Investigation of the Role of Healthy and Sick Equids in the COVID-19 Pandemic through Serological and Molecular Testing
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Sampling
2.2. Quantitative PCR Analyses
2.3. Serology
2.4. Statistical Analyses
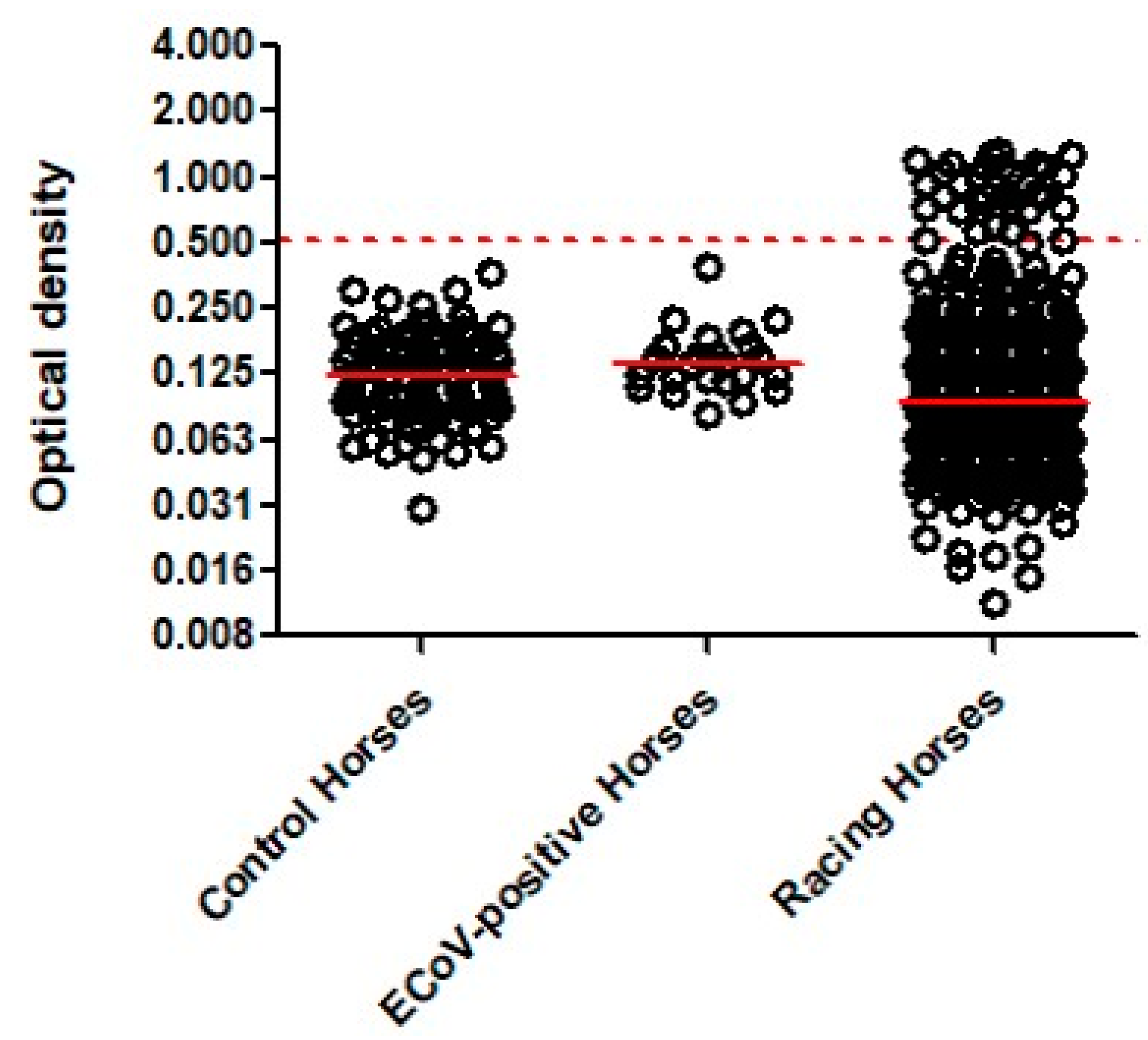
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, J. SARS-CoV-2: An emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020, 16, 1678–1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.P.; Pfenning, A.R.; Zhao, H.; et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, G.; Wang, Q.; Gao, G.F. Bat-to-human: Spike features determining ’host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015, 23, 468–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Moneim, A.S.; Abdelwhab, E.M. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens 2020, 9, 529. [Google Scholar] [CrossRef]
- Hossain, M.G.; Javed, A.; Akter, S.; Saha, S. SARS-CoV-2 host diversity: An update of natural infections and experimental evidence. J. Microbiol. Immunol. Infect. 2021, 54, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Jo, W.K.; de Oliveira-Filho, E.F.; Rasche, A.; Greenwood, A.D.; Osterrieder, K.; Drexler, J.F. Potential zoonotic sources of SARS-CoV-2 infections. Transbound. Emerg. Dis. 2021, 68, 1824–1834. [Google Scholar] [CrossRef] [PubMed]
- Bouricha, E.M.; Hakmi, M.; Akachar, J.; Belyamani, L.; Ibrahimi, A. In silico analysis of ACE2 orthologues to predict animal host range with high susceptibility to SARS-CoV-2. 3 Biotech 2020, 10, 483. [Google Scholar] [CrossRef] [PubMed]
- Buonocore, M.; Marino, C.; Grimaldi, M.; Santoro, A.; Firoznezhad, M.; Paciello, O.; Prisco, F.; D’Ursi, A.M. New putative animal reservoirs of SARS-CoV-2 in Italian fauna: A bioinformatic approach. Heliyon 2020, 6, e05430. [Google Scholar] [CrossRef]
- Pusterla, N.; James, K.; Barnum, S.; Delwart, E. Investigation of three newly identified equine parvoviruses in blood and nasal fluid samples of clinically healthy horses and horses with acute onset of respiratory disease. Animals 2021, 11, 3006. [Google Scholar] [CrossRef] [PubMed]
- Kooijman, L.J.; Mapes, S.M.; Pusterla, N. Development of an equine coronavirus-specific enzyme-linked immunosorbent assay to determine serologic responses in naturally infected horses. J. Vet. Diagn. Investig. 2016, 28, 414–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pusterla, N.; Kass, P.H.; Mapes, S.; Johnson, C.; Barnett, D.C.; Vaala, W.; Gutierrez, C.; McDaniel, R.; Whitehead, B.; Manning, J. Surveillance programme for important equine infectious respiratory pathogens in the USA. Vet. Rec. 2011, 169, 12. [Google Scholar] [CrossRef] [PubMed]
