Effects of Dietary Quinoa Seeds on Cecal Microorganisms and Muscle Fatty Acids of Female Luhua Chickens
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Moral Statement
2.2. Experimental Design
2.3. Experimental Diet and Feeding Management
2.3.1. Composition and Nutritional Level of Experimental Diets
2.3.2. Test Animals
2.3.3. Experimental Animal Feeding and Management
2.4. Collection and Processing of Test Samples
2.4.1. Collection and Processing of Muscle Tissue Samples
2.4.2. Collection of Cecal Microbial Samples
2.5. Determination of Indexes and Methods
2.5.1. Determination of Fatty Acid Content in Muscle
Chromatographic Condition
2.5.2. Genomic DNA Extraction, PCR Amplification and 16S rRNA High-Throughput Sequencing
Genomic DNA Extraction
PCR Amplification
Mixing and Purification of PCR Products
Library Construction and Computer Sequencing
2.5.3. Bioinformatics Analysis
2.5.4. Statistical Analysis
3. Results
3.1. Effect of Quinoa on Fatty Acid Composition and Content in Breast Muscle of Female Luhua Chicken
3.2. Effect of Quinoa on Fatty Acid Composition and Content in Leg Muscle of Female Luhua Chicken
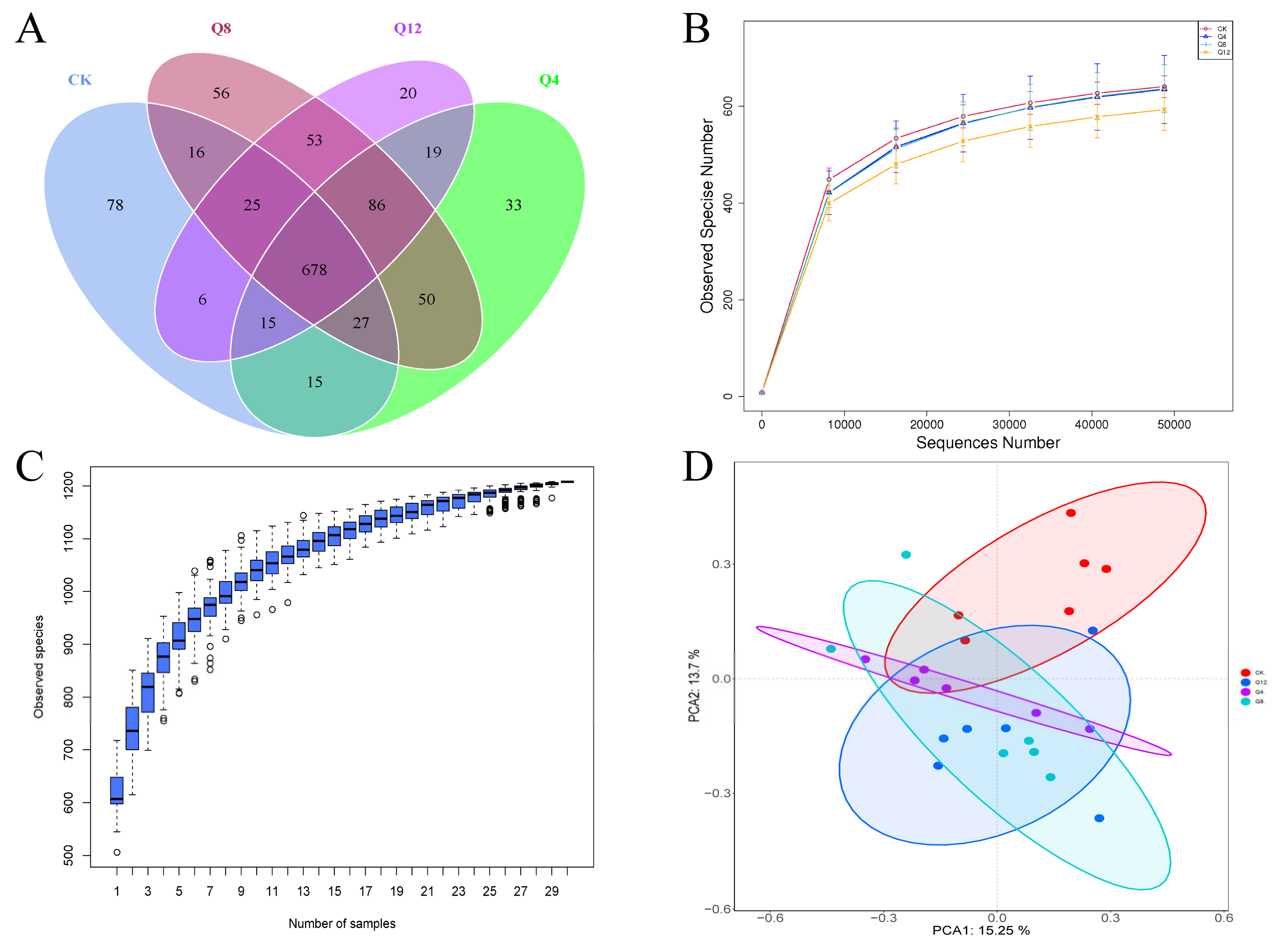
3.3. Cecal Microbial Diversity
3.4. Cecal Microbial Composition
3.5. Species Diversity Analysis
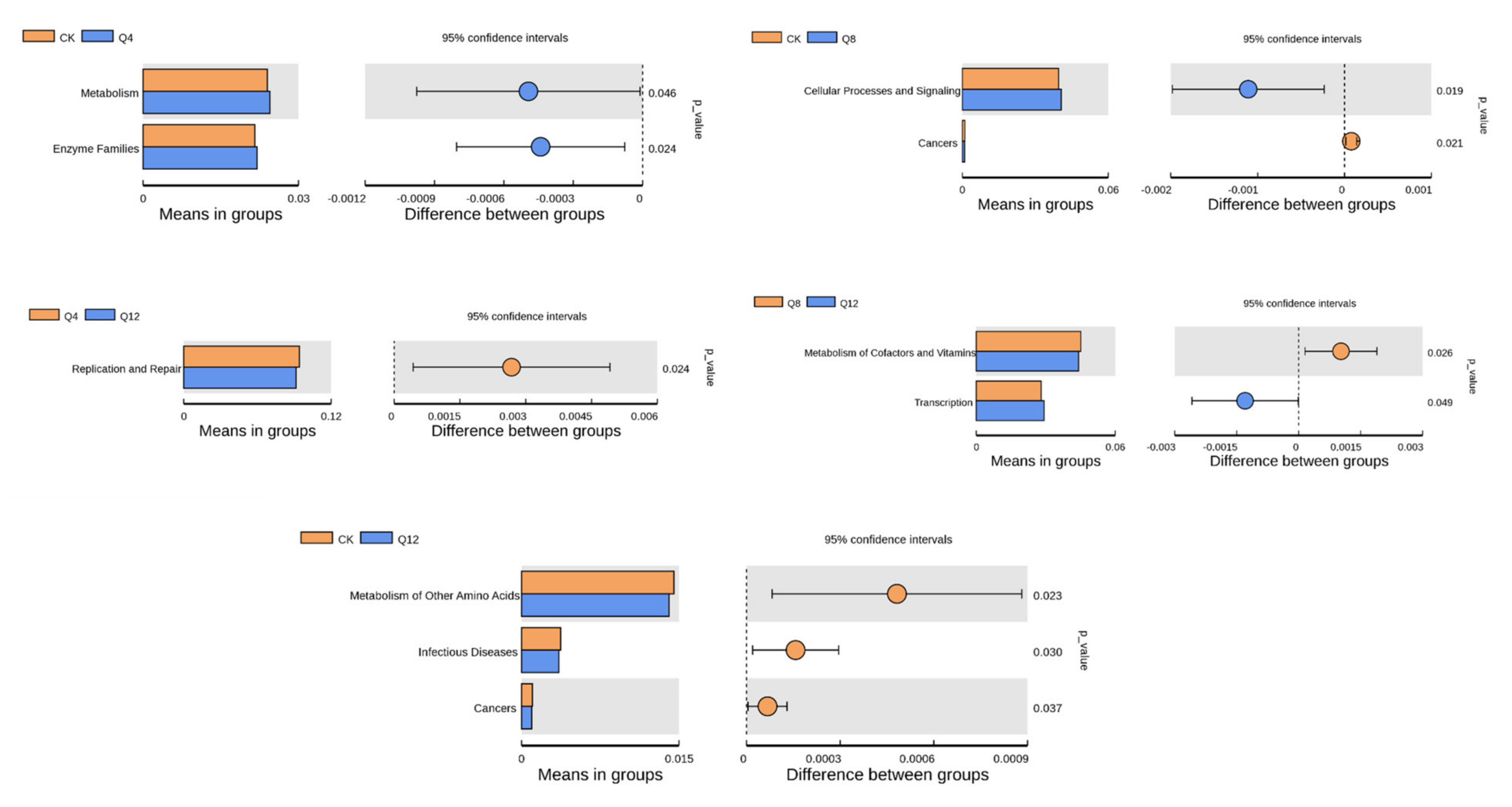
3.6. Prediction of Cecal Microbial Function
3.7. Correlation Analysis
4. Discussion
4.1. Effect of Quinoa on Fatty Acids of Female Luhua Chicken Muscle
4.2. Effect of Quinoa on Cecal Microbiota of Female Luhua Chickens
4.3. Correlation Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kang, J.K. Cultivation technology of organic quinoa in Qinghai plateau. Qinghai Agro-Technol. Ext. 2018, 4, 24–25. [Google Scholar]
- Villa, D.; Russo, L.; Kerbab, K.; Landi, M.; Rastrelli, L. Chemical and nutritional characterization of Chenopodium pallidicaule (caihua) and Chenopodium quinoa (quinoa) seeds. Emir. J. Food Agric. 2014, 26, 609–615. [Google Scholar] [CrossRef]
- Nickel, J.; Spanier, L.P.; Botelho, F.T.; Gularte, M.A.; Helbig, E. Effect of different types of processing on the total phenolic compound content, antioxidant capacity, and saponin content of Chenopodium quinoa Willd grains. Food Chem. 2016, 209, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Nowak, V.; Du, J.; Charrondière, U.R. Assessment of the nutritional composition of quinoa (Chenopodium quinoa Willd). Food Chem. 2016, 193, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Schilick, G.; Bubenheim, D.L. Quinoa: An Emerging New Crop with Potential for CELSS; National Aeronautics and Space Administration, Ames Research Center: Mountain View, CA, USA, 1993. [Google Scholar]
- Quinoa FAO. An Ancient Crop to Contribute to World Food Security; Food and Agriculture Organization: Roma, Italy, 2011. [Google Scholar]
- James, L. Chapter 1 Quinoa (Chenopodium quinoa Willd): Composition, Chemistry, Nutritional, and Functional Properties. Adv. Food Nutr. Res. 2009, 58, 1–31. [Google Scholar]
- Asher, A.; Galili, S.; Whitney, T.; Rubinovich, L. The potential of quinoa (Chenopodium quinoa) cultivation in Israel as a dual-purpose crop for grain production and livestock feed. Sci. Hortic. 2020, 272, 109534. [Google Scholar] [CrossRef]
- Kala, P.; Moudr, J. Composition and nutritinal value of quinoa (Chenopodium quinoa)—A review. Czech J. Food Sci. 2018, 18, 115–119. [Google Scholar] [CrossRef]
