Porcine Intestinal Apical-Out Organoid Model for Gut Function Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Crypts from Porcine Small Intestine
2.2. Culture of Porcine Small Intestinal Organoid
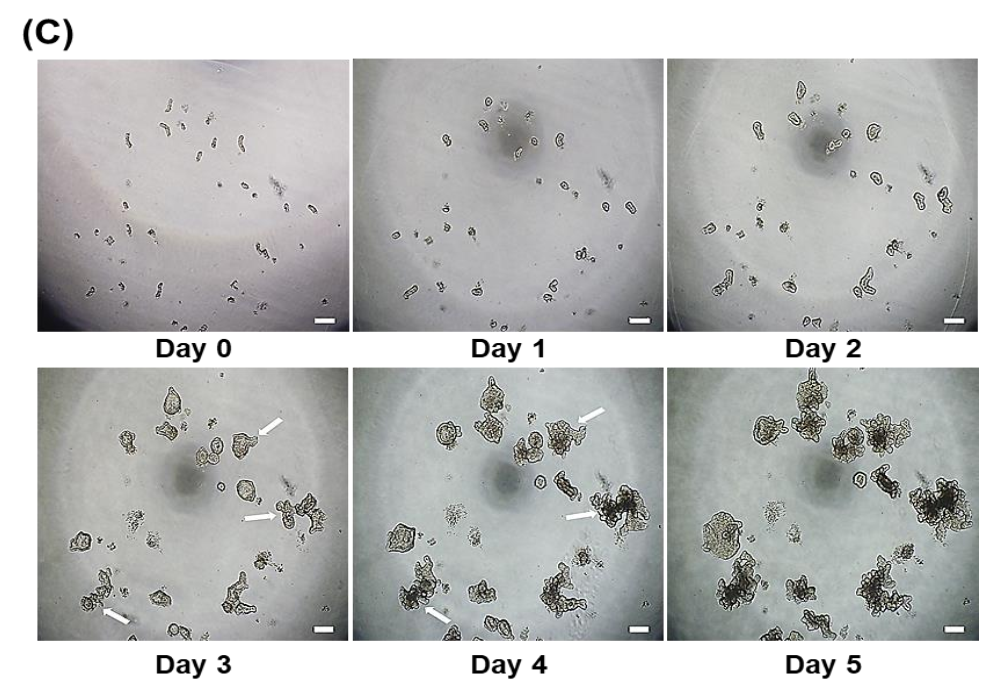
2.3. Sub-Culture Technique and Development of Apical-Out Porcine Organoid
2.4. Imaging of Porcine Organoids
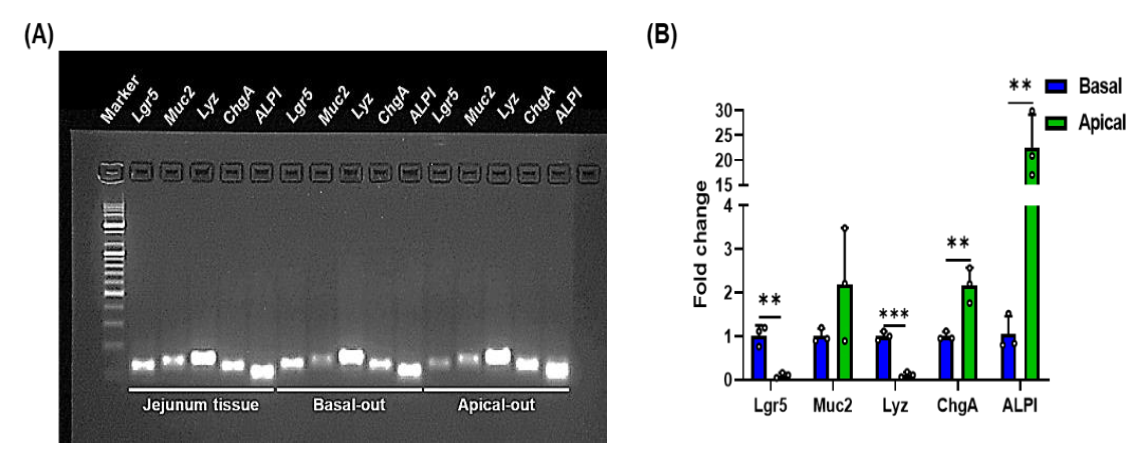
2.5. Quantitative Real-Time PCR (qRT-PCR)
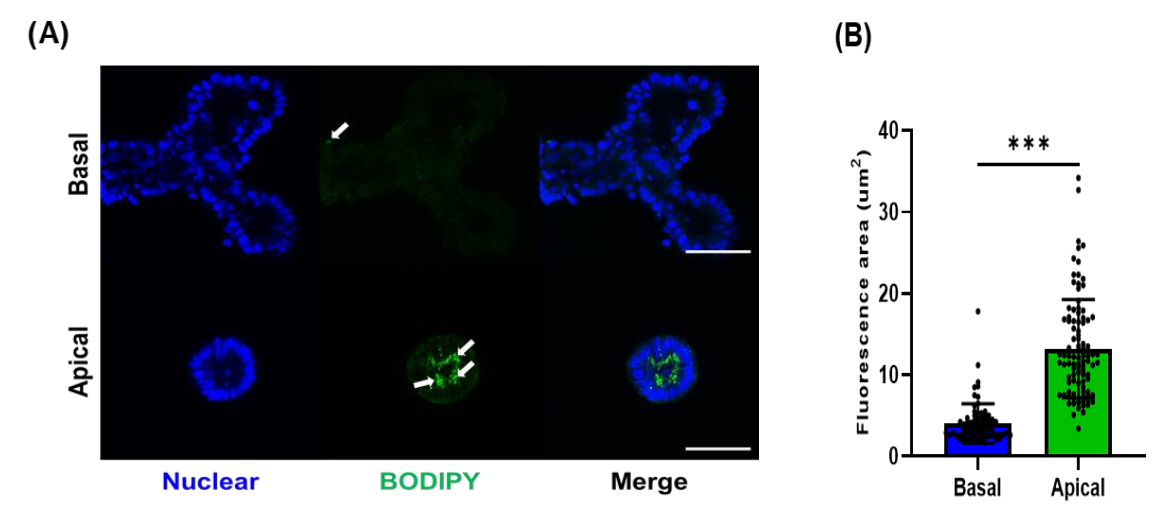
2.6. Assessment for Fatty Acid Uptake
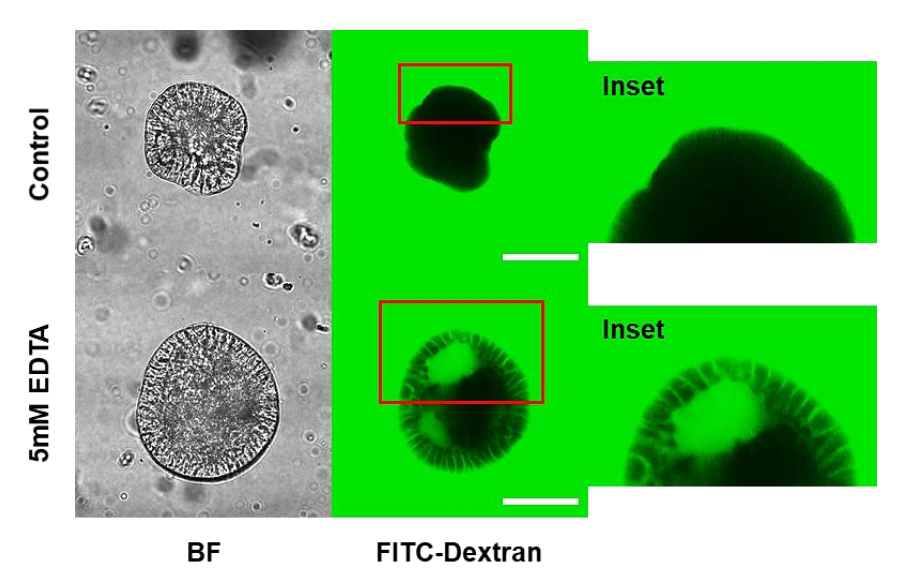
2.7. Breach Epithelial Barrier
2.8. Statistical Analysis
3. Results
3.1. Development of Porcine Small Intestinal Organoids
3.2. Morphological Difference between Porcine Apical-Out and Basal-Out Organoids
3.3. Fatty Acid Analog Uptake by Different Types of Porcine Organoids
3.4. Porcine Apical-Out Organoids Have Gut Barrier Function
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 2018, 50, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizk, P.; Barker, N. Gut stem cells in tissue renewal and disease: Methods, markers, and myths. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 475–496. [Google Scholar] [CrossRef] [PubMed]
- Gehart, H.; Clevers, H. Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 19–34. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Meurens, F.; Summerfield, A.; Nauwynck, H.; Saif, L.; Gerdts, V. The pig: A model for human infectious diseases. Trends Microbiol. 2012, 20, 50–57. [Google Scholar] [CrossRef]
- Kararli, T.T. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm. Drug Dispos. 1995, 16, 351–380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Widmer, G.; Tzipori, S. A pig model of the human gastrointestinal tract. Gut Microbes 2013, 4, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, A.; Gonzalez, L.; Blikslager, A. Large animal models: The key to translational discovery in digestive disease research. CMGH Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 716–724. [Google Scholar] [CrossRef] [Green Version]
- Powell, R.H.; Behnke, M.S. WRN conditioned media is sufficient for in vitro propagation of intestinal organoids from large farm and small companion animals. Biol. Open 2017, 6, 698–705. [Google Scholar] [CrossRef] [Green Version]
