Identification and Antimicrobial Resistance in Klebsiella spp. Isolates from Turkeys in Poland between 2019 and 2022
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacterial Isolation
2.2. Antibiotic Sensitivity Testing and Detection of Extended-Spectrum β-Lactamases
2.3. Statistical Analysis
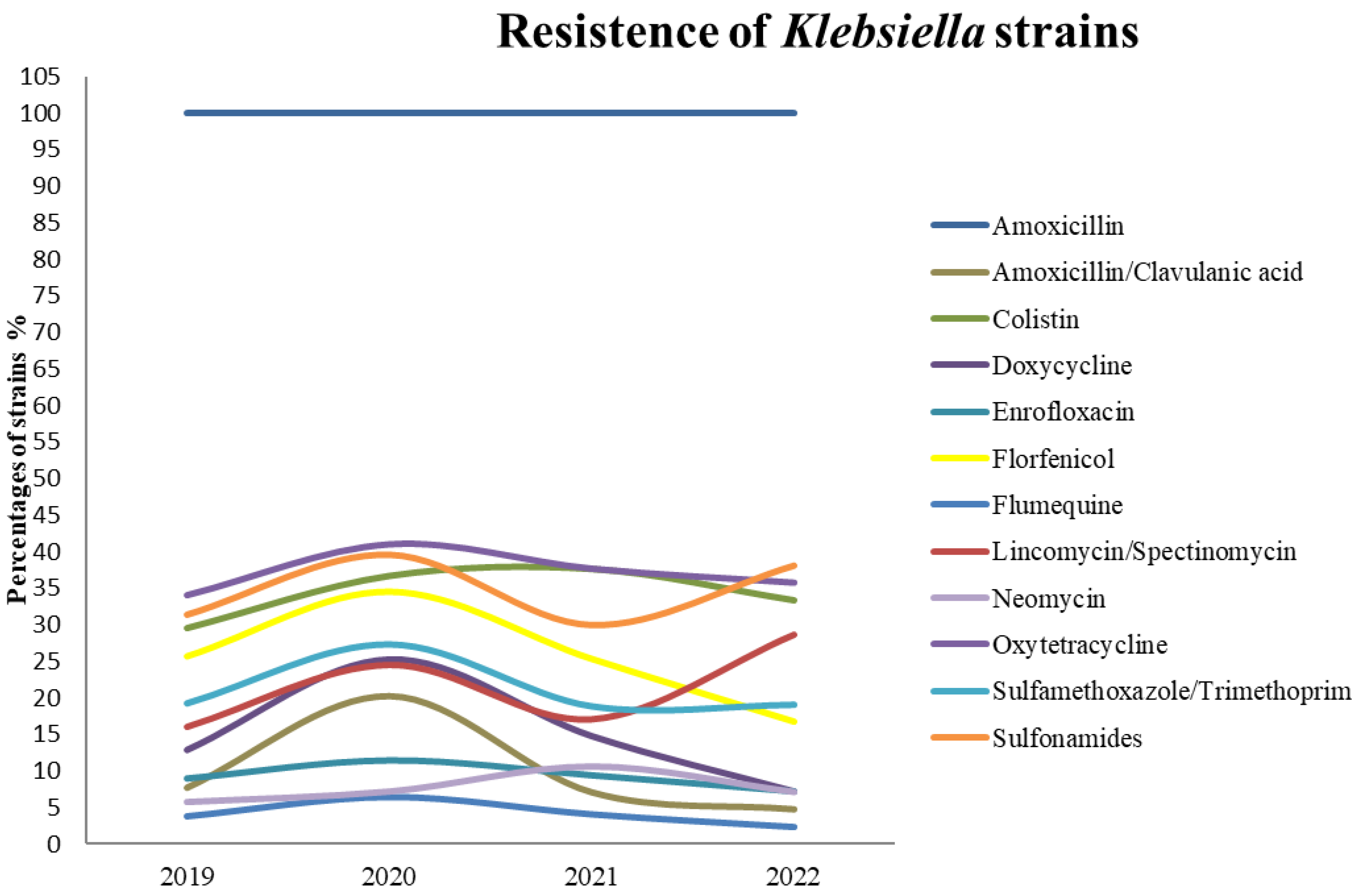
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- European Medicines Agency. European Surveillance of Veterinary Antimicrobial Consumption, 2021. ‘Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2019 and 2020’. (EMA/58183/2021). Available online: https://www.ema.europa.eu/en/documents/report/sales-veterinary-antimicrobial-agents-31-european-countries-2019-2020-trends-2010-2020-eleventh_en.pdf (accessed on 3 October 2022).