- Tremaroli, V.; Backhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Spanogiannopoulos, P.; Bess, E.N.; Carmody, R.N.; Turnbaugh, P.J. The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 2016, 14, 273–287. [Google Scholar] [CrossRef]
- Stephens, W.Z.; Burns, A.R.; Stagaman, K.; Wong, S.; Rawls, J.F.; Guillemin, K.; Bohannan, B.J. The composition of the zebrafish intestinal microbial community varies across development. ISME J. 2016, 10, 644–654. [Google Scholar] [CrossRef]
- Felix, S.; Fredrik, B. The gut microbiota--masters of host development and physiology. Nat. Reviews. Microbiol. 2013, 11, 227–238. [Google Scholar]
- Waite, D.W.; Taylor, M.W. Characterizing the avian gut microbiota: Membership, driving influences, and potential function. Front. Microbiol. 2014, 5, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, E.Y.; Mcbride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; Mccue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.; Ursell, L.; Parfrey, L.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Jacobsen, E.E.; Skadhauge, B.; Jacobsen, S.E. Effect of dietary inclusion of quinoa on broiler growth performance. Anim. Feed. Sci. Technol. 1997, 65, 5–14. [Google Scholar] [CrossRef]
- Johansson, K.R.; Sarles, W.B.; Shapiro, S.K. The Intestinal Microflora of Hens as Influenced by Various Carbohydrates in a Biotin-deficient Ration. J. Bacteriol. 1948, 56, 619–634. [Google Scholar] [CrossRef]
- Degolier, T.; Mahoney, S.; Duke, G. Relationships of Avian Cecal Lengths to Food Habits, Taxonomic Position, and Intestinal Lengths. Condor 1999, 101, 622–634. [Google Scholar] [CrossRef]
- Dragana, S.; Stuart, E.D.; Robert, J.H.; Mark, S.G.; Tamsyn, M.G.; Chen, H.; Volker, R.H.; Robert, J.M. Intestinal microbiota associated with differential feed conversion efficiency in chickens. Appl. Microbiol. Biotechnol. 2012, 96, 1361–1369. [Google Scholar]
- Clench, M.H.; Mathias, J.R. The avian cecum: A review. Wilson Bull. 1995, 107, 93–121. [Google Scholar]
- Chaplin, S.B. Effect of cecectomy on water and nutrient absorption of birds. J. Exp. Zool. 1989, 3, 81–86. [Google Scholar] [CrossRef]
- Li, M.; Zhou, H.; Pan, X.; Xu, T.; Zhang, Z.; Zi, X.; Jiang, Y. Cassava foliage affects the microbial diversity of Chinese indigenous geese caecum using 16S rRNA sequencing. Sci. Rep. 2017, 7, 45697. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Xiong, X.; Su, Y.; Huang, L.; Chen, C. 16S rRNA gene-based association study identified microbial taxa associated with pork intramuscular fat content in feces and cecum lumen. BMC Microbiol. 2017, 17, 162. [Google Scholar] [CrossRef]
- Zhao, X.; Ding, X.; Yang, Z.; Shen, Y.; Zhang, S.; Shi, S. Effects of Clostridium butyricum on thigh muscle lipid metabolism of broilers. Chin. J. Anim. Nutr. 2017, 29, 2884–2892. [Google Scholar]
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.D.; Han, F.L.; Lu, J.T.; Guo, J.L.; Qi, C.L.; Wang, C.L.; Xiao, S.S.; Bu, X.Y.; Wang, X.D.; Qin, J.G.; et al. Influence of dietary phospholipid on growth performance, body composition, antioxidant capacity and lipid metabolism of Chinese mitten crab, Eriocheir sinensis. Aquaculture 2020, 516, 734653. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Chang, E.B. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Kindt, A.; Liebisch, G.; Clavel, T.; Haller, D.; Hörmannsperger, G.; Yoon, H.; Kolmeder, D.; Sigruener, A.; Krautbauer, S.; Seeliger, C.; et al. The gut microbiota promotes hepatic fatty acid desaturation and elongation in mice. Nat. Commun. 2018, 9, 3760. [Google Scholar] [CrossRef]
