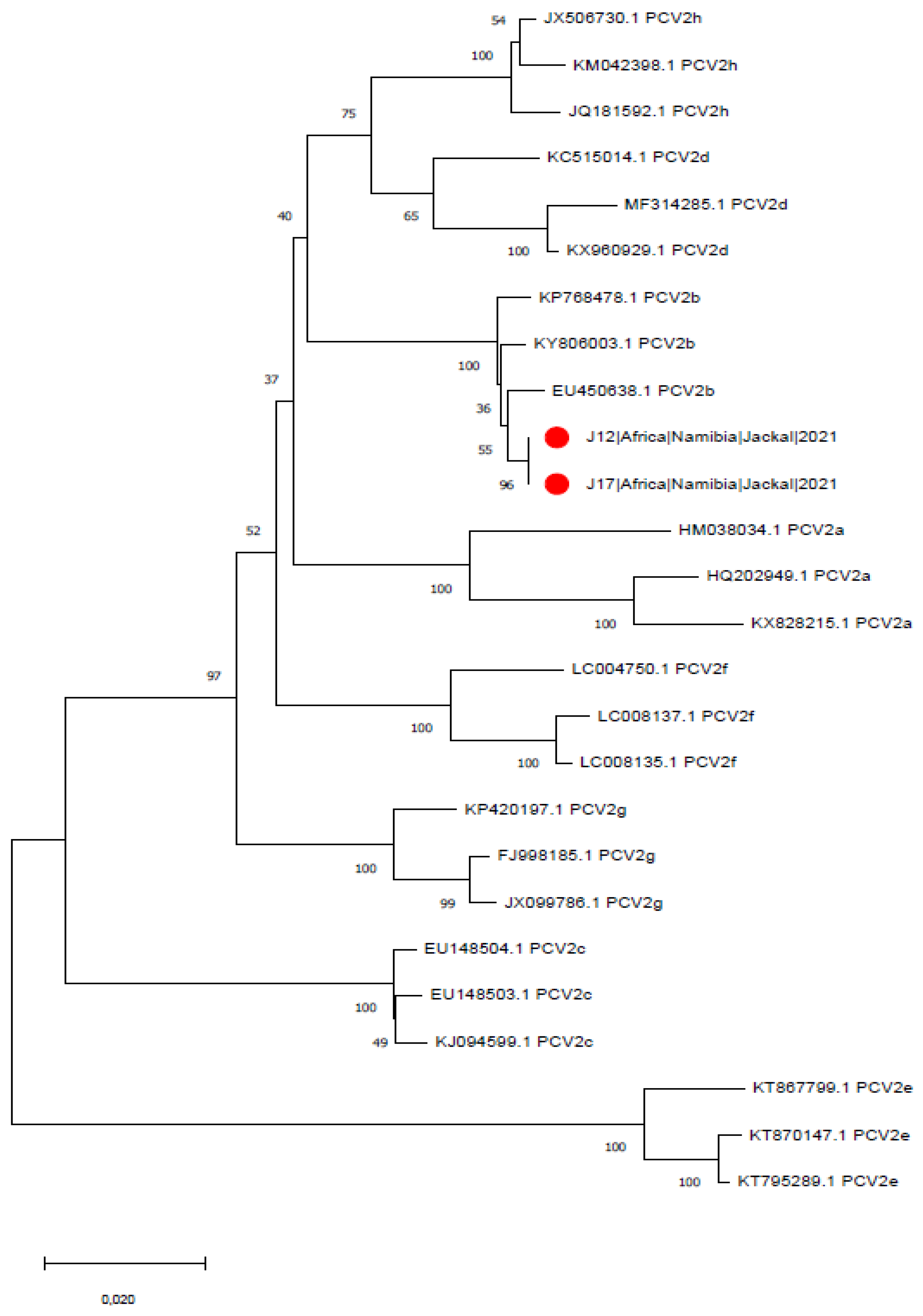
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lv, Q.; Guo, K.; Zhang, Y. Current Understanding of Genomic DNA of Porcine Circovirus Type 2. Virus Genes 2014, 49, 1–10. [Google Scholar] [CrossRef]
- Huang, L.P.; Lu, Y.H.; Wei, Y.W.; Guo, L.J.; Liu, C.M. Identification of One Critical Amino Acid That Determines a Conformational Neutralizing Epitope in the Capsid Protein of Porcine Circovirus Type 2. BMC Microbiol. 2011, 11, 188. [Google Scholar] [CrossRef] [PubMed]
- Kekarainen, T.; Segalés, J. Porcine Circovirus 2 Immunology and Viral Evolution. Porc. Health Manag. 2015, 1, 17. [Google Scholar] [CrossRef] [PubMed]
- Dhindwal, S.; Avila, B.; Feng, S.; Khayat, R. Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites to Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid. J. Virol. 2019, 93, e02222-18. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Cortey, M.; Segalés, J.; Hughes, J.; Drigo, M. Phylodynamic Analysis of Porcine Circovirus Type 2 Reveals Global Waves of Emerging Genotypes and the Circulation of Recombinant Forms. Mol. Phylogenetics Evol. 2016, 100, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Segalés, J. Joaquim Porcine Circovirus 2 (PCV-2) Genotype Update and Proposal of a New Genotyping Methodology. PLoS ONE 2018, 13, e0208585. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.-Y.; Choi, Y.-C.; Park, I.-B.; Lee, C.-H.; Kang, S.-J.; Chun, T. The ORF5 Protein of Porcine Circovirus Type 2 Enhances Viral Replication by Dampening Type I Interferon Expression in Porcine Epithelial Cells. Vet. Microbiol. 2018, 226, 50–58. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Cao, J.; Zhou, N.; Jin, Y.; Wu, J.; Zhou, J. Identification and Functional Analysis of the Novel ORF4 Protein Encoded by Porcine Circovirus Type 2. J. Virol. 2013, 87, 1420–1429. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, J.; Xu, S.; Cai, S.; Ao, C.; Fang, L.; Xiao, S.; Chen, H.; Jiang, Y. Identification and Functional Analysis of the Novel ORF6 Protein of Porcine Circovirus Type 2 in Vitro. Vet. Res. Commun. 2018, 42, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, Y.; Chen, I.; Lau, J.; He, F.; Lau, A.; Wang, Z.; Karuppannan, A.K.; Kwang, J. The ORF3 Protein of Porcine Circovirus Type 2 Interacts with Porcine Ubiquitin E3 Ligase Pirh2 and Facilitates P53 Expression in Viral Infection. J. Virol. 2007, 81, 9560–9567. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, P.; Rushton, J.; Wieland, B. Cost of Post-Weaning Multi-Systemic Wasting Syndrome and Porcine Circovirus Type-2 Subclinical Infection in England–An Economic Disease Model. Prev. Vet. Med. 2013, 110, 88–102. [Google Scholar] [CrossRef]