- Quinn, P.J.; Markey, B.K.; Leonard, F.C.; FitzPatrick, E.S.; Fanning, S. Concise Review of Veterinary Microbiology, 2nd ed.; Wiley Blackwell: Hoboken, NJ, USA, 2016; pp. 58–59. [Google Scholar]
- Martin, R.M.; Bachman, M.A. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Rimoldi, S.G.; Gentile, B.; Pagani, C.; Digregorio, A.; Anselmo, A.; Palozzi, A.M.; Fortunato, A.; Pittigliio, V.; Ridolfo, A.L.; Gismondo, M.R.; et al. Whole genome sequencing for the molecular characterization of carbapenem resistant Klebsiella pneumoniae strains isolated at the Italian ASST Fatebenefratelli Sacco Hospital, 2012–2014. BMC Infect. Dis. 2017, 17, 666. [Google Scholar] [CrossRef] [PubMed]
- Effah, C.Y.; Sun, T.; Liu, S.; Wu, Y. Klebsiella pneumoniae: An increasing threat to public health. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1. [Google Scholar] [CrossRef]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Micro. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Koncicki, A.; Stenzel, T.; Koncicka-Świderska, K.; Tykałowski, B.; Śmiałek, M.; Kowalczyk, J. Algorytmy diagnostyczno-terapeutyczne w przebiegu wybranych bakteryjnych chorób indyków. Available online: https://academica.edu.pl/reading/readMeta?cid=90915165&uid=92924904 (accessed on 16 October 2022).
- Li, Y.; Kumar, S.; Zhang, L.; Wu, H. Klebsiella pneumonia and Its Antibiotic Resistance: A Bibliometric Analysis. BioMed Res. Int. 2022, 2020, 1668789. [Google Scholar] [CrossRef]
- Sweeney, M.T.; Diaz-Campos, D.V.; Bowden, R.; Fritsche, T.R.; Hayes, J.; Langston, C.; Lubbers, B.; Martin-Jimenez, T.; Miller, C.; Pallotta, C. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; VET01; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019; pp. 1–156. [Google Scholar]
- Gniadkowski, M.; Trzciński, K.; Pałucha, A.; Hryniewicz, W. Wykrywanie β-laktamaz o rozszerzonym zakresie działania (ESBL) w izolatach klinicznych Klebsiella pneumoniae: Test dwóch krążków i test ATB BLSE. Diag. Laborator. 1996, 32, 697. [Google Scholar]
- Applegate, T.J.; Klose, V.; Steiner, T.; Ganner, A.; Schatzmayr, G. Probiotics and phytogenics for poultry: Myth or reality? J. Appl. Poult. Res. 2010, 19, 194–210. [Google Scholar] [CrossRef]
- Murugesan, G.R.; Syed, B.; Haldar, S.; Pender, C. Phytogenic feed additives as an alternative to antibiotic growth promoters in broiler chickens. Front. Vet. Sci. 2015, 2, 21. [Google Scholar]
- Singer, R.S.; Porter, L.J.; Thomson, D.U.; Gage, M.; Beaudoin, A.; Wishnie, J.K. Raising Animals Without Antibiotics: U.S. Producer and Veterinarian Experiences and Opinions. Front. Vet. Sci. 2019, 6, 452. [Google Scholar] [CrossRef] [PubMed]
- Mehdi, Y.; Létourneau-Montminy, M.-P.; Gaucher, M.-L.; Chorfi, Y.; Suresh, G.; Rouissi, T.; Brar, S.K.; Côté, C.; Ramirez, A.A.; Godbout, S. Use of antibiotics in broiler production: Global impacts and alternatives. Anim. Nutr. 2018, 4, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Diarra, M.S.; Malouin, F. Antibiotics in canadian poultry productions and anticipated alternatives. Front. Microbiol. 2014, 5, 282. [Google Scholar] [CrossRef] [PubMed]
- Frankic, T.; Voljč, M.; Salobir, J.; Rezar, V. Use of herbs and spices and their extracts in animal nutrition. Acta Agric. Slov. 2009, 94, 95–102. [Google Scholar]
- Li, H.L.; Zhao, P.Y.; Lei, Y.; Hossain, M.M.; Kim, I.H. Phytoncide, phytogenic feed additive as an alternative to conventional antibiotics, improved growth performance and decreased excreta gas emission without adverse effect on meat quality in broiler chickens. Livest. Sci. 2015, 181, 1–6. [Google Scholar] [CrossRef]
