Investigation of Copy Number Variations (CNVs) of the Goat PPP3CA Gene and Their Effect on Litter Size and Semen Quality
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Isolation
2.2. Detection of Various Indicators of Semen Quality
2.2.1. The Determination of Semen Concentration
2.2.2. The Determination of the Percentage of Viable Sperm
2.2.3. The Determination of Sperm Deformity Rate
2.2.4. The Determination of Sperm Plasma Membrane Integrity
2.3. Primer Design and CNV Detection
2.4. Copy Number Analysis and Statistical Analyses
3. Results
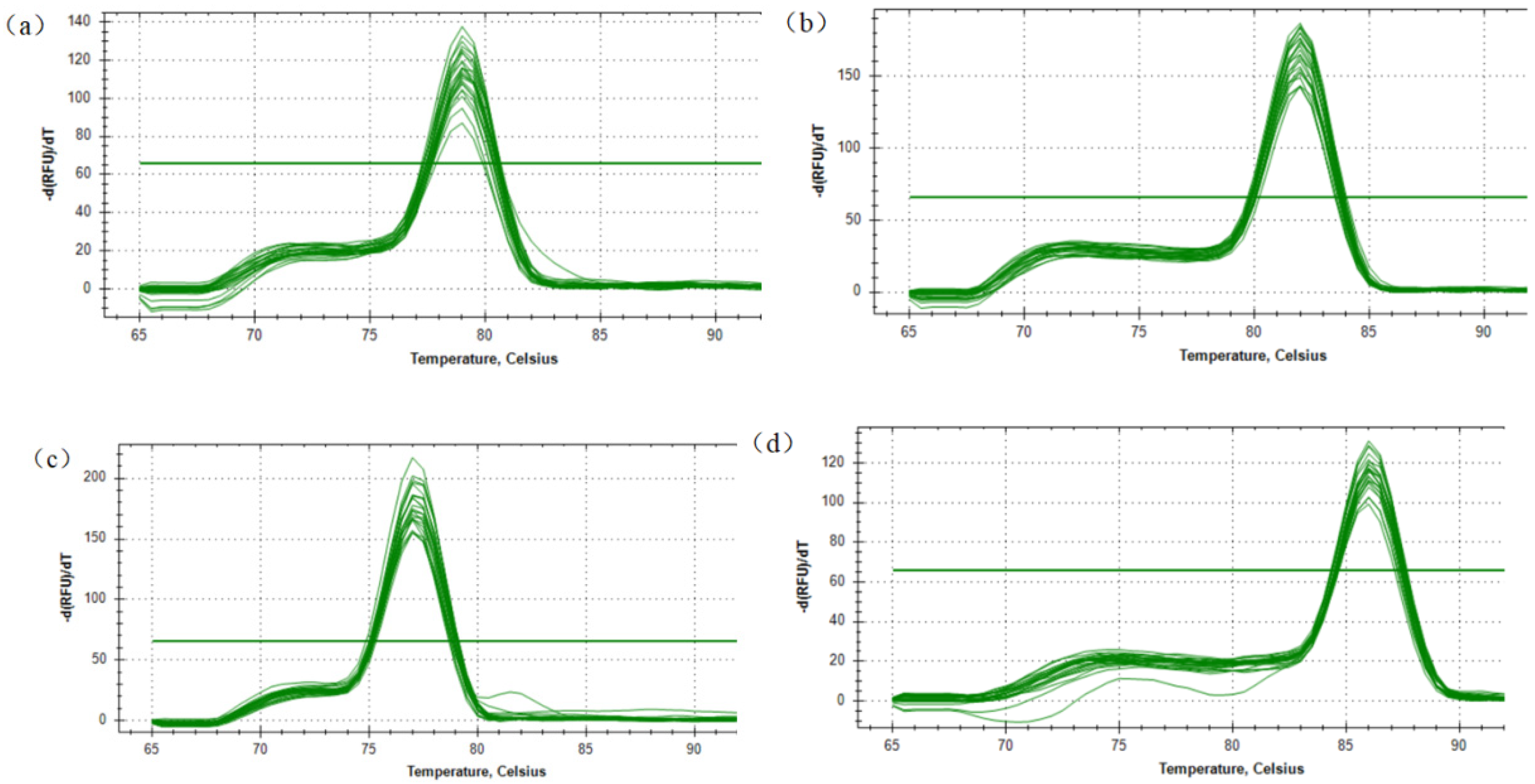
3.1. qPCR Primer Detection
3.2. Distribution of Different CNV Types in Goats
3.3. Association Analysis of CNV Type and Goat Litter Size
3.4. Association Analysis of CNV Type and Goat Semen Quality
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tash, J.S.; Krinks, M.; Patel, J.; Means, R.L.; Klee, C.B.; Means, A.R. Identification, characterization, and functional correlation of calmodulin-dependent protein phosphatase in sperm. J. Cell Biol. 1988, 106, 1625–1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rydzanicz, M.; Wachowska, M.; Cook, E.C.; Lisowski, P.; Kuźniewska, B.; Szymańska, K.; Diecke, S.; Prigione, A.; Szczałuba, K.; Szybińska, A.; et al. Novel calcineurin A (PPP3CA) variant associated with epilepsy, constitutive enzyme activation and downregulation of protein expression. Eur. J. Hum. Genet. 2019, 27, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Daei-Farshbaf, N.; Aflatoonian, R.; Amjadi, F.S.; Nikniyaz, H.; Taleahmad, S.; Bakhtiyari, M. Identification of calcineurin as a predictor of oocyte quality and fertilization competence based on microarray data. Comput. Biol. Chem. 2021, 94, 107561. [Google Scholar] [CrossRef] [PubMed]
- Panneerselvam, S.; Wang, J.; Zhu, W.; Dai, H.; Pappas, J.G.; Rabin, R.; Low, K.J.; Rosenfeld, J.A.; Emrick, L.; Xiao, R.; et al. PPP3CA truncating variants clustered in the regulatory domain cause early-onset refractory epilepsy. Clin Genet. 2021, 100, 227–233. [Google Scholar] [CrossRef]
- Islam, R.; Liu, X.; Gebreselassie, G.; Abied, A.; Ma, Q.; Ma, Y. Genome-wide association analysis reveals the genetic locus for high reproduction trait in Chinese Arbas cashmere goat. Genes Genom. 2020, 42, 893–899. [Google Scholar] [CrossRef]
- Dias, M.M.; Souza, F.R.; Takada, L.; Feitosa, F.L.; Costa, R.B.; Diaz, I.D.; Cardoso, D.F.; Tonussi, R.L.; Baldi, F.; Albuquerque, L.G.; et al. Study of lipid metabolism-related genes as candidate genes of sexual precocity in Nellore cattle. Genet. Mol. Res. 2015, 14, 234–243. [Google Scholar] [CrossRef]