- Fair, K.L.; Colquhoun, J.; Hannan, N.R.F. Intestinal organoids for modelling intestinal development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20170217. [Google Scholar] [CrossRef]
- Gonzalez, L.M.; Williamson, I.; Piedrahita, J.A.; Blikslager, A.T.; Magness, S.T. Cell lineage identification and stem cell culture in a porcine model for the study of intestinal epithelial regeneration. PLoS ONE 2013, 8, e66465. [Google Scholar] [CrossRef] [Green Version]
- Khalil, H.A.; Lei, N.Y.; Brinkley, G.; Scott, A.; Wang, J.; Kar, U.K.; Jabaji, Z.B.; Lewis, M.; Martín, M.G.; Dunn, J.C.Y.; et al. A novel culture system for adult porcine intestinal crypts. Cell Tissue Res. 2016, 365, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Beaumont, M.; Blanc, F.; Cherbuy, C.; Egidy, G.; Giuffra, E.; Lacroix-Lamandé, S.; Wiedemann, A. Intestinal organoids in farm animals. Vet. Res. 2021, 52, 33. [Google Scholar] [CrossRef]
- van der Hee, B.; Loonen, L.M.P.; Taverne, N.; Taverne-Thiele, J.J.; Smidt, H.; Wells, J.M. Optimized procedures for generating an enhanced, near physiological 2D culture system from porcine intestinal organoids. Stem Cell Res. 2018, 28, 165–171. [Google Scholar] [CrossRef]
- Hoffmann, P.; Schnepel, N.; Langeheine, M.; Künnemann, K.; Grassl, G.A.; Brehm, R.; Seeger, B.; Mazzuoli-Weber, G.; Breves, G. Intestinal organoid-based 2D monolayers mimic physiological and pathophysiological properties of the pig intestine. PLoS ONE 2021, 16, e0256143. [Google Scholar] [CrossRef]
- Co, J.Y.; Margalef-Catala, M.; Li, X.; Mah, A.T.; Kuo, C.J.; Monack, D.M.; Amieva, M.R. Controlling epithelial polarity: A human enteroid model for host-pathogen interactions. Cell Rep. 2019, 26, 2509–2520.e4. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-G.; Zhu, M.; Chen, M.-X.; Fan, H.-B.; Fu, H.-L.; Zhou, J.-Y.; Zhai, Z.-Y.; Gao, C.-Q.; Yan, H.-C.; Wang, X.-Q. Acute exposure to deoxynivalenol inhibits porcine enteroid activity via suppression of the Wnt/β-catenin pathway. Toxicol. Lett. 2019, 305, 19–31. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Kim, T.-Y.; Kim, Y.; Lee, S.-H.; Kim, S.; Kang, S.W.; Yang, J.-Y.; Baek, I.-J.; Sung, Y.H.; Park, Y.-Y.; et al. Microbiota-derived lactate accelerates intestinal stem-cell-mediated epithelial development. Cell Host Microbe 2018, 24, 833–846.e6. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Couto, M.R.; Gonçalves, P.; Catarino, T.A.; Martel, F. The effect of inflammatory status on butyrate and folate uptake by tumoral (Caco-2) and non-tumoral (IEC-6) intestinal epithelial cells. Cell J. 2017, 19, 96. [Google Scholar]