- Sudikas, G.; Kulpys, J.; Jerešiunas, A.; Juškiene, V.; Norviliene, J. The influence of probiotics on carcass, meat and fat quality in pigs. Vet. Zootech. 2010, 52, 79–86. [Google Scholar]
- Liu, Y.H.; Li, Y.Y.; Feng, X.C.; Wang, Z.; Xia, Z.F. Dietary supplementation with Clostridium butyricum modulates serum lipid metabolism, meat quality, and the amino acid and fatty acid composition of Peking ducks. Poult. Sci. 2018, 97, 3218–3229. [Google Scholar] [CrossRef]
- Wen, C.L.; Yan, W.; Sun, C.J.; Ji, C.L.; Zhou, Q.Q.; Zhang, D.X.; Zheng, J.X.; Yang, N. The gut microbiota is largely independent of host genetics in regulating fat deposition in chickens. ISME J. 2019, 13, 1422–1436. [Google Scholar] [CrossRef]
- Monk, J.M.; Mcmurray, D.N.; Chapkin, R.S. Clinical Effects of n-3 PUFA Supplementation in Human Health and Inflammatory Diseases. In Omega-3 Oils; AOCS Press: Urbana, IL, USA, 2011; pp. 31–60. [Google Scholar]
- Burdge, G.C.; Calder, P.C. Conversion of α-linolenic acid to longer-chain polyunsaturated fatty acids in human adults. Reprod. Nutr. Dev. 2005, 45, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Konieczka, P.; Czauderna, M.; Smulikowska, S. The enrichment of chicken meat with omega-3 fatty acids by dietary fish oil or its mixture with rapeseed or flaxseed—Effect of feeding duration:Dietary fish oil, flaxseed, and rapeseed and n-3 enriched broiler meat. Anim. Feed. Sci. Technol. 2017, 223, 42–52. [Google Scholar] [CrossRef]
- Marino, R.; Caroprese, M.; Annicchiarico, G.; Ciampi, F.; Ciliberti, M.G.; Malva, A.D.; Santillo, A.; Sevi, A.; Albenzio, M. Effect of Diet Supplementation with Quinoa Seed and/or Linseed on Immune Response, Productivity and Meat Quality in Merinos Derived Lambs. Animals 2018, 8, 204. [Google Scholar] [CrossRef] [PubMed]
- Mago, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ Inc. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; George, M.G.; James, M.T.; James, R.C. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, Y.; Jiang, M. Research progress of polyunsaturated fatty acids. China Oils Fats 2008, 33, 42–46. [Google Scholar]
- Chen, D.; Wang, X.; Guo, Q.; Deng, H.; Luo, J.; Yi, K.; Sun, A.; Chen, K.; Shen, Q. Muscle Fatty Acids, Meat Flavor Compounds and Sensory Characteristics of Xiangxi Yellow Cattle in Comparison to Aberdeen Angus. Animals 2022, 12, 1161. [Google Scholar] [CrossRef] [PubMed]
- Hulan, H.W.; Ackman, R.G.; Ratnayake, W.; Proudfoot, F.G. Omega-3 Fatty Acid Levels and General Performance of Commercial Broilers Fed Practical Levels of Redfish Meal. Poult. Sci. 1989, 68, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Poorghasemi, M.; Seidavi, A.; Qotbi, A.A.A.; Laudadio, V.; Tufarelli, V. Influence of dietary fat source on growth performance responses and carcass traits of broiler chicks. Asian-Australas. J. Anim. Sci. 2013, 26, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.-R.; Huang, J.; Wei, Y.-M.; Li, M.-Q.; He, X.-G.; Zheng, J. Biological characteristics and application of quinoa. Pratacultural Sci. 2017, 34, 607–613. [Google Scholar]
- Filho, A.M.M.; Pirozi, M.R.; Borges, J.T.D.S.; Sant’Ana, H.M.P.; Chaves, J.B.P.; Coimbra, J.S.D.R. Quinoa: Nutritional, functional, and antinutritional aspects. Crit. Rev. Food Sci. Nutr. 2017, 57, 1618–1630. [Google Scholar] [CrossRef]