- E, G.X.; Zhao, Y.J.; Huang, Y.F. Selection signatures of litter size in Dazu black goats based on a whole genome sequencing mixed pools strategy. Mol. Biol. Rep. 2019, 46, 5517–5523. [Google Scholar] [CrossRef]
- Wan, L.; Ma, J.; Xu, G.; Wang, D.; Wang, N. Molecular cloning, structural analysis and tissue expression of protein phosphatase 3 catalytic subunit alpha isoform (PPP3CA) gene in Tianfu goat muscle. Int. J. Mol. Sci. 2014, 15, 2346–2358. [Google Scholar] [CrossRef] [Green Version]
- Bär, L.; Großmann, C.; Gekle, M.; Föller, M. Calcineurin inhibitors regulate fibroblast growth factor 23 (FGF23) synthesis. Naunyn Schmiedebergs Arch. Pharmacol. 2017, 390, 1117–1123. [Google Scholar] [CrossRef]
- Bai, Y.; Li, J.; Zhu, H.; Liu, J.; Dong, S.; Li, L.; Qu, L.; Chen, H.; Song, X.; Lan, X. Deletion mutation within the goat PPP3CA gene identified by GWAS significantly affects litter size. Reprod. Fertil. Dev. 2021, 33, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.J.; Chen, H.; Liao, X.; Allayee, H.; Shih, D.M.; Lee, G.S.; Hovland, D.N.; Robbins, W.A., Jr.; Carnes, K.; Hess, R.A.; et al. FK506, a calcineurin inhibitor, prevents cadmium-induced testicular toxicity in mice. Toxicol. Sci. 2007, 100, 474–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, A.T.; Lord, T.; Stanger, S.J.; Roman, S.D.; McCluskey, A.; Robinson, P.J.; Aitken, R.J.; Nixon, B. Dynamin regulates specific membrane fusion events necessary for acrosomal exocytosis in mouse spermatozoa. J. Biol. Chem. 2012, 287, 37659–37672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redgrove, K.A.; Bernstein, I.R.; Pye, V.J.; Mihalas, B.P.; Sutherland, J.M.; Nixon, B.; McCluskey, A.; Robinson, P.J.; Holt, J.E.; McLaughlin, E.A. Dynamin 2 is essential for mammalian spermatogenesis. Sci. Rep. 2016, 6, 35084. [Google Scholar] [CrossRef] [PubMed]
- Di Gerlando, R.; Mastrangelo, S.; Moscarelli, A.; Tolone, M.; Sutera, A.M.; Portolano, B.; Sardina, M.T. Genomic structural diversity in local goats: Analysis of copy-number variations. Animals 2020, 10, 1040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, S.; Tang, Q.; Jiang, E.; Wang, K.; Lan, X.; Pan, C. Goat sperm associated antigen 17 protein gene (SPAG17): Small and large fragment genetic variation detection, association analysis, and mRNA expression in gonads. Genomics 2020, 112, 5115–5121. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Niu, Q.; Zhang, T.; Zhao, G.; Zhu, B.; Chen, Y.; Zhang, L.; Gao, X.; Gao, H.; Liu, G.E.; et al. Genomic sequencing analysis reveals copy number variations and their associations with economically important traits in beef cattle. Genomics 2021, 113, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Sebat, J.; Lakshmi, B.; Troge, J.; Alexander, J.; Young, J.; Lundin, P.; Månér, S.; Massa, H.; Walker, M.; Chi, M.; et al. Large-scale copy number polymorphism in the human genome. Science 2004, 305, 525–528. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Woodward-Greene, J.; Kang, X.; Pan, M.G.; Rosen, B.; Van Tassell, C.P.; Chen, H.; Liu, G.E. Genome-wide CNV analysis revealed variants associated with growth traits in African indigenous goats. Genomics 2020, 112, 1477–1480. [Google Scholar] [CrossRef]
- Kang, X.; Li, M.; Liu, M.; Liu, S.; Pan, M.G.; Wiggans, G.R.; Rosen, B.D.; Liu, G.E. Copy number variation analysis reveals variants associated with milk production traits in dairy goats. Genomics 2020, 112, 4934–4937. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, P.; Shi, S.; Zhang, Z.; Shi, Q.; Xu, J.; He, H.; Lei, C.; Wang, E.; Chen, H.; et al. Association analysis to copy number variation (CNV) of Opn4 gene with growth traits of goats. Animals 2020, 10, 441. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, C.; Guo, Y.; She, S.; Wang, B.; Jiang, Y.; Bai, Y.; Song, X.; Li, L.; Shi, L.; et al. Screening of deletion variants within the goat PRDM6 gene and its effects on growth traits. Animals 2020, 10, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Wang, X.; Zhang, Z.; An, Q.; Wen, Y.; Wang, D.; Liu, X.; Li, Z.; Lyu, S.; Li, L.; et al. Copy number variation of CADM2 gene revealed its association with growth traits across Chinese Capra hircus (goat) populations. Gene 2020, 741, 144519. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Feng, W.; Kang, Y.; Wang, K.; Yang, Y.; Qu, L.; Chen, H.; Lan, X.; Pan, C. Detection of mRNA expression and copy number variations within the goat FecB gene associated with litter size. Front. Vet. Sci. 2021, 8, 758705. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Kang, Z.; Jiang, E.; Yan, H.; Zhu, H.; Liu, J.; Qu, L.; Lan, X.; Pan, C. Genetic effects of DSCAML1 identified in genome-wide association study revealing strong associations with litter size and semen quality in goat (Capra hircus). Theriogenology 2020, 146, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Hui, Y.; Zhang, Y.; Wang, K.; Pan, C.; Chen, H.; Qu, L.; Song, X.; Lan, X. Goat DNMT3B: An indel mutation detection, association analysis with litter size and mRNA expression in gonads. Theriogenology 2020, 147, 108–115. [Google Scholar] [CrossRef]
