Description of a New Cobra (Naja Laurenti, 1768; Squamata, Elapidae) from China with Designation of a Neotype for Naja atra
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Molecular Phylogenetic Analysis
2.3. Morphological Analysis
3. Results
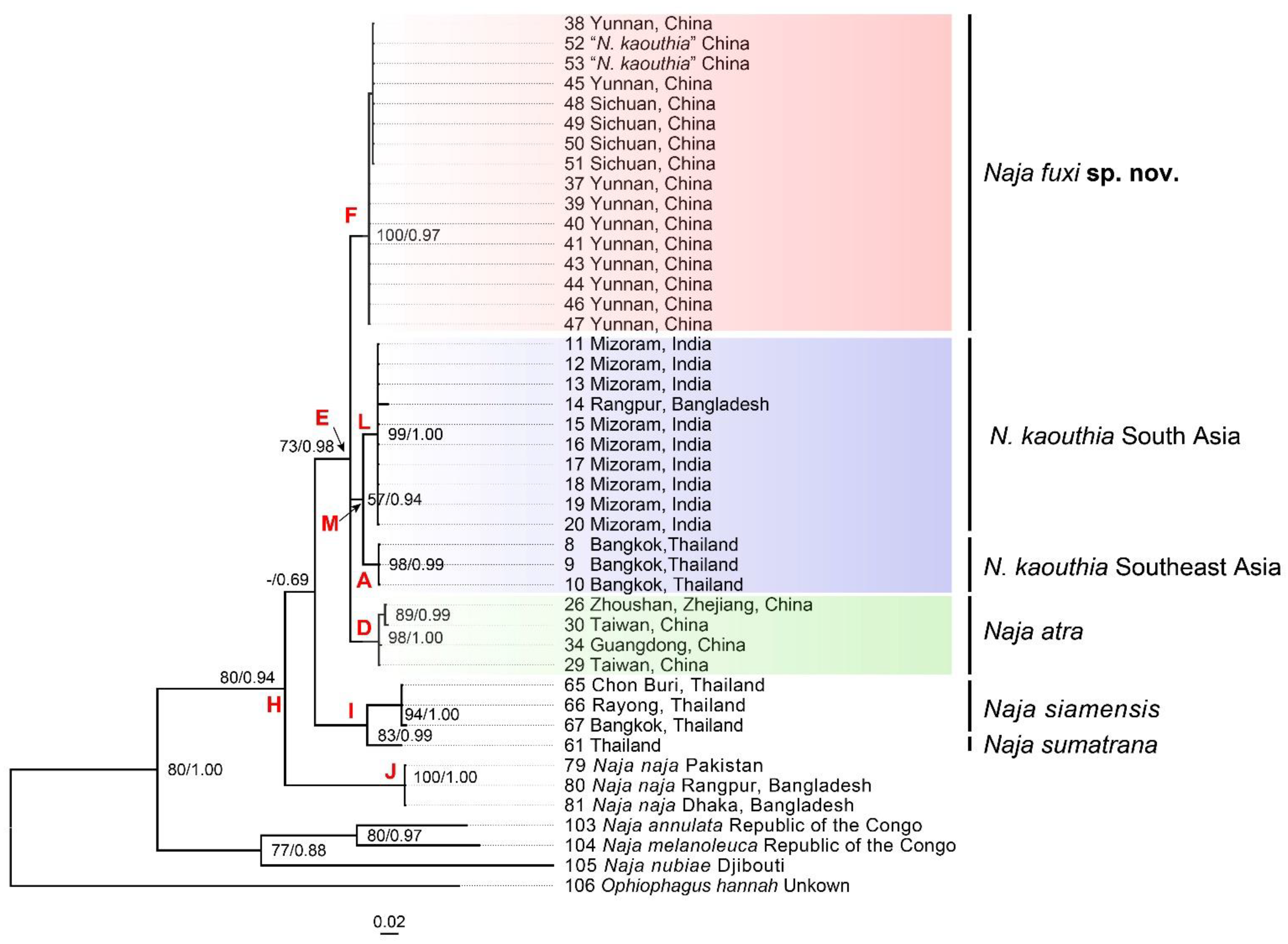
3.1. Molecular Results
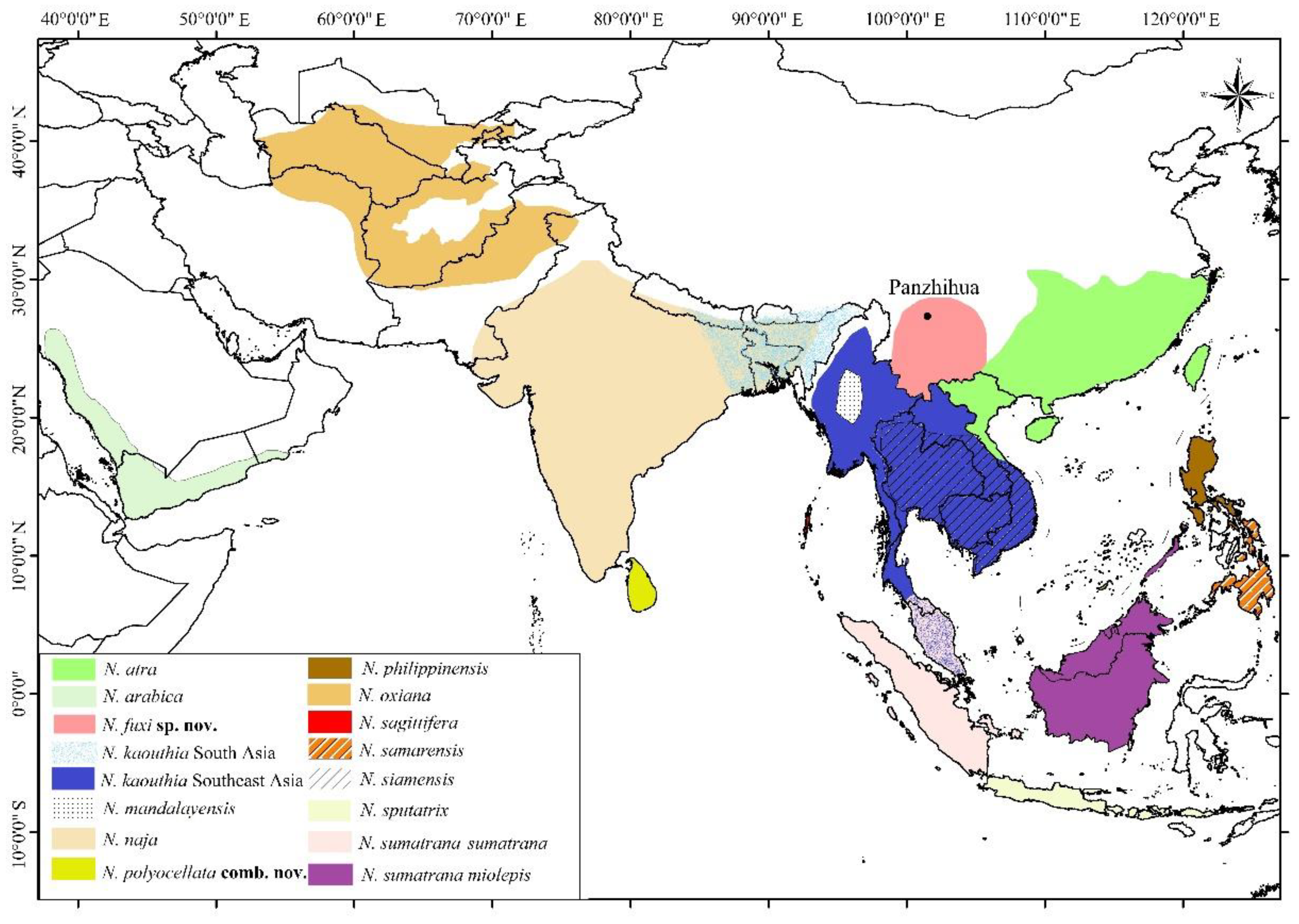
3.2. Morphological Results and Taxonomic Conclusion
3.3. Taxonomic Account
3.3.1. Naja atra Cantor, 1842
3.3.2. Naja kaouthia Lesson, 1831
3.3.3. Naja fuxi sp. nov. Shi, Vogel, Chen, Ding
4. Discussion
4.1. The Taxonomy of Some Asian Cobras
4.2. Geographical Variations of N. kaouthia
4.3. Spitting Behavior and Venom Discharge Orifice
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alirol, E.; Sharma, S.K.; Bawaskar, H.S.; Kuch, U.; Chappuis, F. Snake bite in South Asia: A review. PLoS Negl.Trop. Dis. 2010, 4, e603. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.C.; Yang, W.F. Epidemiology of venomous snakebites in China. J. Emerg. Tradit. Chin. Med. 2012, 21, 778–780. [Google Scholar]
- Wei, J.-F.; Lü, Q.-M.; Jin, Y.; Li, D.-S.; Xiong, Y.-L.; Wang, W.-Y. Alpha-neurotoxins of Naja atra and Naja kaouthia snakes in different regions. Acta Biochim. Biophys. Sin 2003, 35, 683–688. [Google Scholar] [PubMed]
- Sintiprungrat, K.; Watcharatanyatip, K.; Senevirathne, W.D.S.T.; Chaisuriya, P.; Chokchaichamnankit, D.; Srisomsap, C.; Ratanabanangkoon, K. A comparative study of venomics of Naja naja from India and Sri Lanka, clinical manifestations and antivenomics of an Indian polyspecific antivenom. J. Proteom. 2015, 132, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Ratnarathorn, N.; Harnyuttanakorn, P.; Chanhome, L.; Evans, S.E.; Day, J.J. Geographical differentiation and cryptic diversity in the monocled cobra, Naja kaouthia (Elapidae), from Thailand. Zool. Scr. 2019, 48, 711–726. [Google Scholar] [CrossRef]
- Senji Laxme, R.R.; Khochare, S.; de Souza, H.F.; Ahuja, B.; Suranse, V.; Martin, G.; Whitaker, R.; Sunagar, K. Beyond the ‘big four’: Venom profiling of the medically important yet neglected Indian snakes reveals disturbing antivenom deficiencies. PLoS Negl. Trop. Dis. 2019, 13, e0007899. [Google Scholar] [CrossRef]
- Chen, Z.N.; Shi, S.C.; Vogel, G.; Ding, L.; Shi, J.S. Multiple lines of evidence reveal a new species of Krait (Squamata, Elapidae, Bungarus) from Southwestern China and Northern Myanmar. ZooKeys 2021, 1025, 35–71. [Google Scholar] [CrossRef]
- Deraniyagala, P.E.P. A new colour variety of cobra from Ceylon & South India. Ceylon J. Sci. 1939, 20, 233–235. [Google Scholar]
- Deraniyagala, P.E.P. The taxonomy of the cobras of southeastern Asia, Part 2. Spolia Zeylan. 1961, 29, 205–232. [Google Scholar]
- Deraniyagala, P.E.P. Some new races of the Python, Chrysopelea, binocellate cobra and Tith-Polonga inhabiting Ceylon and India. Spoloa Zeylan. 1945, 24, 103–113. [Google Scholar]
- Deraniyagala, P.E.P. The taxonomy of the cobras of southeastern Asia. Spolia Zeylan. 1960, 29, 41–63. [Google Scholar]
- Slowinski, J.B.; Wüster, W. A new cobra (Elapidae: Naja) from Myanmar (Burma). Herpetologica 2000, 56, 257–270. [Google Scholar]
- Wüster, W. Taxonomic changes and toxinology: Systematic revisions of the asiatic cobras (Naja naja complex). Toxicon 1995, 34, 99–406. [Google Scholar] [CrossRef] [PubMed]