- Altuna, J.L.; Silva, M.; Álvarez, M.; Quinteros, M.F.; Morales, D.; Carrillo, W. Ecuadorian quinoa (Chenopodium quinoa Willd) fatty acids profile. Asian J. Pharm. Clin. Res. 2018, 11, 109. [Google Scholar]
- Peiretti, P.G.; Gai, F.; Tassone, S. Fatty acid profile and nutritive value of quinoa (Chenopodium quinoa Willd.) seeds and plants at different growth stages. Anim. Feed. Sci. Technol. 2013, 183, 56–61. [Google Scholar] [CrossRef]
- Alvarez-Jubete, L.; Wijngaard, H.; Arendt, E.K.; Gallagher, E. Polyphenol composition and in vitro antioxidant activity of amaranth, quinoa buckwheat and wheat as affected by sprouting and baking. Food Chem. 2010, 119, 770–778. [Google Scholar] [CrossRef]
- Simopoulos, A.P. Evolutionary aspects of diet, the omega-6/omega-3 ratio and genetic variation: Nutritional implications for chronic diseases. Biomed. Pharmacother. 2006, 60, 502–507. [Google Scholar] [CrossRef]
- Zotte, A.D.; Szendroe, Z. The role of rabbit meat as functional food. Meat Sci. 2011, 88, 319–331. [Google Scholar] [CrossRef]
- Cortinas, L.; Barroeta, A.; Villaverde, C.; Galobart, J.; Guardiola, F.; Baucells, M.D. Influence of the dietary polyunsaturation level on chicken meat quality: Lipid oxidation. Poult. Sci. 2005, 84, 48–55. [Google Scholar] [CrossRef]
- Xu, J.; Feng, L.; Jiang, W.D.; Wu, P.; Liu, Y.; Jiang, J.; Kuang, S.Y.; Tang, L.; Zhou, X.Q. Different dietary protein levels affect flesh quality, fatty acids and alter gene expression of Nrf2-mediated antioxidant enzymes in the muscle of grass carp (Ctenophar-yngodon idella). Aquaculture 2018, 493, 272–282. [Google Scholar] [CrossRef]
- Xue, T.; Wang, Y.Z.; Pan, Q.H.; Wang, Q.; Yuan, J.F.; Che, T.S. Establishment of a cell line from the kidney of black carp and its susceptibility to spring viremia of carp virus. J. Fish Dis. 2018, 41, 365–374. [Google Scholar] [CrossRef]
- Scaife, J.R.; Moyo, J.; Galbraith, H.; Michie, W.; Campbell, V. Effect of different dietary supplemental fats and oils on the tissue fatty acid composition and growth of female broilers. Br. Poult. Sci. 2007, 35, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Abm, R.B.; Mun, H.S.; Yang, C.J. Breast and Thigh Meat Chemical Composition and Fatty Acid Profile in BroilersFed Diet with Dietary Fat Sources. J. Food Process. Technol. 2017, 8, 672. [Google Scholar]
- Saleh, A.A.; Alharthi, A.S.; Alhotan, R.A.; Atta, M.S.; Abdel-Moneim, A.M.E. Soybean Oil Replacement by Poultry Fat in Broiler Diets: Performance, Nutrient Digestibility, Plasma Lipid Profile and Muscle Fatty Acids Content. Animals 2021, 11, 2609. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Re: Role of the gut microbiota in nutrition and health. BMJ 2018, 361, 17–22. [Google Scholar]
- Ahir, V.B.; Koringa, P.G.; Bhatt, V.D.; Ramani, U.V.; Tripathi, A.K.; Singh, K.M.; Dhagat, U.M.; Patel, J.S.; Patel, M.M.; Katudia, K.H.; et al. Metagenomic analysis of poultry gut microbes. Indian J. Poult. Sci. 2010, 45, 111–114. [Google Scholar]
- Kaakoush, N.O. Insights into the role of erysipelotrichaceae in the human host. Front. Cell. Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef]
- Fernando, S.C.; Purvis, H.T.; Najar, F.Z.; Sukharnikov, L.O.; Krehbiel, C.R.; Nagaraja, T.G.; Roe, B.A.; Desilva, U. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 2010, 76, 7482–7490. [Google Scholar] [CrossRef]