- Pusterla, N.; Mapes, S.; Wademan, C.; White, A.; Hodzic, E. Investigation of the role of lesser characterised respiratory viruses associated with upper respiratory tract infections in horses. Vet. Rec. 2013, 172, 315. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Schuurman, N.; Li, W.; Wang, C.; Smit, L.A.M.; Broens, E.M.; Wagenaar, J.A.; van Kuppeveld, F.J.M.; Bosch, B.J.; Egberink, H. Serologic screening of severe acute respiratory syndrome coronavirus 2 infection in cats and dogs during first coronavirus disease wave, the Netherlands. Emerg. Infect. Dis. 2021, 27, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Okba, N.M.A.; Müller, M.A.; Li, W.; Wang, C.; GeurtsvanKessel, C.H.; Corman, V.M.; Lamers, M.M.; Sikkema, R.S.; de Bruin, E.; Chandler, F.D.; et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020, 26, 1478–1488. [Google Scholar] [CrossRef]
- Dróżdż, M.; Krzyżek, P.; Dudek, B.; Makuch, S.; Janczura, A.; Paluch, E. Current state of knowledge about role of pets in zoonotic transmission of SARS-CoV-2. Viruses 2021, 13, 1149. [Google Scholar] [CrossRef]
- Rashedi, J.; Mahdavi Poor, B.; Asgharzadeh, V.; Pourostadi, M.; Samadi Kafil, H.; Vegari, A.; Tayebi-Khosroshahi, H.; Asgharzadeh, M. Risk factors for COVID-19. Infez. Med. 2020, 28, 469–474. [Google Scholar]
- Saba, L.; Gerosa, C.; Fanni, D.; Marongiu, F.; La Nasa, G.; Caocci, G.; Barcellona, D.; Balestrieri, A.; Coghe, F.; Orru, G.; et al. Molecular pathways triggered by COVID-19 in different organs: ACE2 receptor-expressing cells under attack? A review. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12609–12622. [Google Scholar]
- Deng, J.; Jin, Y.; Liu, Y.; Sun, J.; Hao, L.; Bai, J.; Huang, T.; Lin, D.; Jin, Y.; Tian, K. Serological survey of SARS-CoV-2 for experimental, domestic, companion and wild animals excludes intermediate hosts of 35 different species of animals. Transbound. Emerg. Dis. 2020, 67, 1745–1749. [Google Scholar] [CrossRef]
- Cerino, P.; Buonerba, C.; Brambilla, G.; Atripaldi, L.; Tafuro, M.; Concilio, D.D.; Vassallo, L.; Conte, G.L.; Cuomo, M.C.; Maiello, I.; et al. No detection of SARS-CoV-2 in animals exposed to infected keepers: Results of a COVID-19 surveillance program. Future Sci. OA 2021, 7, FSO711. [Google Scholar] [CrossRef]