- Wüster, W. The cobras of the genus Naja in Inida. Hamadryad 1998, 23, 15–32. [Google Scholar]
- Wüster, W.; Thorpe, R.S. Population affinities of the Asiatic cobra (Naja naja) species complex in south-east Asia: Reliability and random resampling. Linn. Soc. Lond. Biol. J. 1989, 36, 391–409. [Google Scholar] [CrossRef]
- Wüster, W.; Thorpe, R.S. Asiatic cobras: Population systematics of the Naja naja species complex (Serpentes: Elapidae) in India and Central Asia. Herpetologica 1992, 48, 69–85. [Google Scholar]
- Wüster, W.; Thorpe, R.S. Dentitional phenomena in cobras revisited: Spitting and fang structure in the Asiatic species of Naja (Serpentes: Elapidae). Herpetologica 1992, 48, 424–434. [Google Scholar]
- Wüster, W.; Warrell, D.A.; Jintakune, P. Redescription of Naja siamensis (Serpentes: Elapidae), a widely overlooked spitting cobra from S.E. Asia: Geographic variation, medical importance and designation of a neotype. J. Zool. 1997, 243, 771–788. [Google Scholar] [CrossRef]
- Boulenger, G.A. Colubridae (Opisthoglyphae and Proteroglyphae), Amblycephalidae, and Viperidae. Catalogue of the snakes in the British Museum (Natural History). In Order of the Trustees; 1896; Volume 3. [Google Scholar]
- Nicholson, E. Indian Snakes: An Elementary Treatise on Ophiology with a Descriptive Catalogue of the Snakes Found in India and the Adjoining Countries; Higgibotham and Company: Fort Worth, TX, USA, 1874. [Google Scholar]
- Linnaeus, C. Holmiae: Laurentii Salvii. In Systemae Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis; Emanuel, B., Ed.; Impensis Georg: Leipzig, Germany, 1758. [Google Scholar]
- Wallach, V.; Williams, K.L.; Boundy, J. Snakes of the World. A Catalogue of Living and Extinct Species; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Uetz, P. The reptile database turns 20. Herpetol. Rev. 2006, 47, 330–334. [Google Scholar]
- Wallach, V.; Wüster, W.; Broadley, D.G. In praise of subgenera: Taxonomic status of cobras of the genus Naja Laurenti (Serpentes: Elapidae). Zootaxa 2009, 2236, 26–36. [Google Scholar] [CrossRef]
- Uetz, P.; Freed, P.; Hošek, J. The Reptile Database. 2022. Available online: http://www.reptile-database.org (accessed on 26 October 2022).
- Attarde, S.; Khochare, S.; Iyer, A.; Dam, P.; Martin, G.; Sunagar, K. Venomics of the enigmatic Andaman Cobra (Naja sagittifera) and the preclinical failure of Indian antivenoms in Andaman and Nicobar islands. Front. Pharm. 2021, 12, 768210. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, E.; Nazarizadeh, M.; Fatemizadeh, F.; Khani, A.; Kaboli, M. The phylogeny, phylogeography, and diversification history of the westernmost Asian cobra (Serpentes: Elapidae: Naja oxiana) in the Trans-Caspian region. Ecol. Evol. 2020, 11, 2024–2039. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S.; Lalremsanga, H.T.; Rahman, M.M.M.; Ahsan, F.; Biakzuala, L.; Kumar, V.; Chandra, K.; Siddiki, A.M.A.M.Z. DNA barcoding elucidates the population genetic diversity of venomous cobra species (Reptilia: Elapidae) in Indo-Bangladesh region. Mitochondrial DNA B 2020, 5, 2525–2530. [Google Scholar] [CrossRef]