- Peng, L.; Li, Z.-R.; Green, R.S.; Holzman, I.R.; Lin, J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 2009, 139, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Diesing, A.-K.; Nossol, C.; Panther, P.; Walk, N.; Post, A.; Kluess, J.; Kreutzmann, P.; Dänicke, S.; Rothkötter, H.-J.; Kahlert, S. Mycotoxin deoxynivalenol (DON) mediates biphasic cellular response in intestinal porcine epithelial cell lines IPEC-1 and IPEC-J2. Toxicol. Lett. 2011, 200, 8–18. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Deng, J.; Hu, X.; Zhou, S.; Wu, J.; Xiao, D.; Darko, K.O.; Huang, Y.; Tao, T.; Peng, M.; et al. Vitamin A inhibits the action of LPS on the intestinal epithelial barrier function and tight junction proteins. Food Funct. 2019, 10, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Yin, J.; Chen, J.; Ma, X.; Wu, M.; Li, X.; Yao, K.; Tan, B.; Yin, Y. 4-Phenylbutyric acid accelerates rehabilitation of barrier function in IPEC-J2 cell monolayer model. Anim. Nutr. 2021, 7, 1061–1069. [Google Scholar] [CrossRef]
- Kaur, G.; Dufour, J.M. Cell lines: Valuable tools or useless artifacts. Spermatogenesis 2012, 2, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Fujii, M.; Matano, M.; Toshimitsu, K.; Takano, A.; Mikami, Y.; Nishikori, S.; Sugimoto, S.; Sato, T. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell 2018, 23, 787–793.e6. [Google Scholar] [CrossRef] [Green Version]
- Derricott, H.; Luu, L.; Fong, W.Y.; Hartley, C.S.; Johnston, L.J.; Armstrong, S.D.; Randle, N.; Duckworth, C.A.; Campbell, B.J.; Wastling, J.M.; et al. Developing a 3D intestinal epithelium model for livestock species. Cell Tissue Res. 2019, 375, 409–424. [Google Scholar] [CrossRef] [Green Version]
- Wilson, S.S.; Tocchi, A.; Holly, M.K.; Parks, W.C.; Smith, J.G. A small intestinal organoid model of non-invasive enteric pathogen–epithelial cell interactions. Mucosal Immunol. 2015, 8, 352–361. [Google Scholar] [CrossRef]
- Zietek, T.; Giesbertz, P.; Ewers, M.; Reichart, F.; Weinmüller, M.; Urbauer, E.; Haller, D.; Demir, I.E.; Ceyhan, G.O.; Kessler, H.; et al. Organoids to study intestinal nutrient transport, drug uptake and metabolism–update to the human model and expansion of applications. Front. Bioeng. Biotechnol. 2020, 8, 1065. [Google Scholar] [CrossRef]