- Jami, E.; White, B.A.; Mizrahi, I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 2017, 9, e85423. [Google Scholar] [CrossRef] [PubMed]
- Meital, N.O.; Hadar, N.; Omry, K. Microbial Changes during Pregnancy, Birth, and Infancy. Front. Microbiol. 2016, 7, 1031–1033. [Google Scholar]
- Ruth, E.L.; Turnbaugh, P.J.; Samuel, K.; Jeffrey, I.G. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar]
- Marcobal, A.; Barboza, M.; Froehlich, J.W.; Block, D.E.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 2010, 58, 5334–5340. [Google Scholar] [CrossRef]
- Wexler, A.G.; Goodman, A.L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2017, 2, 17026. [Google Scholar] [CrossRef]
- Venegas, D.P.; Fuente, M.; Landskron, G.; González, M.J.; Hermoso, M.A. Corrigendum:Short chain fatty acids(SCFAs)-mediated gut epithelial and immune regulation and Its relevance for inflammatory bowel diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Vallès, Y.; Artacho, A.; Pascual-García, A.; Ferrús, M.L.; Gosalbes, M.J.; Abellán, J.J.; Francino, M.P. Microbial succession in the gut: Directional trends of taxonomic and functional change in a birth cohort of Spanish infants. PLoS Genet. 2014, 10, e1004406. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Filippidou, S.; Wunderlin, T.; Junier, T.; Junier, T.; Jeanneret, N.; Dorador, C.; Molina, V.; Johnson, D.; Junier, P. A combination of extreme environmental conditions favor the prevalence of Endospore-forming Firmicutes. Front. Microbiol. 2016, 7, 1707. [Google Scholar] [CrossRef]
- Ley, R.E.; Bckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Natl. Acad. Sci. 2005, 102, 11070–11075. [Google Scholar] [CrossRef]
- Komaroff, A.L. The Microbiome and Risk for Obesity and Diabetes. JAMA J. Am. Med. Assoc. 2017, 317, 355–356. [Google Scholar] [CrossRef]
- Wang, H.; Ni, X.; Lei, L.; Dong, Z.; Jing, L.; Qing, X.; Li, G.; Pan, K.; Bo, J. Controlling of growth performance, lipid deposits and fatty acid composition of chicken meat through a probiotic, Lactobacillus johnsonii during subclinical Clostridium perfringens infection. Lipids Health Dis. 2017, 16, 1–10. [Google Scholar] [CrossRef]
- Madigan, M.T.; Martinko, J.M.; Parker, J. Brock Biology of Microorganisms, 11th ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2005. [Google Scholar]
- Hossain, M.E.; Kim, G.M.; Lee, S.K.; Yang, C.J. Growth Performance, Meat Yield, Oxidative Stability, and Fatty Acid Composition of Meat from Broilers Fed Diets Supplemented with a Medicinal Plant and Probiotics. Asian-Australas. J. Anim. Sci. 2012, 25, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.H.; Wang, Y.; Rui, D.U.; Luo, Y.L.; Liu, C.; Yao, D.; Jin, Y. Research of Fatty Acid Deposition Mechanism in Adipose Tissue of Sunit Sheep. Sci. Technol. Food Ind. 2019, 40, 11–16. [Google Scholar]
- Yu, M.; Li, Z.; Rong, T.; Wang, G.; Liu, Z.; Chen, W.L.J.; Li, J.; Ma, X. Different dietary starch sources alter the carcass traits, meat quality, and the profile of muscle amino acid and fatty acid in finishing pigs. J. Anim. Sci. Biotechnol. 2020, 11, 78. [Google Scholar] [CrossRef] [PubMed]
- Yang, F. Effects of Paper Mulberry Silage on the Growth Performance, Rumen Microbiota and Muscle Fatty Acid Composition in Hu Lambs. Fermentation 2021, 7, 286. [Google Scholar]