- Supikamolseni, A.; Ngaoburanawit, N.; Sumontha, M.; Chanhome, L.; Suntrarachun, S.; Peyachoknagul, S.; Srikulnath, K. Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand. Genet. Mol. Res 2015, 14, 13981–13997. [Google Scholar] [CrossRef] [PubMed]
- Zhao, E.M.; Huang, M.H.; Zong, Y. Fauna Sinica, Reptilia, Squamata. Serpentes; Science Press: Beijing, China, 1998. [Google Scholar]
- Zhao, E.M. Coloured Atlas of Reptiles of Sichuan; China Forestry Press: Beijing, China, 2003. [Google Scholar]
- Muansanga, L.; Biakzuala, L.; Rinsanga, L.; Vanlalchhuana, M.; Hmar, Z.G.; Vanlalhrima; Romalsawma; Lianzela, S.; Vanlalhriatzuala; Laltlanchhuaha, H.; et al. Natural history notes. Naja kaouthia (Monocled cobra): Diet and reproduction. Herpetol. Rev. 2020, 51, 356. [Google Scholar]
- Lin, L.H.; Qu, Y.F.; Li, H.; Zhou, K.Y.; Ji, X. Genetic structure and demographic history should inform conservation: Chinese cobras currently treated as homogenous show population divergence. PLoS ONE 2012, 7, e36334. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burbrink, F.T.; Lawson, R.; Slowinski, J.B. Mitochondrial DNA phylogeography of the polytypic North American rat snake (Elaphe obsoleta): A critique of the subspecies concept. Evolution 2000, 54, 2107–2118. [Google Scholar] [PubMed]
- Arevalo, E.; Davis, S.K.; Sites Jr, J.W. Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in central Mexico. Syst. Biol. 1994, 43, 387–418. [Google Scholar] [CrossRef]
- Che, J.; Chen, H.M.; Yang, J.X.; Jin, J.Q.; Jiang, K.; Yuan, Z.Y.; Murphy, R.W.; Zhang, Y.P. Universal COI primers for DNA barcoding amphibians. Mol. Ecol. Resour. 2012, 12, 247–258. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Collet, M.; Jean-Francois, T. Une nouvelle et remarquable espèce de naja semi-aquatique (Elapidae, sous-genre Boulengerina Dollo, 1886) de la République Démocratique du Congo. Bull. Société Herpétologique Fr. 2020, 173, 41–52. [Google Scholar]
- Li, J.N.; Liang, D.; Wang, Y.; Guo, P.; Zhang, P.; Huang, S.; Zhang, P. A large-scale systematic framework of Chinese snakes based on a unified multilocus marker system. Mol. Phylogenet. Evol. 2020, 148, 106807. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Calcott, B.; Ho, S.Y.; Guindon, S. Partition Finder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 2001, 294, 2310–2314. [Google Scholar] [CrossRef] [PubMed]
- Slowinski, J.B. A Phylogenetic Analysis of Bungarus (Elapidae) Based on Morphological Characters. J. Herpetol. 1994, 28, 440–446. [Google Scholar] [CrossRef]
- Vogel, G.; David, P.; Pauwels, O.S. A review of morphological variation in Trimeresurus popeiorum (Serpentes: Viperidae: Crotalinae), with the description of two new species. Zootaxa 2004, 727, 1–63. [Google Scholar] [CrossRef]
- Kuch, U.; Kizirian, D.; Truong, N.Q.; Lawson, R.; Donnelly, M.A.; Mebs, D. A new species of Krait (Squamata: Elapidae) from the Red River System of Northern Vietnam. Copeia 2005, 2005, 818–833. [Google Scholar] [CrossRef]
- Dowling, H.G. A proposed standard system of counting ventrals in snakes. Br. J. Herpetol. 1951, 1, 97–99. [Google Scholar]
- Jiang, K. A method for evaginating the hemipenis of Preserved snakes. Sich. J. Zool. 2010, 29, 122–123. [Google Scholar]
- Young, B.A.; Dunlap, K.; Koenig, K.; Singer, M. The buccal buckle: The functional morphology of venom spitting in cobras. J. Exp. Biol. 2004, 207, 3483–3494. [Google Scholar] [CrossRef] [PubMed]