- Daehre, K.; Projahn, M.; Friese, A.; Semmler, T.; Guenther, S.; Roesler, U.H. ESBL-Producing Klebsiella pneumoniae in the Broiler Production Chain and the First Description of ST3128. Front. Microbiol. 2018, 3, 2302. [Google Scholar] [CrossRef]
- Hiroi, M.; Yamazaki, F.; Harada, T.; Takahashi, N.; Iida, N.; Noda, Y.; Yagi, M.; Nishio, T.; Kanda, T.; Kawamori, F.; et al. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in food-producing animals. J. Vet. Med. Sci. 2012, 74, 189–195. [Google Scholar] [CrossRef]
- Mahanti, A.; Ghosh, P.; Samanta, I.; Joardar, S.N.; Bandyopadhyay, S.; Bhattacharyya, D.; Banerjee, J.; Batabyal, S.; Kumar Sar, T.; Kumar Dutta, T. Prevalence of CTX-M-Producing Klebsiella spp. in broiler, kuroiler, and indigenous poultry in West Bengal State, India. Microb. Drug Resist. 2018, 24, 299–306. [Google Scholar] [CrossRef]
- Zhao, C.; Ge, B.; De Villena, J.; Sudler, R.; Yeh, E.; Zhao, S.; White, D.G.; Wagner, D.; Meng, J. Prevalence of Campylobacter spp., Escherichia Coli, and Salmonella serovars in retail chicken, Turkey, pork, and beef from the greater Washington, dc, area. Appl. Environ. Microbiol. 2001, 67, 5431–5436. [Google Scholar] [CrossRef]
- Theocharidi, N.A.; Balta, I.; Houhoula, D.; Tsantes, A.G.; Lalliotis, G.P.; Polydera, A.C.; Stamatis, H.; Halvatsiotis, P. High Prevalence of Klebsiella pneumoniae in Greek Meat Products: Detection of Virulence and Antimicrobial Resistance Genes by Molecular Techniques. Foods 2022, 11, 708. [Google Scholar] [CrossRef]
- Hartantyo, S.H.P.; Chau, M.L.; Koh, T.H.; Yap, M.; Yi, T.; Cao, D.Y.H.; Gutiérrez, R.A.; Ng, L.C. Foodborne Klebsiella pneumoniae: Virulence Potential, Antibiotic Resistance, and Risks to Food Safety. J. Food Prot. 2020, 83, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Fielding, B.C.; Mnabisa, A.; Gouws, P.A.; Morris, T. Antimicrobial-resistant Klebsiella species isolated from free-range chicken samples in an informal settlement. Arch. Med. Sci. 2012, 8, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Projahn, M.; von Tippelskirch, P.; Semmler, T.; Guenther, S.; Alter, T.; Roesler, U. Contamination of chicken meat with extended-spectrum beta-lactamase producing-Klebsiella pneumoniae and Escherichia coli during scalding and defeathering of broiler carcasses. Food Microbiol. 2019, 77, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, M.; Liu, Y.; Wang, X.; Wang, Y.; Lu, J.; Xu, H. Characterization of antimicrobial resistance in Klebsiella species isolated from chicken broilers. Int. J. Food Microbiol. 2016, 232, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Majewski, M.; Józefiak, A.; Kimsa-Furdzik, M.; Dziubdziela, L.; Hudak-Nowak, M.; Wilczyński, J.; Anusz, K. Antimicrobial resistance of Escherichia coli and Klebsiella spp. conventionally sampled from factory-farmed chickens—Clinical submissions. Ann. Agric. Environ. Med. 2021, 28, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S.; Giguère, S.; Guardabassi, L.; Morley, P.S.; Papich, M.; Ricciuto, D.R.; Sykes, J.E. ACVIM Consensus Statement on Therapeutic Antimicrobial Use in Animals and Antimicrobial Resistance. J. Vet. Intern. Med. 2015, 29, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Jankowski, J. (Department of Poultry Science and Apiculture, Univeristy of Warmia and Mazury, Olsztyn, Poland); Śmiałek, M. (Department of Avian Diseases, University of Warmia and Mazury, Olsztyn, Poland). Personal communication, 2022.