- Bosco-Lauth, A.M.; Walker, A.; Guilbert, L.; Porter, S.; Hartwig, A.; McVicker, E.; Bielefeldt-Ohmann, H.; Bowen, R.A. Non susceptibility of livestock to SARS-CoV-2 infection. Emerg. Microbes Infect. 2021, 10, 2199–2201. [Google Scholar] [CrossRef] [PubMed]
- Sailleau, C.; Dumarest, M.; Vanhomwegen, J.; Delaplace, M.; Caro, V.; Kwasiborski, A.; Hourdel, V.; Chevaillier, P.; Barbarino, A.; Comtet, L.; et al. First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound. Emerg. Dis. 2020, 67, 2324–2328. [Google Scholar] [CrossRef] [PubMed]
- Barrs, V.R.; Peiris, M.; Tam, K.W.S.; Law, P.Y.T.; Brackman, C.J.; To, E.M.W.; Yu, V.Y.T.; Chu, D.K.W.; Perera, R.A.P.M.; Sit, T.H.C. SARS-CoV-2 in quarantined domestic cats from COVID-19 households or close contacts, Hong Kong, China. Emerg. Infect. Dis. 2020, 26, 3071–3074. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.I.; Elia, G.; Grassi, A.; Giordano, A.; Desario, C.; Medardo, M.; Smith, S.L.; Anderson, E.R.; Prince, T.; Patterson, G.T.; et al. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat. Commun. 2020, 11, 6231. [Google Scholar] [CrossRef]
- Calvet, G.A.; Pereira, S.A.; Ogrzewalska, M.; Pauvolid-Corrêa, A.; Resende, P.C.; Tassinari, W.S.; Costa, A.P.; Keidel, L.O.; da Rocha, A.S.B.; da Silva, M.F.B.; et al. Investigation of SARS-CoV-2 infection in dogs and cats of humans diagnosed with COVID-19 in Rio de Janeiro, Brazil. PLoS ONE 2021, 16, e0250853. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Arrondo, I.; Portillo, A.; Palomar, A.M.; Santibáñez, S.; Santibáñez, P.; Cervera, C.; Oteo, J.A. Detection of SARS-CoV-2 in pets living with COVID-19 owners diagnosed during the COVID-19 lockdown in Spain: A case of an asymptomatic cat with SARS-CoV-2 in Europe. Transbound. Emerg. Dis. 2021, 68, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Hamer, S.A.; Pauvolid-Corrêa, A.; Zecca, I.B.; Davila, E.; Auckland, L.D.; Roundy, C.M.; Tang, W.; Torchetti, M.K.; Killian, M.L.; Jenkins-Moore, M.; et al. SARS-CoV-2 infections and viral isolations among serially tested cats and dogs in households with infected owners in Texas, USA. Viruses 2021, 13, 938. [Google Scholar] [CrossRef] [PubMed]