- Kazandjian, T.D.; Petras, D.; Robinson, S.D.; van Thiel, J.; Greene, H.W.; Arbuckle, K.; Casewell, N.R. Convergent evolution of pain-inducing defensive venom components in spitting cobras. Science 2021, 371, 386–390. [Google Scholar] [CrossRef]
- Trape, J.F.; Chirio, L.; Broadley, D.G.; Wüster, W. Phylogeography and systematic revision of the Egyptian cobra (Serpentes: Elapidae: Naja haje) species complex, with the description of a new species from West Africa. Zootaxa 2009, 2236, 1–25. [Google Scholar] [CrossRef]
- Wüster, W.; Thorpe, R.S.; Cox, M.J.; Jintakune, P.; Nabhitabhata, J. Population systematics of the snake genus Naja (Reptilia: Serpentes: Elapidae in Indochina: Multivariate morphomertrics and comparative mitochondrial DNA sequencing (cytochrome oxidase I). J. Evol. Biol. 1995, 8, 493–510. [Google Scholar] [CrossRef]
- Huang, S. Sinoophis; The Strait Publishing & Distribution Group: Fuzhou, China, 2021. [Google Scholar]
- Cantor, T.E. Zoology of Chusan; Bennett, J., Ed.; Society for the Study of Amphibians and Reptiles: Calcutta, India, 1842. [Google Scholar]
- Paterna, A. Spitting behaviour in the Chinese cobra Naja atra. Herpetol. Bull. 2019, 148, 22–25. [Google Scholar] [CrossRef]
- Lesson, R.P. Catalogue des Reptiles qui font partie d’une Collection zoologique recueille dans l’Inde continentale ou en Afrique, et apportée en France par M. Lamare-Piquot. Bull. Sci. Nat. Géologie 1831, 25, 119–123. [Google Scholar]
- Lalremsanga, H.T.; Senthil Kumar, N. Annexure—IX Project Completion Report: Large-Scale DNA Barcoding Assessment of Snakes in Mizoram, Indo-Myanmar Biodiversity Hotspot; Mizoram University: Mizoram, India, 2020. [Google Scholar]
- Santra, V.; Wüster, W. Natural history notes. Naja kaouthia (Monocled cobra): Behavior/Spitting. Herpetol. Rev. 2017, 48, 455–456. [Google Scholar]
- Ashaharraza, K.; Lalremsanga, H.T. Notes on the diet of the Monocled Cobra Naja kaouthia Lesson, 1831 with a report on a predator of the King Cobra Ophiophagus hannah (Cantor, 1836). Sauria 2020, 42, 45–48. [Google Scholar]
- Yang, D.T.; Rao, D.Q. Amphibia and Reptilia of Yunnan; Yunnan Science and Technology Press: Kuming, China, 2008. [Google Scholar]
- Rao, S.; Bai, J. Experience of observation and nursing of 126 cases of snake bites. Med. Pharm. Yunnan 2016, 37, 127–129. [Google Scholar]
- Nutaphand, W. Cobra; Pata Zoo Publications: Bangkok, Thailand, 1986. [Google Scholar]
- Giri, R.; Baral, R.; Giri, R.; Shah, K.B.; Tillack, F. First records of the spitting behaviour of monocled cobra (Naja kaouthia) from Nepal. Russ. J. Herpetol. 2021, 28, 122–124. [Google Scholar] [CrossRef]
- Tumwipat, B.; Nutaphand, W. The Treatment of Poisonous Snakebite Victims and the Poisonous Snakes of Thailand; Peekhanet Print Shop: Bangkok, Thailand, 1982. [Google Scholar]
- Wüster, W.; Thorpe, R.S. Naja siamensis, a cryptic species of venomous snake revealed by mtDNA sequencing. Experientia 1994, 50, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Slowinski, J.B.; Keogh, J.S. Phylogenetic relationships of elapid snakes based on cytochrome b mtDNA sequences. Mol. Phylogenet. Evol 2000, 15, 157–164. [Google Scholar] [CrossRef]