- Castro, A.M.M.G.; Castro, J.G.; Budiño, F.E.L.; Baldin, C.M.; Silva, S.O.S.; Brandão, P.E.; Richtzenhain, L.J. Detection of Genetic Characterization of Porcine Circovirus 2 (PCV2) in Brazilian Wildlife Boars. Braz. J. Microbiol. 2012, 43, 1022–1025. [Google Scholar] [CrossRef][Green Version]
- Hälli, O.; Ala-Kurikka, E.; Nokireki, T.; Skrzypczak, T.; Raunio-Saarnisto, M.; Peltoniemi, O.A.T.; Heinonen, M. Prevalence of and Risk Factors Associated with Viral and Bacterial Pathogens in Farmed European Wild Boar. Vet. J. 2012, 194, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Turcitu, M.A.; Wellenberg, G.J.; Barboi, G.; Codreanu, M.D.; Vuta, V.B.; Nicolae, S.; Barbuceanu, F.; Coste, H.; Cioranu, R. Genetic Diversity of Porcine Circovirus Type 2 (PCV2) in the Romanian Wild Boar Population. Res. Vet. Sci. 2011, 91, e103–e106. [Google Scholar] [CrossRef]
- Cságola, A.; Cadar, D.; Tuboly, T. Replication and Transmission of Porcine Circovirus Type 2 in Mice. Acta Vet. Hung. 2008, 56, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yuan, X.; Zhang, C.; Miao, L.; Wu, J.; Shi, J.; Xu, S.; Cui, S.; Wang, J.; Ai, H. A Mouse Model to Study Infection against Porcine Circovirus Type 2: Viral Distribution and Lesions in Mouse. Virol. J. 2010, 7, 158. [Google Scholar] [CrossRef]
- Halami, M.Y.; Freick, M.; Shehata, A.A.; Müller, H.; Vahlenkamp, T.W. Susceptibility of Calves to Porcine Circovirus-2 (PCV2). Vet. Microbiol. 2014, 173, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Kappe, E.C.; Halami, M.Y.; Schade, B.; Alex, M.; Hoffmann, D.; Gangl, A.; Meyer, K.; Dekant, W.; Schwarz, B.; Johne, R.; et al. Bone Marrow Depletion with Haemorrhagic Diathesis in Calves in Germany: Characterization of the Disease and Preliminary Investigations on Its Aetiology. Berl. Munch. Tierarztl. Wochenschr. 2010, 123, 31–41. [Google Scholar] [CrossRef]
- Nayar, G.P.; Hamel, A.L.; Lin, L.; Sachvie, C.; Grudeski, E.; Spearman, G. Evidence for Circovirus in Cattle with Respiratory Disease and from Aborted Bovine Fetuses. Can. Vet. J. 1999, 40, 277. [Google Scholar]
- Lorincz, M.; Cságola, A.; Biksi, I.; Szeredi, L.; Dán, Á.; Tuboly, T. Detection of Porcine Circovirus in Rodents–Short Communication. Acta Vet. Hung. 2010, 58, 265–268. [Google Scholar] [CrossRef]