- Frutos, P.; Hervás, G.; Natalello, A.; Luciano, G.; Fondevila, M.; Priolo, A.; Toral, P.G. Ability of tannins to modulate ruminal lipid metabolism and milk and meat fatty acid profiles. Anim. Feed. Sci. Technol. 2020, 269, 114623. [Google Scholar] [CrossRef]




| Items | Diets (%) | |||
|---|---|---|---|---|
| CK | Q4 | Q8 | Q12 | |
| Corn | 64.00 | 64.00 | 64.00 | 64.00 |
| Wheat middling | 12.00 | 8.00 | 4.00 | 0.00 |
| Quinoa seeds | 0.00 | 4.00 | 8.00 | 12.00 |
| Soybean meal | 20.00 | 20.00 | 20.00 | 20.00 |
| Limestone | 1.20 | 1.20 | 1.20 | 1.20 |
| CaHPO4 | 1.20 | 1.20 | 1.20 | 1.20 |
| NaCl | 0.30 | 0.30 | 0.30 | 0.30 |
| 1% Premix | 1.00 | 1.00 | 1.00 | 1.00 |
| Zeolite powder | 0.30 | 0.30 | 0.30 | 0.30 |
| Total | 100 | 100 | 100 | 100 |
| Nutrient levels | ||||
| Metabolic energy, ME (MJ/Kg) | 12.24 | 12.32 | 12.40 | 12.48 |
| Dry matter, DM | 84.20 | 84.43 | 84.66 | 84.88 |
| Crude protein, CP | 16.12 | 16.18 | 16.25 | 16.31 |
| Crude fiber, CF | 2.47 | 2.60 | 2.74 | 2.88 |
| Ether extract, EE | 2.86 | 2.98 | 3.10 | 3.22 |
| Calcium, Ca | 0.87 | 0.88 | 0.88 | 0.88 |
| Total phosphorus, TP | 0.59 | 0.59 | 0.58 | 0.57 |
| Available phosphorus, AP | 0.20 | 0.20 | 0.20 | 0.20 |
| Lysine, Lys | 0.81 | 0.83 | 0.84 | 0.86 |
| Methionine, Met | 0.26 | 0.30 | 0.33 | 0.36 |
| Methionine + Cystine, Met + Cys | 0.56 | 0.59 | 0.61 | 0.64 |
| Threonine, Thr | 0.62 | 0.62 | 0.62 | 0.62 |
| Tryptophan, Trp | 0.19 | 0.19 | 0.18 | 0.18 |
| Items | Group | |||
|---|---|---|---|---|
| CK | Q4 | Q8 | Q12 | |
| Butyric acid (C4:0) | 0.44 ± 0.14 a | 0.40 ± 0.06 a | 0.18 ± 0.06 c | 0.29 ± 0.08 b |
| Octanoic acid (C8:0) | 0.35 ± 0.07 a | 0.20 ± 0.02 ab | 0.16 ± 0.03 b | 0.18 ± 0.004 b |
| Myristic acid (C14:0) | 0.69 ± 0.05 a | 0.50 ± 0.03 d | 0.64 ± 0.09 b | 0.56 ± 0.05 c |
| Myristoleic acid (C14:1) | 0.21 ± 0.03 c | 0.27 ± 0.02 b | 0.21 ± 0.03 c | 0.30 ± 0.04 a |
| Pentadecanoic acid (C15:0) | 3.04 ± 0.63 | 2.87 ± 0.37 | 2.83 ± 0.62 | 2.56 ± 0.65 |
| Palmitic acid (C16:0) | 26.96 ± 1.54 a | 24.84 ± 0.30 b | 24.88 ± 3.03 b | 25.43 ± 0.79 b |
| Palmitoleic acid (C16:1) | 4.64 ± 0.90 b | 5.82 ± 0.44 a | 5.46 ± 0.92 a | 5.35 ± 0.92 a |
| Heptadecanoic acid (C17:0) | 0.69 ± 0.17 a | 0.36 ± 0.09 b | 0.42 ± 0.10 b | 0.40 ± 0.05 b |
| Heptadecenoic acid (C17:1) | 0.56 ± 0.13 c | 0.83 ± 0.18 a | 0.69 ± 0.06 b | 0.67 ± 0.17 b |
| Stearic acid (C18:0) | 10.90 ± 1.11 a | 8.44 ± 0.30 b | 8.40 ± 1.21 b | 8.21 ± 0.63 b |
| Oleic acid (C18:1n9c) | 29.07 ± 5.50 b | 31.95 ± 2.16 a | 31.70 ± 4.22 a | 31.89 ± 2.32 a |
| Elaidic acid (C18:1n9t) | 0.29 ± 0.04 c | 0.51 ± 0.04 b | 0.49 ± 0.07 b | 1.04 ± 0.11 a |
| Linoleic acid (C18:2n6c) | 13.55 ± 1.25 a | 12.62 ± 1.50 b | 12.23 ± 1.39 b | 11.99 ± 1.01 b |
| Linolenic acid (C18:3n3) | 0.55 ± 0.13 b | 0.70 ± 0.04 a | 0.69 ± 0.08 a | 0.71 ± 0.06 a |
| γ-linoleic acid (C18:3n6) | 0.16 ± 0.05 | 0.17 ± 0.01 | 0.16 ± 0.03 | 0.14 ± 0.001 |
| Eicosenoic acid (C20:1) | 0.26 ± 0.03 c | 0.49 ± 0.02 a | 0.42 ± 0.04 b | 0.41 ± 0.03 b |
| Eicosadienoic acid (C20:2) | 0.21 ± 0.04 c | 0.75 ± 0.02 a | 0.45 ± 0.04 b | 0.46 ± 0.05 b |
| Eicostrienoic acid (C20:3n6) | 0.56 ± 0.17 b | 0.74 ± 0.05 a | 0.56 ± 0.13 b | 0.64 ± 0.07 b |
| Arachidonic acid (AA,C20:4n6) | 4.73 ± 1.40 | 4.24 ± 1.04 | 4.76 ± 1.53 | 5.22 ± 1.35 |
| Eicosapentaenoic acid (EPA,C20:5n3) | 0.86 ± 0.17 c | 1.08 ± 0.09 b | 1.07 ± 0.12 b | 1.27 ± 0.06 a |