- Venter, M.; Richter, K. Towards effective diagnostic assays for COVID-19: A review. J. Clin. Pathol. 2020, 73, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Wernike, K.; Aebischer, A.; Michelitsch, A.; Hoffmann, D.; Freuling, C.; Balkema-Buschmann, A.; Graaf, A.; Müller, T.; Osterrieder, N.; Rissmann, M.; et al. Multi-species ELISA for the detection of antibodies against SARS-CoV-2 in animals. Transbound. Emerg. Dis. 2021, 68, 1779–1785. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, H.; Gao, J.; Huang, K.; Yang, Y.; Hui, X.; He, X.; Li, C.; Gong, W.; Zhang, Y.; et al. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg. Microbes Infect. 2020, 9, 2013–2019. [Google Scholar] [CrossRef] [PubMed]
- Fritz, M.; Rosolen, B.; Krafft, E.; Becquart, P.; Elguero, E.; Vratskikh, O.; Denolly, S.; Boson, B.; Vanhomwegen, J.; Gouilh, M.A.; et al. High prevalence of SARS-CoV-2 antibodies in pets from COVID-19+ households. One Health 2021, 11, 100192. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreenivasan, C.C.; Thomas, M.; Wang, D.; Li, F. Susceptibility of livestock and companion animals to COVID-19. J. Med. Virol. 2021, 93, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
| Target Gene (GenBank) | Oligonucleotides |
|---|---|
| Spike gene (MT773134) | SARS-CoV-2-forward primer: GGCACAGGTGTTCTTACTGAGTCTAAC SARS-CoV-2-reverse primer: CAAGTGTCTGTGGATCACGGAC SARS-CoV-2-probe: FAM-TGGCAGAGACATTGCTGA-MGB Plasmid positive control: TTCAACTTCAATGGTTTAACAGGCACAG GTGTTCTTA CTGAGTCTAACAAAAAGTTTCTGCCTTTCCAACAAT TTGGCAGAGACATTGCTGACACTACTGATGCTGTCCGTGATCCACAGACACTTGAGATTCTTGACATTACACCATGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lawton, K.O.Y.; Arthur, R.M.; Moeller, B.C.; Barnum, S.; Pusterla, N. Investigation of the Role of Healthy and Sick Equids in the COVID-19 Pandemic through Serological and Molecular Testing. Animals 2022, 12, 614. https://doi.org/10.3390/ani12050614
Lawton KOY, Arthur RM, Moeller BC, Barnum S, Pusterla N. Investigation of the Role of Healthy and Sick Equids in the COVID-19 Pandemic through Serological and Molecular Testing. Animals. 2022; 12(5):614. https://doi.org/10.3390/ani12050614
Chicago/Turabian StyleLawton, Kaila O. Y., Rick M. Arthur, Benjamin C. Moeller, Samantha Barnum, and Nicola Pusterla. 2022. "Investigation of the Role of Healthy and Sick Equids in the COVID-19 Pandemic through Serological and Molecular Testing" Animals 12, no. 5: 614. https://doi.org/10.3390/ani12050614
APA StyleLawton, K. O. Y., Arthur, R. M., Moeller, B. C., Barnum, S., & Pusterla, N. (2022). Investigation of the Role of Healthy and Sick Equids in the COVID-19 Pandemic through Serological and Molecular Testing. Animals, 12(5), 614. https://doi.org/10.3390/ani12050614