- Gangwar, C.; Mishra, A.K.; Gururaj, K.; Kumar, A.; Kharche, S.D.; Saraswat, S.; Kumar, R.; Ramachandran, N. Semen quality and total microbial load: An association study in important Indian Goat breeds during different seasons. Andrologia 2021, 53, e13995. [Google Scholar] [CrossRef]
- Campbell, R.C.; Dott, H.M.; Glover, T.D. Nigrosin eosin as a stain for differentiating live and dead spermatozoa. J. Agric. Sci. 1956, 48, 1–8. [Google Scholar] [CrossRef]
- Abril-Sánchez, S.; Crosignani, N.; Freitas-de-Melo, A.; Terrazas, A.; Damián, J.P.; Beracochea, F.; Silveira, P.; Ungerfeld, R. Sedation or anaesthesia decrease the stress response to electroejaculation and improve the quality of the collected semen in goat bucks. Animal 2018, 12, 2598–2608. [Google Scholar] [CrossRef]
- Jeyendran, R.S.; Van der Ven, H.H.; Perez-Pelaez, M.; Crabo, B.G.; Zaneveld, L.J. Development of an assay to assess the functional integrity of the human sperm membrane and its relationship to other semen characteristics. J. Reprod. Fertil. 1984, 70, 219–228. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Fu, W.; Su, R.; Tian, X.; Du, D.; Zhao, Y.; Zheng, Z.; Chen, Q.; Gao, S.; Cai, Y.; et al. Towards the complete goat pan-genome by recovering missing genomic segments from the reference genome. Front. Genet. 2019, 10, 1169. [Google Scholar] [CrossRef]
- Shi, S.Y.; Li, L.J.; Zhang, Z.J.; Wang, E.Y.; Wang, J.; Xu, J.W.; Liu, H.B.; Wen, Y.F.; He, H.; Lei, C.Z.; et al. Copy number variation of MYLK4 gene and its growth traits of Capra hircus (goat). Anim. Biotechnol. 2020, 31, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zheng, Z.; Cai, Y.; Chen, T.; Li, C.; Fu, W.; Jiang, Y. CNVcaller: Highly efficient and widely applicable software for detecting copy number variations in large populations. Gigascience 2017, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Su, P.; Akhatayeva, Z.; Pan, C.; Zhang, Q.; Lan, X. Novel InDel variations of the Cry2 gene are associated with litter size in Australian White sheep. Theriogenology 2022, 179, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Bai, Y.; Lan, X.; Zhao, H. Goat AKAP12: Indel mutation detection, association analysis with litter size and alternative splicing variant expression. Front. Genet. 2021, 12, 648256. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yuan, R.; Luo, Y.; Kang, Z.; Zhu, H.; Qu, L.; Lan, X.; Song, X. Exploration of genetic variants within the goat A-Kinase anchoring protein 12 (AKAP12) gene and their effects on growth traits. Animals 2021, 11, 2090. [Google Scholar] [CrossRef] [PubMed]