| Behenic acid (C22:0) | 0.34 ± 0.12 | 0.30 ± 0.05 | 0.36 ± 0.07 | 0.35 ± 0.10 |
| Docosahexaenoic acid (DHA, C22:6n3) | 0.37 ± 0.11 c | 0.68 ± 0.08 b | 0.67 ± 0.14 b | 0.81 ± 0.08 a |
| Nervonic acid (C24:1) | 1.12 ± 0.26 b | 1.76 ± 0.16 a | 1.55 ± 0.41 a | 1.74 ± 0.35 a |
| Saturated fatty acid (SFA) | 42.18 ± 4.33 a | 37.84 ± 0.62 b | 38.55 ± 1.62 b | 37.79 ± 1.22 b |
| Unsaturated fatty acid (UFA) | 57.82 ± 4.33 b | 61.90 ± 0.88 a | 62.50 ± 4.65 a | 62.10 ± 1.29 a |
| Monounsaturated fatty acid (MUFA) | 37.93 ± 3.60 a | 41.36 ± 2.01 b | 42.47 ± 6.69 b | 41.21 ± 2.72 b |
| Polyunsaturated fatty acid (PUFA) | 20.22 ± 2.51 | 20.54 ± 1.65 | 20.03 ± 2.48 | 20.88 ± 2.09 |
| n-3 Polyunsaturated fatty acids (n-3 PUFAs) | 1.83 ± 0.23 c | 2.46 ± 0.19 b | 2.45 ± 0.24 b | 2.79 ± 0.12 a |
| n-6 Polyunsaturated fatty acids (n-6 PUFAs) | 20.29 ± 2.63 a | 17.58 ± 1.87 b | 17.44 ± 1.93 b | 17.79 ± 1.90 b |
| n-6PUFA/n-3PUFA | 9.88 ± 1.59 a | 7.18 ± 0.91 b | 6.81 ± 1.37 b | 6.39 ± 0.65 c |
| PUFA/SFA | 0.53 ± 0.12 | 0.54 ± 0.04 | 0.54 ± 0.06 | 0.55 ± 0.05 |
| Items | Group | |||
|---|---|---|---|---|
| CK | Q4 | Q8 | Q12 | |
| Lauric acid (C12:0) | 0.04 ± 0.008 | 0.04 ± 003 | 0.04 ± 0.002 | |
| Tridecylic acid (C13:0) | 0.51 ± 0.11 | 0.37 ± 0.02 | 0.46 ± 0.08 | |
| Myristic acid (C14:0) | 1.25 ± 0.09 a | 0.63 ± 0.06 b | 0.63 ± 0.11 b | 0.65 ± 0.07 b |
| Myristoleic acid (C14:1) | 0.24 ± 0.02 b | 0.42 ± 0.03 a | 0.44 ± 0.03 a | 0.42 ± 0.02 a |
| Pentadecanoic acid (C15:0) | 0.10 ± 0.005 | 0.09 ± 0.008 | 0.10 ± 0.01 | 0.90 ± 0.01 |
| Pentadecenic acid (C15:1) | 0.20 ± 0.06 b | 0.35 ± 0.07 a | 0.35 ± 0.05 a | 0.36 ± 0.04 a |
| Palmitic acid (C16:0) | 27.41 ± 1.53 a | 25.76 ± 0.59 b | 26.06 ± 1.29 b | 26.03 ± 1.41 b |
| Palmitoleic acid (C16:1) | 5.50 ± 0.82 b | 7.63 ± 0.78 a | 7.63 ± 1.02 a | 7.45 ± 0.93 a |
| Heptadecanoic acid (C17:0) | 0.14 ± 0.01 a | 0.13 ± 0.02 b | 0.12 ± 0.02 b | 0.12 ± 0.02 b |
| Heptadecenoic acid (C17:1) | 0.13 ± 0.04 b | 0.28 ± 0.03 a | 0.28 ± 0.02 a | 0.26 ± 0.02 a |
| Stearic acid (C18:0) | 11.22 ± 1.55 a | 7.05 ± 1.55 b | 7.19 ± 1.02 b | 7.21 ± 0.61 b |
| Oleic acid (C18:1 n9c) | 29.89 ± 2.59 b | 34.14 ± 3.37 a | 33.12 ± 2.80 a | 34.19 ± 2.28 a |
| Elaidic acid (C18:1n9t) | 0.31 ± 0.03 c | 0.43 ± 0.04 b | 0.42 ± 0.03 b | 0.88 ± 0.05 a |
| Linoleic acid (C18:2n6c) | 14.37 ± 1.51 b | 15.44 ± 1.95 a | 14.93 ± 1.13 b | 14.31 ± 1.64 b |
| Linolelaidic acid (C18:2n6t) | 0.13 ± 0.01 a | 0.09 ± 0.02 b | 0.09 ± 0.02 b | 0.09 ± 0.02 b |
| Linolenic acid (C18:3n3) | 1.08 ± 0.04 d | 1.21 ± 0.09 c | 1.38 ± 0.07 a | 1.29 ± 0.06 b |
| γ-linoleic acid (C18:3n6) | 0.10 ± 0.03 d | 0.12 ± 0.02 c | 0.19 ± 0.04 a | 0.16 ± 0.03 b |
| Eicosenoic acid (C20:1) | 0.25 ± 0.04 b | 0.42 ± 0.04 a | 0.41 ± 0.05 a | 0.40 ± 0.03 a |
| Eicosadienoic acid (C20:2) | 0.19 ± 0.05 b | 0.31 ± 0.06 a | 0.16 ± 0.03 bc | 0.12 ± 0.06 c |
| Eicostrienoic acid (C20:3n6) | 0.35 ± 0.11 a | 0.25 ± 0.08 b | 0.36 ± 0.10 a | 0.30 ± 0.05 ab |
| Arachidonic acid (AA,C20:4n6) | 2.79 ± 0.72 a | 2.52 ± 0.62 ab | 2.18 ± 0.46 b | 2.48 ± 0.56 ab |
| Eicosapentaenoic acid (EPA,C20:5n3) | 0.70 ± 0.12 c | 0.81 ± 0.09 b | 0.92 ± 0.07 a | 0.84 ± 0.07 b |
| Heneicosan oic acid (C21:0) | 0.29 ± 0.05 a | 0.04 ± 0.002 b | 0.04 ± 0.002 b | 0.09 ± 0.05 b |
| Behenic acid (C22:0) | 0.24 ± 0.07 a | 0.15 ± 0.08 b | 0.15 ± 0.05 b | 0.20 ± 0.08 ab |
| Docosahexaenoic acid (DHA,C22:6n3) | 0.75 ± 0.10 c | 0.80 ± 0.04 b | 0.84 ± 0.03 a | 0.84 ± 0.06 a |