- Xu, P.; Becker, H.; Elizalde, M.; Masclee, A.; Jonkers, D. Intestinal organoid culture model is a valuable system to study epithelial barrier function in IBD. Gut 2018, 67, 1905–1906. [Google Scholar] [CrossRef]
- van der Hee, B.; Madsen, O.; Vervoort, J.; Smidt, H.; Wells, J.M. Congruence of transcription programs in adult stem cell-derived jejunum organoids and original tissue during long-term culture. Front. Cell Dev. Biol. 2020, 8, 375. [Google Scholar] [CrossRef]
- Zhu, M.; Qin, Y.-C.; Gao, C.-Q.; Yan, H.-C.; Li, X.-G.; Wang, X.-Q. Extracellular glutamate-induced mTORC1 activation via the IR/IRS/PI3K/Akt pathway enhances the expansion of porcine intestinal stem cells. J. Agric. Food Chem. 2019, 67, 9510–9521. [Google Scholar] [CrossRef]
- Han, X.; Mslati, M.; Davies, E.; Chen, Y.; Allaire, J.M.; Vallance, B.A. Creating a more perfect union: Modeling intestinal bacteria-epithelial interactions using organoids. CMGH Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 769–782. [Google Scholar] [CrossRef]
- Li, Y.; Yang, N.; Chen, J.; Huang, X.; Zhang, N.; Yang, S.; Liu, G.; Liu, G. Next-generation porcine intestinal organoids: An apical-out organoid model for swine enteric virus infection and immune response investigations. J. Virol. 2020, 94, e01006-20. [Google Scholar] [CrossRef]
- Nash, T.J.; Morris, K.M.; Mabbott, N.A.; Vervelde, L. Inside-out chicken enteroids with leukocyte component as a model to study host–pathogen interactions. Commun. Biol. 2021, 4, 377. [Google Scholar] [CrossRef]
- Gajda, A.M.; Storch, J. Enterocyte fatty acid-binding proteins (FABPs): Different functions of liver and intestinal FABPs in the intestine. Prostaglandins Leukot. Essent. Fatty Acids 2015, 93, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Co, J.Y.; Margalef-Català, M.; Monack, D.M.; Amieva, M.R. Controlling the polarity of human gastrointestinal organoids to investigate epithelial biology and infectious diseases. Nat. Protoc. 2021, 16, 5171–5192. [Google Scholar] [CrossRef]
- Suzuki, T. Regulation of the intestinal barrier by nutrients: The role of tight junctions. Anim. Sci. J. 2020, 91, e13357. [Google Scholar] [CrossRef] [Green Version]