- Miyata, H.; Satouh, Y.; Mashiko, D.; Muto, M.; Nozawa, K.; Shiba, K.; Fujihara, Y.; Isotani, A.; Inaba, K.; Ikawa, M. Sperm calcineurin inhibition prevents mouse fertility with implications for male contraceptive. Science 2015, 350, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Rusnak, F.; Mertz, P. (2000). Calcineurin: Form and function. Physiol. Rev. 2000, 80, 1483–1521. [Google Scholar] [CrossRef]
- Dey, S.; Eisa, A.; Kline, D.; Wagner, F.F.; Abeysirigunawardena, S.; Vijayaraghavan, S. Roles of glycogen synthase kinase 3 alpha and calcineurin in regulating the ability of sperm to fertilize eggs. FASEB J. 2020, 34, 1247–1269. [Google Scholar] [CrossRef] [Green Version]
- Akhatayeva, Z.; Mao, C.; Jiang, F.; Pan, C.; Lin, C.; Hao, K.; Lan, T.; Chen, H.; Zhang, Q.; Lan, X. Indel variants within the PRL and GHR genes associated with sheep litter size. Reprod. Domest. Anim. 2020, 55, 1470–1478. [Google Scholar] [CrossRef]
- Kang, Z.; Jiang, E.; Wang, K.; Pan, C.; Chen, H.; Yan, H.; Zhu, H.; Liu, J.; Qu, L.; Lan, X. Goat membrane associated ring-CH-type finger 1 (MARCH1) mRNA expression and association with litter size. Theriogenology 2019, 128, 8–16. [Google Scholar] [CrossRef]
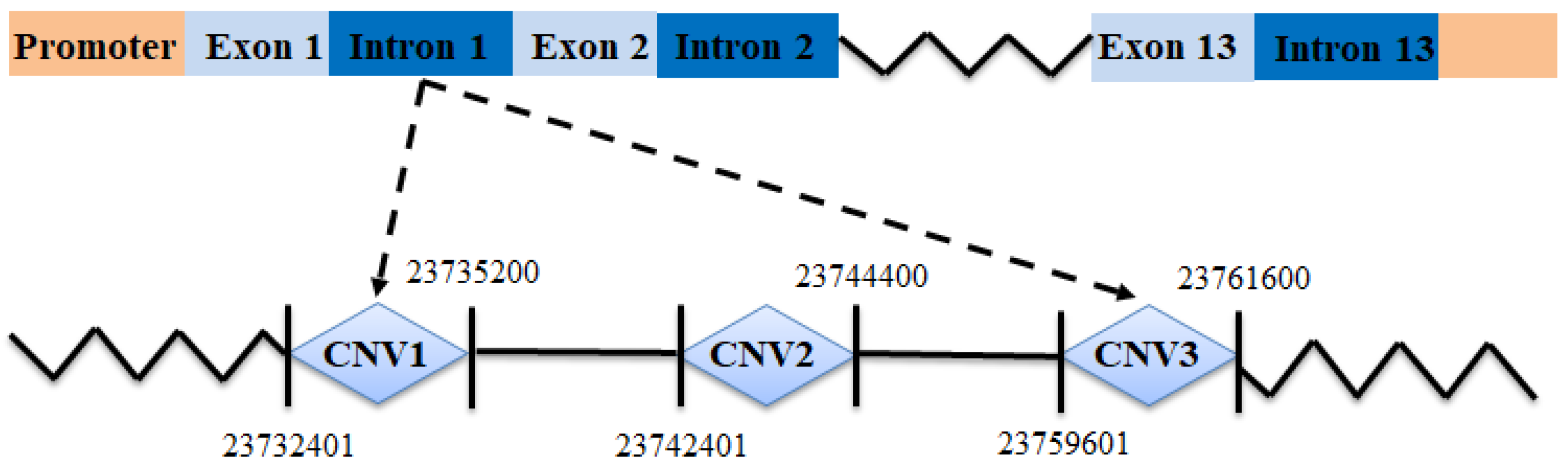
| Primers | Start | End | Length | Location | Reference Sequence |
|---|---|---|---|---|---|
| CNV1 | 23732401 | 23735200 | 2799 | intron | NC_030813.1 |
| CNV2 | 23742401 | 23744400 | 1999 | intron | NC_030813.1 |
| CNV3 | 23759601 | 23761600 | 1999 | intron | NC_030813.1 |
| Primers | Sequences (5′-3′) | Sizes (bp) |
|---|---|---|
| CNV1 | F: CCTCTGCCTACTCTTTCCTGC | 106 |
| R: GGGGGAAATGGTCCCCAAA | ||
| CNV2 | F: AGAGGGAACCTCCATGGTCA | 108 |
| R: CCAAGCCTCAACTCACTCCA | ||
| CNV3 | F: CCGACTCCCCCTGAAGTAGT | 199 |
| R: TCTCCAGCAATGTTAGGCTGT | ||
| MC1R | F: GGCCTGAGAGGGGAATCACA | 126 |
| R: AGTGGGTCTCTGGATGGAGG |
| Breed | Loci | Sizes | Typic Frequencies | ||
|---|---|---|---|---|---|
| Gain | Medium | Loss | |||
| SBWC goat (female) | CNV1 | 300 | 0.363 (n = 109) | 0.187 (n = 56) | 0.450 (n = 135) |
| CNV2 | 307 | 0.861 (n = 206) | 0.153 (n = 47) | 0.176 (n = 54) | |
| CNV3 | 301 | 0.781 (n = 235) | 0.056 (n = 17) | 0.163 (n = 49) | |
| SBWC goat (male) | CNV1 | 46 | 0.370 (n = 17) | 0.522 (n = 24) | 0.108 (n = 5) |
| CNV2 | 46 | 0.783 (n = 36) | 0.130 (n = 6) | 0.087 (n = 4) | |
| CNV3 | 46 | 1.000 (n = 46) | - | - | |
| GZHM goat (female) | CNV1 | 64 | 0.047 (n = 3) | 0.734 (n = 47) | 0.219 (n = 14) |
| CNV2 | 64 | 0.094 (n = 6) | 0.703 (n = 45) | 0.203 (n = 13) | |
| CNV3 | 64 | 0.875 (n = 56) | 0.109 (n = 7) | 0.016 (n = 1) | |
| Mutations | Sizes | Gain | Medium | Loss | p-Values |
|---|---|---|---|---|---|
| CNV1 | 300 | 1.40 B ± 0.05 (n = 109) | 1.25 B ± 0.06 (n = 56) | 1.72 A ± 0.04 (n = 135) | 7.6802 × 10−11 |