| Nervonic acid (C24:1) | 0.48 ± 0.16 b | 0.70 ± 0.14 a | 0.60 ± 0.12 a | 0.68 ± 0.11 a |
| Saturated fatty acid (SFA) | 42.85 ± 3.85 a | 34.27 ± 1.65 b | 34.76 ± 1.43 b | 34.73 ± 1.38 b |
| Unsaturated fatty acid (UFA) | 57.02 ± 4.11 b | 65.72 ± 1.66 a | 65.22 ± 1.44 a | 65.32 ± 1.42 a |
| Monounsaturated fatty acids (MUFA) | 36.65 ± 3.08 b | 44.34 ± 3.77 a | 43.24 ± 2.93 a | 44.56 ± 2.57 a |
| Polyunsaturated fatty acids (PUFA) | 19.85 ± 2.26 b | 21.38 ± 2.79 ab | 21.98 ± 2.38 a | 20.76 ± 2.76 ab |
| n-3 polyunsaturated fatty acids (n-3 PUFA) | 2.42 ± 0.37 d | 2.82 ± 0.17 c | 3.14 ± 0.13 a | 2.98 ± 0.14 b |
| n-6 polyunsaturated fatty acids (n-6 PUFA) | 17.43 ± 1.83 | 18.36 ± 2.73 | 18.69 ± 2.35 | 17.68 ± 2.69 |
| n-6PUFA/n-3PUFA | 8.10 ± 1.60 a | 6.52 ± 0.94 b | 5.96 ± 0.75 b | 5.94 ± 0.88 b |
| PUFA/SFA | 0.46 ± 0.07 b | 0.62 ± 0.08 a | 0.63 ± 0.07 a | 0.60 ± 0.09 a |
| Items | Sample Group | |||
|---|---|---|---|---|
| CK | Q4 | Q8 | Q12 | |
| Raw PE | 91,767 | 88,872 | 91,771 | 88,116 |
| Raw Tags | 85,365 | 84,815 | 88,060 | 84,872 |
| Clean Tags | 83,636 | 83,125 | 86,507 | 83,432 |
| Effective Tags | 63,247 | 66,096 | 67,843 | 65,090 |
| Base | 26,377,047 | 27,643,822 | 28,334,530 | 27,092,227 |
| AvgLen | 417 | 418.33 | 417.5 | 416 |
| Q20 | 98.44 | 98.36 | 98.45 | 98.515 |
| Q30 | 94.95 | 94.72 | 94.935 | 95.11 |
| GC (%) | 53.2 | 52.90 | 53.22 | 53.08 |
| Effective (%) | 68.77 | 74.38 | 73.98 | 73.95 |
| Items | Group | |||
|---|---|---|---|---|
| CK | Q4 | Q8 | Q12 | |
| observed_species | 640.67 ± 24.53 | 635.00 ± 76.96 | 636.50 ± 54.56 | 593.17 ± 47.69 |
| Shannon | 6.54 ± 0.30 a | 5.98 ± 0.07 b | 6.17 ± 0.18 b | 6.10 ± 0.24 b |
| Simpson | 0.97 ± 0.01 a | 0.95 ± 0.01 b | 0.96 ± 0.01 ab | 0.96 ± 0.01 ab |
| chao1 | 668.93 ± 18.73 | 682.35 ± 89.63 | 674.81 ± 56.79 | 631.91 ± 52.24 |
| ACE | 675.88 ± 20.52 | 681.89 ± 88.39 | 686.06 ± 55.86 | 636.88 ± 48.49 |
| PD_whole_tree | 43.46 ± 5.24 | 43.88 ± 9.36 | 39.70 ± 2.49 | 38.58 ± 3.01 |
| OTU catalogue | 1209 | Annotated on Class level | 93.05% |
| Annotated on Database | 1179 (97.52%) | Annotated on Class level | 89.00% |
| Annotated on Unclassified | 30 (2.48%) | Annotated on Family level | 80.23% |
| Annotated on Kingdom level | 97.52% | Annotated on Genus level | 40.03% |
| Annotated on Phylum level | 95.70% | Annotated on Species level | 12.49% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, T.; Jiang, X.; Yang, F.; Wei, Y.; Zhao, S.; Jiao, T. Effects of Dietary Quinoa Seeds on Cecal Microorganisms and Muscle Fatty Acids of Female Luhua Chickens. Animals 2022, 12, 3334. https://doi.org/10.3390/ani12233334
Wu T, Jiang X, Yang F, Wei Y, Zhao S, Jiao T. Effects of Dietary Quinoa Seeds on Cecal Microorganisms and Muscle Fatty Acids of Female Luhua Chickens. Animals. 2022; 12(23):3334. https://doi.org/10.3390/ani12233334
Chicago/Turabian StyleWu, Tao, Xiaofan Jiang, Farong Yang, Yuming Wei, Shengguo Zhao, and Ting Jiao. 2022. "Effects of Dietary Quinoa Seeds on Cecal Microorganisms and Muscle Fatty Acids of Female Luhua Chickens" Animals 12, no. 23: 3334. https://doi.org/10.3390/ani12233334
APA StyleWu, T., Jiang, X., Yang, F., Wei, Y., Zhao, S., & Jiao, T. (2022). Effects of Dietary Quinoa Seeds on Cecal Microorganisms and Muscle Fatty Acids of Female Luhua Chickens. Animals, 12(23), 3334. https://doi.org/10.3390/ani12233334