- Han, F.; Bahain, J.-J.; Deng, C.; Boëda, E.; Hou, Y.; Wei, G.; Huang, W.; Garcia, T.; Shao, Q.; He, C.; et al. The earliest evidence of hominid settlement in China: Combined electron spin resonance and uranium series (ESR/U-series) dating of mammalian fossil teeth from Longgupo cave. Quat. Int. 2017, 434, 75–83. [Google Scholar] [CrossRef]
- Auffenberg, W. The herpetofauna of Komodo, with notes on adjacent areas. Bull. Fla. State Mus. Biol. Sci. 1980, 25, 39–156. [Google Scholar]
- Leviton, A.E.; Brown, R.M.; Siler, C.D. The dangerously venomous snakes of the Philippine Archipelago. In The Coral Triangle: The 2011 Hearst Biodiversity Philippine Expedition; Williams, G.C., Gosliner, T.M., Eds.; California Academy of Sciences: San Francisco, CA, USA, 2014. [Google Scholar]








| Sample ID | Taxa | Voucher Numbers | Locality | cyt b | ND4 | COI |
|---|---|---|---|---|---|---|
| 1 | Naja kaouthia | WW 585 | Chumphon Province, Thailand | GQ359507 | EU624209 | |
| 2 | N. kaouthia | isolate 818 | Thailand | MT346707 | MT346903 | |
| 3 | N. kaouthia | Unknown | LR880477 | |||
| 4 | N. kaouthia | isolate 813 | Vietnam | MT346706 | MT346902 | |
| 5 | N. kaouthia | isolate 812 | Vietnam | MT346705 | MT346901 | |
| 6 | N. kaouthia | CAS 206602 | Ayeyarwady, Myanmar | AF217835 | AY058982 | |
| 7 | N. kaouthia | isolate 839 | Myanmar | MT346708 | MT346904 | |
| 8 | N. kaouthia | NKA3 | Bangkok, Thailand | AB920185 | ||
| 9 | N. kaouthia | NKA2 | Bangkok, Thailand | AB920184 | ||
| 10 | N. kaouthia | NKA1 | Bangkok, Thailand | AB920183 | ||
| 11 | N. kaouthia | PUCZM/X/SL787 | Mizoram, India | MH107858 | ||
| 12 | N. kaouthia | PUCZM/X/SL774 | Mizoram, India | MH107857 | ||
| 13 | N. kaouthia | PUCZM/X/SL773 | Mizoram, India | MH107856 | ||
| 14 | N. kaouthia | CUZSI81 | Badarganj, Rangpur, Bangladesh | KM521202 | ||
| 15 | N. kaouthia | MZMU1163 | Aizawl, Mizoram, India | MT348385 | ||
| 16 | N. kaouthia | MZMU982 | Mizoram University Campus, India | MT348386 | ||
| 17 | N. kaouthia | MZMU1170 | Aizawl, Mizoram, India | MT348387 | ||
| 18 | N. kaouthia | MZMU998 | Mizoram University Campus, India | MT348388 | ||
| 19 | N. kaouthia | MZMU1426 | Mizoram University Campus, India | MT348389 | ||
| 20 | N. kaouthia | MZMU1195 | Mizoram University Campus, India | MT348390 | ||
| 21 | N. sagittifera | isolate 400 | Unknown | MT346720 | MT346916 | |
| 22 | N. sagittifera | isolate 1815 | Unknown | MT346721 | MT346917 | |
| 23 | N. oxiana | isolate 838 | Unknown | MT346714 | MT346910 | |
| 24 | N. oxiana | isolate 832 | Unknown | MT346713 | MT346909 | |
| 25 | N. atra | LJT-FJ2020060 | Dongping Mountain, Xiamen, Fujian, China | ON221325 | ON221392 | |
| 26 | N. atra | GXNU 2021070801 | Zhoushan Island, Zhejiang, China | ON221326 | ||
| 27 | N. atra | C29 | Eastern China | JN160670 | ||
| 28 | N. atra | isolate 793 | Taiwan, China | MT346704 | MT346900 | |
| 29 | N. atra | RN1570 | Taiwan, China | KP749826 | ||
| 30 | N. atra | RN1298 | Taiwan, China | KP749825 | ||
| 31 | N. atra | CIB 20190430 | Wuyishan, Fujian, China | ON221327 | ||
| 32 | N. atra | CIB 2021042007 | Luofushan, Guangdong, China | ON221328 | ||
| 33 | N. atra | CIB CR430 | Guangzhou, Guangdong, China | ON221329 | ON237986 | ON221393 |
| 34 | N. atra | CIB 093930 | Purchased from Guangzhou, Guangdong Province, China | EU913475 | EU913475 | EU913475 |