| CNV2 | 307 | 1.40 B ± 0.03 (n = 206) | 1.79 A ± 0.06 (n = 47) | 1.74 A ± 0.06 (n = 54) | 5.0895 × 10−9 |
| CNV3 | 301 | 1.50 ± 0.03 (n = 235) | 1.41 ± 0.12 (n = 17) | 1.57 ± 0.07 (n = 49) | 0.487 |
| Mutations | Parameters | Genotypes | p-Values | ||
|---|---|---|---|---|---|
| Gain | Medium | Loss | |||
| CNV1 | EV | 0.94 ± 0.09 ab (n = 17) | 0.75 b ± 0.05 (n = 24) | 1.12 a ± 0.18 (n = 5) | 0.035 |
| SC | 292.94 b ± 3.46 (n = 17) | 271.10 b ± 3.37 (n = 24) | 465.20 a ± 6.03 (n = 5) | 0.047 | |
| SV | 50.90 B ± 0.03 (n = 17) | 54.60 b ± 0.04 (n = 24) | 74.00 Aa ± 0.04 (n = 5) | 0.026 | |
| SdR | 34.00 ± 0.04 (n = 17) | 27.50 ± 0.04 (n = 24) | 21.10 ± 0.05 (n = 5) | 0.320 | |
| SaR | 82.90 ± 0.02 (n = 15) | 91.60 ± 0.01 (n = 22) | 92.40 ± 0.03 (n = 5) | 0.169 | |
| LS | 151.30 B ± 2.18 (n = 17) | 147.20 B ± 1.77 (n = 24) | 343.30 A ± 5.01 (n = 5) | 2.3 × 10−4 | |
| CNV2 | EV | 0.82 ± 0.57 (n = 36) | 1.00 ± 0.12 (n = 6) | 0.83 ± 0.14 (n = 4) | 0.141 |
| SC | 269.50 b ± 2.64 (n = 36) | 373.10 ab ± 5.08 (n = 6) | 468.10 a ± 6.41 (n = 4) | 0.030 | |
| SV | 53.30 ± 0.27 (n = 36) | 60.80 ± 0.07 (n = 6) | 65.00 ± 0.12 (n = 4) | 0.315 | |
| SdR | 31.30 ± 0.03 (n = 36) | 19.90 ± 0.05 (n = 6) | 24.80 ± 0.05 (n = 4) | 0.343 | |
| SPR | 90.70 ± 0.02 (n = 33) | 82.80 ± 0.03 (n = 5) | 77.80 ± 0.12 (n = 4) | 0.157 | |
| LS | 146.30 B ± 1.54 (n = 36) | 215.80 AB ± 2.82 (n = 6) | 314.10 A ± 5.14 (n = 4) | 0.005 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, Y.; Zhang, T.; Liu, N.; Wang, C.; Guo, Z.; Pan, C.; Zhu, H.; Lan, X. Investigation of Copy Number Variations (CNVs) of the Goat PPP3CA Gene and Their Effect on Litter Size and Semen Quality. Animals 2022, 12, 445. https://doi.org/10.3390/ani12040445
Bai Y, Zhang T, Liu N, Wang C, Guo Z, Pan C, Zhu H, Lan X. Investigation of Copy Number Variations (CNVs) of the Goat PPP3CA Gene and Their Effect on Litter Size and Semen Quality. Animals. 2022; 12(4):445. https://doi.org/10.3390/ani12040445
Chicago/Turabian StyleBai, Yangyang, Taiyuan Zhang, Nuan Liu, Congliang Wang, Zhengang Guo, Chuanying Pan, Haijing Zhu, and Xianyong Lan. 2022. "Investigation of Copy Number Variations (CNVs) of the Goat PPP3CA Gene and Their Effect on Litter Size and Semen Quality" Animals 12, no. 4: 445. https://doi.org/10.3390/ani12040445
APA StyleBai, Y., Zhang, T., Liu, N., Wang, C., Guo, Z., Pan, C., Zhu, H., & Lan, X. (2022). Investigation of Copy Number Variations (CNVs) of the Goat PPP3CA Gene and Their Effect on Litter Size and Semen Quality. Animals, 12(4), 445. https://doi.org/10.3390/ani12040445