- Radzikowska, U.; Rinaldi, A.O.; Çelebi Sözener, Z.; Karaguzel, D.; Wojcik, M.; Cypryk, K.; Akdis, M.; Akdis, C.A.; Sokolowska, M. The influence of dietary fatty acids on immune responses. Nutrients 2019, 11, 2990. [Google Scholar] [CrossRef] [Green Version]
- Bartholome, A.L.; Albin, D.M.; Baker, D.H.; Holst, J.J.; Tappenden, K.A. Supplementation of total parenteral nutrition with butyrate acutely increases structural aspects of intestinal adaptation after an 80% jejunoileal resection in neonatal piglets. JPEN J. Parenter. Enter. Nutr. 2004, 28, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Hornef, M.W.; Dupont, A. The intestinal epithelium as guardian of gut barrier integrity. Cell. Microbiol. 2015, 17, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Li, B.-R.; Wu, J.; Li, H.-S.; Jiang, Z.-H.; Zhou, X.-M.; Xu, C.-H.; Ding, N.; Zha, J.-M.; He, W.-Q. In vitro and in vivo approaches to determine intestinal epithelial cell permeability. J. Vis. Exp. 2018, 140, 57032. [Google Scholar]


| Gene | Forward | Reverse | Product Size (bp) 1 |
|---|---|---|---|
| Lgr5 | CCTTGGCCCTGAACAAAATA | ATTTCTTTCCCAGGGAGTGG | 110 |
| Muc 2 | GCTGGCCGACAACAAGAAGA | TGGTGGGAGGATGGTTGGAA | 126 |
| Lyz | GCAAGACACCCAAAGCAGTT | ATGCCACCCATGCTTTAACG | 132 |
| ChgA | TGAAGTGCATCGTCGAGGTC | GAGGATCCGTTCATCTCCTCG | 104 |
| ALPI | AGGAACCCAGAGGGACCATTC | CACAGTGGCTGAGGGACTTAGG | 83 |
| GAPDH | ATTCCACCCACGGCAAGTTC | CACCAGCATCACCCCATTTG | 126 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joo, S.-S.; Gu, B.-H.; Park, Y.-J.; Rim, C.-Y.; Kim, M.-J.; Kim, S.-H.; Cho, J.-H.; Kim, H.-B.; Kim, M. Porcine Intestinal Apical-Out Organoid Model for Gut Function Study. Animals 2022, 12, 372. https://doi.org/10.3390/ani12030372
Joo S-S, Gu B-H, Park Y-J, Rim C-Y, Kim M-J, Kim S-H, Cho J-H, Kim H-B, Kim M. Porcine Intestinal Apical-Out Organoid Model for Gut Function Study. Animals. 2022; 12(3):372. https://doi.org/10.3390/ani12030372
Chicago/Turabian StyleJoo, Sang-Seok, Bon-Hee Gu, Yei-Ju Park, Chae-Yun Rim, Min-Ji Kim, Sang-Ho Kim, Jin-Ho Cho, Hyeun-Bum Kim, and Myunghoo Kim. 2022. "Porcine Intestinal Apical-Out Organoid Model for Gut Function Study" Animals 12, no. 3: 372. https://doi.org/10.3390/ani12030372
APA StyleJoo, S.-S., Gu, B.-H., Park, Y.-J., Rim, C.-Y., Kim, M.-J., Kim, S.-H., Cho, J.-H., Kim, H.-B., & Kim, M. (2022). Porcine Intestinal Apical-Out Organoid Model for Gut Function Study. Animals, 12(3), 372. https://doi.org/10.3390/ani12030372