- Pinheiro, A.L.B.C.; Bulos, L.H.S.; Onofre, T.S.; de Paula Gabardo, M.; de Carvalho, O.V.; Fausto, M.C.; Guedes, R.M.C.; de Almeida, M.R.; Silva, A. Verification of Natural Infection of Peridomestic Rodents by PCV2 on Commercial Swine Farms. Res. Vet. Sci. 2013, 94, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Truong, Q.L.; Seo, T.W.; Yoon, B.I.; Kim, H.C.; Han, J.H.; Hahn, T.W. Prevalence of Swine Viral and Bacterial Pathogens in Rodents and Stray Cats Captured around Pig Farms in Korea. J. Vet. Med. Sci. 2013, 75, 1647–1650. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Hao, J.; Zhang, R.; Tang, M.; Li, W.; Hui, W.; Fu, Q.; Wang, C.; Xin, S.; Zhang, S.; et al. First Detection and Phylogenetic Analysis of Porcine Circovirus Type 2 in Raccoon Dogs. BMC Vet. Res. 2019, 15, 107. [Google Scholar] [CrossRef]
- Song, T.; Zhang, S.; Hao, J.; Xin, S.; Hui, W.; Tang, M.; Li, W.; Tian, R.; Liu, X.; Rui, P.; et al. First Detection and Genetic Analysis of Fox-Origin Porcine Circovirus Type 2. Transbound. Emerg. Dis. 2019, 66, 1–6. [Google Scholar] [CrossRef]
- Wang, G.S.; Sun, N.; Tian, F.L.; Wen, Y.J.; Xu, C.; Li, J.; Chen, Q.; Wang, J.B. Genetic Analysis of Porcine Circovirus Type 2 from Dead Minks. J. Gen. Virol. 2016, 97, 2316–2322. [Google Scholar] [CrossRef] [PubMed]
- Molini, U.; Franzo, G.; Gous, L.; Moller, S.; Hemberger, Y.M.; Chiwome, B.; Marruchella, G.; Khaiseb, S.; Cattoli, G.; Dundon, W.G. Three Different Genotypes of Porcine Circovirus 2 (PCV-2) Identified in Pigs and Warthogs in Namibia. Arch. Virol. 2021, 166, 1723–1728. [Google Scholar] [CrossRef] [PubMed]
- Molini, U.; Coetzee, L.M.; Hemberger, M.Y.; Khaiseb, S.; Cattoli, G.; Dundon, W.G.; Franzo, G. The Oryx Antelope (Oryx gazella): An Unexpected Host for Porcine Circovirus-2 (PCV-2). Pathogens 2021, 10, 1402. [Google Scholar] [CrossRef] [PubMed]
- An, D.J.; Roh, I.S.; Song, D.S.; Park, C.K.; Park, B.K. Phylogenetic Characterization of Porcine Circovirus Type 2 in PMWS and PDNS Korean Pigs between 1999 and 2006. Virus Res. 2007, 129, 115–122. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple Alignment of Nucleotide Sequences Guided by Amino Acid Translations. Nucleic Acids Res. 2010, 38, W7–W13. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Franzo, G.; Segalés, J. Circoviruses (Circoviridae). In Encyclopedia of Virology; Bamford, D.H., Zuckerman, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 182–192. ISBN 9780128145166. [Google Scholar]
- Zhai, S.-L.; Lu, S.-S.; Wei, W.-K.; Lv, D.-H.; Wen, X.-H.; Zhai, Q.; Chen, Q.-L.; Sun, Y.-W.; Xi, Y. Reservoirs of Porcine Circoviruses: A Mini Review. Front. Vet. Sci. 2019, 6, 319. [Google Scholar] [CrossRef] [PubMed]