| 35 | N. atra | C17 | Southcentral China | JN160658 | ||
| 36 | N. atra | LJT-GD2020103 | Dinghu Mountain, Zhaoqing, Guangdong, China | ON221330 | ||
| 37 | N. fuxi sp. nov. | KIZ 090071 | Simao, Pu’er, Yunnan, China | ON221331 | ON237987 | |
| 38 | N. fuxi sp. nov. | KIZ F20180066 | Simao, Pu’er, Yunnan, China | ON221332 | ON237988 | |
| 39 | N. fuxi sp. nov. | KIZ 20180801 | Simao, Pu’er, Yunnan, China | ON221333 | ON237989 | |
| 40 | N. fuxi sp. nov. | KIZ 2020090301 | Menglian, Pu’er, Yunnan, China | ON221334 | ON221394 | ON237990 |
| 41 | N. fuxi sp. nov. | KIZ 2020091201 | Ximeng, Pu’er, Yunnan, China | ON221335 | ON221395 | ON237991 |
| 42 | N. fuxi sp. nov. (“N. atra”) | C31 | Pingbian, Yunnan, China, China | JN160672 | ||
| 43 | N. fuxi sp. nov. | CIB YNJC0022 | Jiangcheng, Pu’er, Yunnan, China | ON221336 | ON221396 | ON237992 |
| 44 | N. fuxi sp. nov. | CIB DL000020 | Jiangcheng, Pu’er, Yunnan, China | ON221337 | ON221397 | ON237993 |
| 45 | N. fuxi sp. nov. | CIB DL000070 | Jiangcheng, Pu’er, Yunnan, China | ON221338 | ON221398 | ON237994 |
| 46 | N. fuxi sp. nov. | CIB DL000096 | Jiangcheng, Pu’er, Yunnan, China | ON221339 | ON237995 | |
| 47 | N. fuxi sp. nov. | CIB DL000249 | Jiangcheng, Pu’er, Yunnan, China | ON221340 | ON221399 | ON237996 |
| 48 | N. fuxi sp. nov. | CIB 2018053147 | Panchizhua, Sichuan, China | ON221341 | ON237997 | |
| 49 | N. fuxi sp. nov. | CIB 098874 | Miyi, Panzhihua, Sichuan, China | ON221342 | ON237998 | |
| 50 | N. fuxi sp. nov. | CIB 098875 | Miyi, Panzhihua, Sichuan, China | ON221343 | ON237999 | |
| 51 | N. fuxi sp. nov. | CIB YS2 | Yanyuan, Panzhihua, Sichuan, China | ON221344 | ON221400 | ON238000 |
| 52 | N. fuxi sp. nov. (“N. kaouthia”) | CHS026 | China | MK064598 | ||
| 53 | N. fuxi sp. nov. (“N. kaouthia”) | CHS726 | China | MK064840 | ||
| 54 | N. sumatrana sumatrana | isolate 589 | Indonesia | MT346737 | MT346933 | |
| 55 | N. sumatrana sumatrana | isolate 587 | Malaysia | MT346736 | MT346932 | |
| 56 | N. sumatrana sumatrana | isolate 586 | Malaysia | MT346735 | MT346931 | |
| 57 | N. sumatrana sumatrana | isolate 295 | Indonesia | MT346734 | MT346930 | |
| 58 | N. sumatrana sumatrana | isolate 294 | Indonesia | MT346733 | MT346929 | |
| 59 | N. sumatrana miolepis | isolate 1827 | Philippines | MT346738 | MT346934 | |
| 60 | N. sumatrana miolepis | isolate 188 | Malaysia | MT346732 | MT346928 | |
| 61 | N. sumatrana | NSU1 | Thailand | AB920186 | ||
| 62 | N. siamensis | isolate 811 | Viet Nam | MT346728 | MT346926 | |
| 63 | N. siamensis | isolate 810 | Viet Nam | MT346727 | MT346925 | |
| 64 | N. siamensis | isolate 26 | Thailand | MT346725 | MT346923 | |
| 65 | N. siamensis | SC01f | Bangkok, Thailand | LC086063 | ||
| 66 | N. siamensis | NSI3 | Rayong, Thailand | AB920188 | ||
| 67 | N. siamensis | NSI2 | Bangkok, Thailand | AB920187 | ||
| 68 | N. mandalayensis | CAS 204375-6 | Monywa, Myanmar | AF155211 | ||
| 69 | N. samarensis | isolate 1803 | Philippines | MT346723 | MT346919 | |
| 70 | N. samarensis | isolate 1806 | Philippines | MT346724 | MT346920 | |
| 71 | N. samarensis | isolate 841 | Philippines | MT346722 | MT346918 | |
| 72 | N. philippinensis | isolate 1828 | Philippines | MT346719 | MT346915 | |