- Misinzo, G.; Kwavi, D.E.; Sikombe, C.D.; Makange, M.; Peter, E.; Muhairwa, A.P.; Madege, M.J. Molecular Characterization of African Swine Fever Virus from Domestic Pigs in Northern Tanzania during an Outbreak in 2013. Trop. Anim. Health Prod. 2014, 46, 1199. [Google Scholar] [CrossRef] [PubMed]
- Costard, S.; Wieland, B.; de Glanville, W.; Jori, F.; Rowlands, R.; Vosloo, W.; Roger, F.; Pfeiffer, D.U.; Dixon, L.K. African Swine Fever: How Can Global Spread Be Prevented? Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2683–2696. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vizcaíno, J.M.; Mur, L.; Martínez-López, B. African Swine Fever (ASF): Five Years around Europe. Vet. Microbiol. 2013, 165, 45–50. [Google Scholar] [CrossRef]
- Bergmann, H.; Schulz, K.; Conraths, F.J.; Sauter-Louis, C. A Review of Environmental Risk Factors for African Swine Fever in European Wild Boar. Animals 2021, 11, 2692. [Google Scholar] [CrossRef] [PubMed]
- Ćirović, D.; Penezić, A.; Krofel, M. Jackals as Cleaners: Ecosystem Services Provided by a Mesocarnivore in Human-Dominated Landscapes. Biol. Conserv. 2016, 199, 51–55. [Google Scholar] [CrossRef]
- Keyes, C.A.; Myburgh, J.; Brits, D. Scavenger Activity in a Peri-Urban Agricultural Setting in the Highveld of South Africa. Int. J. Leg. Med. 2021, 135, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Patterson, A.R.; Meng, X.J.; Halbur, P.G. Porcine Circovirus Type 2 in Muscle and Bone Marrow Is Infectious and Transmissible to Naïve Pigs by Oral Consumption. Vet. Microbiol. 2009, 133, 54–64. [Google Scholar] [CrossRef] [PubMed]
- López-Lorenzo, G.; Díaz-Cao, J.M.; Prieto, A.; López-Novo, C.; López, C.M.; Díaz, P.; Rodríguez-Vega, V.; Díez-Baños, P.; Fernández, G. Environmental Distribution of Porcine Circovirus Type 2 (PCV2) in Swine Herds with Natural Infection. Sci. Rep. 2019, 9, 14816. [Google Scholar] [CrossRef] [PubMed]
- O’Dea, M.A.; Hughes, A.P.; Davies, L.J.; Muhling, J.; Buddle, R.; Wilcox, G.E. Thermal Stability of Porcine Circovirus Type 2 in Cell Culture. J. Virol. Methods 2008, 147, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.; le Potier, M.F.; Maris, P. Virucidal Efficacy of Nine Commercial Disinfectants against Porcine Circovirus Type 2. Vet. J. 2008, 177, 388–393. [Google Scholar] [CrossRef] [PubMed]

| Farm | Collecting Data | Number of Jackals | Age | Sex | PCR |
|---|---|---|---|---|---|
| A | 20 February 2021 | 8 | adult | F | - |
| adult | F | - | |||
| adult | M | - | |||
| juvenile | F | + | |||
| adult | F | - | |||
| juvenile | M | - | |||
| juvenile | F | - | |||
| adult | F | + | |||
| 23 May 2021 | 2 | juvenile | F | - | |
| adult | F | - | |||
| 11 July 2021 | 9 | adult | M | - | |
| adult | M | - | |||
| adult | F | - | |||
| adult | M | - | |||
| adult | F | - | |||
| adult | F | - | |||
| adult | F | - | |||
| adult | F | - | |||
| adult | F | - | |||
| B | 23 May 2021 | 3 | adult | F | - |
| adult | F | - | |||
| adult | M | - | |||
| 30 May 2021 | 4 | adult | M | - | |
| adult | F | - | |||
| adult | F | - | |||
| adult | M | - | |||
| 13 June 2021 | 4 | adult | F | - | |
| juvenile | F | - | |||
| adult | F | - | |||
| adult | M | - | |||
| 4 July 2021 | 2 | adult | F | - | |
| adult | M | - |
| Primer | Sequences | Amplicon Size (bp) |
|---|---|---|
| P1 | −5′-TAA TCC TTC CGA AGA CGA GC-3′ | 629 |
| P2 | 3′-CGA TCA CAC AGT CTC AGT AG-5′ | |
| P3 | 5′-CAG AAG CGT GAT TGG AAG AC-3′ | 630 |
| P4 | 3′-ATG TAG ACC ACG TAG GCC TC-5′ | |
| P5 | 5′-AGA AGC TCT TTA TCG GAG GA-3′ | 701 |
| P6 | 3′-AAG CGA ACC ACA GTC AGA AC-5′ | |
| P7 | 5′-CTA GAA TAA CAG CAC TGG AG-3′ | 621 |
| P8 | 3′-GTT CGT CCT TCC TCA TTA CC-5′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molini, U.; Coetzee, L.M.; Van Zyl, L.; Khaiseb, S.; Cattoli, G.; Dundon, W.G.; Franzo, G. Molecular Detection and Genetic Characterization of Porcine Circovirus 2 (PCV-2) in Black-Backed Jackal (Lupulella mesomelas) in Namibia. Animals 2022, 12, 620. https://doi.org/10.3390/ani12050620
Molini U, Coetzee LM, Van Zyl L, Khaiseb S, Cattoli G, Dundon WG, Franzo G. Molecular Detection and Genetic Characterization of Porcine Circovirus 2 (PCV-2) in Black-Backed Jackal (Lupulella mesomelas) in Namibia. Animals. 2022; 12(5):620. https://doi.org/10.3390/ani12050620
Chicago/Turabian StyleMolini, Umberto, Lauren Michelle Coetzee, Leandra Van Zyl, Siegfried Khaiseb, Giovanni Cattoli, William G. Dundon, and Giovanni Franzo. 2022. "Molecular Detection and Genetic Characterization of Porcine Circovirus 2 (PCV-2) in Black-Backed Jackal (Lupulella mesomelas) in Namibia" Animals 12, no. 5: 620. https://doi.org/10.3390/ani12050620
APA StyleMolini, U., Coetzee, L. M., Van Zyl, L., Khaiseb, S., Cattoli, G., Dundon, W. G., & Franzo, G. (2022). Molecular Detection and Genetic Characterization of Porcine Circovirus 2 (PCV-2) in Black-Backed Jackal (Lupulella mesomelas) in Namibia. Animals, 12(5), 620. https://doi.org/10.3390/ani12050620