| 73 | N. philippinensis | isolate 1825 | Philippines | MT346718 | MT346914 | |
| 74 | N. sputatrix | isolate 584 | Indonesia | MT346730 | MT346922 | |
| 75 | N. sputatrix | isolate 583 | Indonesia | MT346729 | MT346921 | |
| 76 | N. naja | ZMUVAS8 | Pakistan | MK936173 | ||
| 77 | N. naja | isolate 579 | Nepal | MT346711 | MT346907 | |
| 78 | N. polyocellata comb. nov. | isolate 581 | Sri Lanka | MT346712 | MT346908 | |
| 79 | N. naja | ZMUVAS21 | Pakistan | MK941841 | ||
| 80 | N. naja | BDS80/CUZSI80 | Pirganj, Rangpur, Bangladesh | KM521201 | ||
| 81 | N. naja | DUZM_S052.1 | Dhaka, Bangladesh | MT215095 | ||
| 82 | N. arabica | isolate 1678/WW 1678 | Taif, Saudi Arabia | GQ387104 | GQ387075 | |
| 83 | N. haje | isolate 1653 | northern Nigeria | GQ387099 | GQ387070 | |
| 84 | N. senegalensis | isolate 2018 | Mali | GQ387112 | GQ387083 | |
| 85 | N. anchietae | isolate 1892 | Botswana | MT346741 | GQ387087 | |
| 86 | N. annulifera | isolate 881 | Zimbabwe | GQ359504 | GQ359586 | |
| 87 | N. nivea | isolate 1482 | Unknown | MT346760 | MT346936 | |
| 88 | N. mossambica | isolate 882 | Zimbabwe | MT346655 | MT346848 | |
| 89 | N. nigricincta | isolate 877 | Namibia | MT346661 | MT346854 | |
| 90 | N. ashei | isolate 1394 | Kenya | MT346647 | MT346842 | |
| 91 | N. nigricollis | isolate 3103 | Kenya | MT346680 | MT346874 | |
| 92 | N. katiensis | isolate 2022 | Senegal | MT346653 | MT346845 | |
| 93 | N. nubiae | isolate 837 | Egypt | GQ359497 | GQ359579 | |
| 94 | N. pallida | isolate 1080 | Tanzania | GQ359496 | GQ359578 | |
| 95 | N. annulata | isolate 2717 | Republic of the Congo | MT346700 | MT346896 | |
| 96 | N. christyi | isolate PM147 | Democratic Republic of the Congo | MT346701 | MT346897 | |
| 97 | N. savannula | isolate 2046 | Benin | MH337600 | MH337406 | |
| 98 | N. subfulva | isolate 2654 | South Africa | MH337633 | MH337439 | |
| 99 | N. melanoleuca | isolate 2721 | Democratic Republic of the Congo | MH337589 | MH337395 | |
| 100 | N. peroescobari | isolate 1197 | Sao Tome and Principe | MH337634 | MH337440 | |
| 101 | N. guineensis | isolate 2491 | Togo | MH337580 | MH337386 | |
| 102 | N. multifasciata | isolate 4313 | Democratic Republic of the Congo | MT346702 | MT346898 | |
| 103 | N. annulata | UPRP:558270 | Impongui, Likouala, Republic of the Congo | MH274482 | ||
| 104 | N. melanoleuca | UPRP:558271 | Impongui, Likouala, Republic of the Congo | MH274485 | ||
| 105 | N. nubiae | USNM:Herp:589595 | Tadjourah, Day (village), Djibouti | MG700028 | ||
| 106 | Ophiophagus hannah | CIB093929 | Unkown | EU921899 | EU921899 | |
| No. | Species (Sample Numbers in Table 1) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Naja kaouthia Southeast Asia (Myanmar; Thailand; Vietnam) (1–7) | 0.0–1.2 | ||||||||||||
| 2 | N. sagittifera presumed Andaman Islands (21–22) | 2.4–2.9 | 0.0 | |||||||||||
| 3 | N. oxiana Unkown locality (23–24) | 5.0–5.5 | 5.3–5.4 | 0.0 | ||||||||||
| 4 | N. atra Southern and Southeastern China (25–28, 31–36) | 4.6–6.3 | 6.1–6.7 | 5.2–5.8 | 0.1–2.1 | |||||||||
| 5 | N. fuxisp. nov. Southwestern China (Sichuan and Yunnan) (37–51) | 5.3–6.7 | 6.2–6.4 | 4.3–4.4 | 4.1–5.0 | 0.0–0.3 | ||||||||
| 6 | N. sumatrana sumatrana Indonesia and Malaysia (54–58) | 8.2–9.2 | 8.5–8.9 | 7.4–7.8 | 6.9–7.6 | 6.7–7.5 | 0.0–1.2 | |||||||
| 7 | N. sumatrana miolepis (Malaysia and Philippines (59–60) | 8.4–9.1 | 7.8–8.3 | 7.5 | 7.6–8.4 | 7.6–7.8 | 2.9–3.4 | 0.2 | ||||||
| 8 | N. siamensis Viet Nam and Thailand (62–64) | 9.7–0.9 | 9.1–9.7 | 8.4–9.1 | 8.8–10.1 | 8.1–8.9 | 4.3–5.0 | 4.7–5.0 | 0.0–1.7 | |||||
| 9 | N. mandalayensis Monywa, Myanmar (68) | 8.7–9.0 | 8.4 | 7.7 | 8.0–8.8 | 7.4 | 3.2–3.5 | 3.9 | 5.0–5.4 | / | ||||
| 10 | N. samarensis Philippines (68–71) | 9.7–10.4 | 9.4–9.5 | 8.4 | 8.8–9.7 | 8.1–8.5 | 5.3–6.0 | 5.8–6.0 | 7.1–7.6 | 5.7 | 0.0–0.3 | |||
| 11 | N. philippinensis Philippines (72–73) | 9.7–10.1 | 9.4–9.5 | 8.4 | 9.0–9.4 | 8.1–8.2 | 5.0–5.4 | 4.7–5.0 | 6.5–6.8 | 5.4 | 4.3 | 0.0 | ||
| 12 | N. sputatrix Indonesia (74–75) | 9.2–10.0 | 8.6–8.8 | 8.3–8.5 | 7.7–8.5 | 7.6–8.2 | 4.3–4.8 | 5.4–5.7 | 6.2–6.6 | 4.9–5.4 | 6.3–6.9 | 6.2–6.5 | 0.5 | |
| 13 | N. naja Pakistan and Nepal (76–77) | 9.5–11.5 | 9.2–10.9 | 7.9–9.4 | 8.7–10.4 | 9.8–10.4 | 9.2–107 | 9.8–107 | 10.0–11.3 | 10.5–10.7 | 10.6–11.5 | 11.4–11.8 | 8.1–9.9 | 1.4 |
| 14 | N. polyocellatacomb. nov. Sri Lanka (78) | 10.7–11.3 | 10.4 | 8.7 | 9.5–10.4 | 9.1–9.3 | 9.4–9.8 | 9.6–9.8 | 10.0–10.6 | 10.0 | 10.8–11.1 | 11.7 | 9.7–10.2 | 4.3–4.9 |
| No. | Species (Sample Numbers in Table 1) | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|---|
| 1 | Naja kaouthia Southeast Asia (Bangkok, Thailand) (8–10) | 0.0 | ||||||
| 2 | N. kaouthia South Asia (Mizoram, India and Rangpur, Bangladesh) (11–20) | 1.8–2.4 | 0.0–0.6 | |||||
| 3 | N. atra southeastern China (Zhejiang, Taiwan, Guangdong) (26, 29, 30, 34) | 2.7–3.4 | 3.1–4.2 | 0.2–0.6 | ||||
| 4 | N. fuxisp. nov. Southwestern China (Sichuan and Yunnan) (37–41, 43–53) | 2.5–2.9 | 2.0–3.0 | 2.5–3.2 | 0.0–0.2 | |||
| 5 | N. sumatrana Thailand (61) | 6.5 | 6.2–6.9 | 6.1–6.3 | 6.0–6.4 | |||
| 6 | N. siamensis Thailand (Chon Buri, Rayong, Bangkok) (65–67) | 5.6 | 5.6–6.2 | 5.7–6.5 | 5.4–5.8 | 3.7–4.0 | 0.0–0.3 | |
| 7 | N. naja Pakistan and Bangladesh (79–81) | 7.7 | 8.1–8.9 | 7.5–8.2 | 7.4–8.0 | 8.5 | 8.6–8.7 | 0.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, S.-C.; Vogel, G.; Ding, L.; Rao, D.-Q.; Liu, S.; Zhang, L.; Wu, Z.-J.; Chen, Z.-N. Description of a New Cobra (Naja Laurenti, 1768; Squamata, Elapidae) from China with Designation of a Neotype for Naja atra. Animals 2022, 12, 3481. https://doi.org/10.3390/ani12243481
Shi S-C, Vogel G, Ding L, Rao D-Q, Liu S, Zhang L, Wu Z-J, Chen Z-N. Description of a New Cobra (Naja Laurenti, 1768; Squamata, Elapidae) from China with Designation of a Neotype for Naja atra. Animals. 2022; 12(24):3481. https://doi.org/10.3390/ani12243481
Chicago/Turabian StyleShi, Sheng-Chao, Gernot Vogel, Li Ding, Ding-Qi Rao, Shuo Liu, Liang Zhang, Zheng-Jun Wu, and Ze-Ning Chen. 2022. "Description of a New Cobra (Naja Laurenti, 1768; Squamata, Elapidae) from China with Designation of a Neotype for Naja atra" Animals 12, no. 24: 3481. https://doi.org/10.3390/ani12243481
APA StyleShi, S.-C., Vogel, G., Ding, L., Rao, D.-Q., Liu, S., Zhang, L., Wu, Z.-J., & Chen, Z.-N. (2022). Description of a New Cobra (Naja Laurenti, 1768; Squamata, Elapidae) from China with Designation of a Neotype for Naja atra. Animals, 12(24), 3481. https://doi.org/10.3390/ani12243481