| Antibiotic | n | R | R% | I | I% | S | S% |
|---|---|---|---|---|---|---|---|
| Amoxicillin | 507 | 507 | 100.00 | 0 | 0.00 | 0 | 0 |
| Amoxicillin/Clavulanic acid | 507 | 54 | 10.65 | 34 | 6.71 | 419 | 82.64 |
| Colistin | 507 | 23 | 4.54 | 13 | 2.56 | 471 | 92.90 |
| Doxycycline | 507 | 175 | 34.52 | 13 | 2.56 | 319 | 62.92 |
| Enrofloxacin | 507 | 83 | 16.37 | 104 | 20.51 | 320 | 63.12 |
| Florfenicol | 507 | 49 | 9.66 | 9 | 1.78 | 449 | 88.56 |
| Flumequine | 507 | 138 | 27.22 | 104 | 20.51 | 265 | 52.27 |
| Lincomycin/Spectinomycin | 507 | 100 | 19.72 | 22 | 4.34 | 385 | 75.94 |
| Neomycin | 507 | 40 | 7.89 | 10 | 1.97 | 457 | 90.14 |
| Oxytetracycline | 507 | 189 | 37.28 | 3 | 0.59 | 315 | 62.13 |
| Sulfamethoxazole/Trimethoprim | 507 | 108 | 21.30 | 3 | 0.59 | 396 | 78.11 |
| Sulfonamides | 507 | 171 | 33.73 | 5 | 0.99 | 331 | 65.29 |
| Antibiotic | 2019 (n = 156) | 2020 (n = 139) | 2021 (n = 170) | 2022 (n = 42) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | % | I | % | R | % | S | % | I | % | R | % | S | % | I | % | R | % | S | % | I | % | R | % | |
| AMX 1 | 0 | 0.00 | 0 | 0.00 | 156 | 100.00 | 0 | 0.00 | 0 | 0.00 | 139 | 100.00 | 0 | 0.00 | 0 | 0.00 | 170 | 100.00 | 0 | 0.00 | 0 | 0.00 | 42 | 100.00 |
| AMC | 136 | 87.18 | 8 | 5.13 | 12 | 7.69 | 99 | 71.22 | 12 | 8.63 | 28 | 20.14 | 148 | 87.06 | 10 | 5.88 | 12 | 7.06 | 36 | 85.71 | 4 | 9.52 | 2 | 4.76 |
| COL | 148 | 94.87 | 2 | 1.28 | 6 | 3.85 | 122 | 87.77 | 8 | 5.76 | 9 | 6.47 | 160 | 94.12 | 3 | 1.76 | 7 | 4.12 | 41 | 97.62 | 0 | 0.00 | 1 | 2.38 |
| DOX | 104 | 66.67 | 6 | 3.85 | 46 | 29.49 | 84 | 60.43 | 4 | 2.88 | 51 | 36.69 | 104 | 61.18 | 2 | 1.18 | 64 | 37.65 | 27 | 64.29 | 1 | 2.38 | 14 | 33.33 |
| ENR | 103 | 66.03 | 33 | 21.15 | 20 | 12.82 | 79 | 56.83 | 25 | 17.99 | 35 | 25.18 | 107 | 62.94 | 38 | 22.35 | 25 | 14.71 | 31 | 73.81 | 8 | 19.05 | 3 | 7.14 |
| FLO | 131 | 83.97 | 1 | 0.64 | 14 | 8.97 | 118 | 84.89 | 5 | 3.60 | 16 | 11.51 | 151 | 88.82 | 3 | 1.76 | 16 | 9.41 | 39 | 92.86 | 0 | 0.00 | 3 | 7.14 |
| UB | 86 | 55.13 | 30 | 19.23 | 40 | 25.64 | 64 | 46.04 | 27 | 19.42 | 48 | 34.53 | 86 | 50.59 | 41 | 24.12 | 43 | 25.29 | 29 | 69.05 | 6 | 14.29 | 7 | 16.67 |
| L/SPE | 123 | 78.85 | 8 | 5.13 | 25 | 16.03 | 102 | 73.38 | 3 | 2.16 | 34 | 24.46 | 133 | 78.24 | 8 | 4.71 | 29 | 17.06 | 27 | 64.29 | 3 | 7.14 | 12 | 28.57 |
| NEO | 143 | 91.67 | 4 | 2.56 | 9 | 5.77 | 127 | 91.37 | 2 | 1.44 | 10 | 7.19 | 148 | 87.06 | 4 | 2.35 | 18 | 10.59 | 39 | 92.86 | 0 | 0.00 | 3 | 7.14 |
| OT | 103 | 66.03 | 0 | 0.00 | 53 | 33.97 | 80 | 57.55 | 2 | 1.44 | 57 | 41.01 | 105 | 61.76 | 1 | 0.59 | 64 | 37.65 | 27 | 64.29 | 0 | 0.00 | 15 | 35.71 |
| SXT | 126 | 80.77 | 0 | 0.00 | 30 | 19.23 | 100 | 71.94 | 1 | 0.72 | 38 | 27.34 | 136 | 80.00 | 2 | 1.18 | 32 | 18.82 | 34 | 80.95 | 0 | 0.00 | 8 | 19.05 |
| SSS | 100 | 64.10 | 0 | 0.00 | 49 | 31.41 | 81 | 58.27 | 3 | 2.16 | 55 | 39.57 | 117 | 68.82 | 2 | 1.18 | 51 | 30.00 | 26 | 61.90 | 0 | 0.00 | 16 | 38.10 |
| Antibiotic | Amoxicillin n = 507 | Amoxicillin/ Clavulanic acid n = 54 | Colistin n = 23 | Doxycycline n = 175 | Enrofloxacin n = 83 | Florfenicol n = 49 | Flumequine n = 138 | Lincomycin/ Spectinomycin n = 100 | Neomycin n = 40 | Oxytetracycline n = 189 | Sulfamethoxazole/ Trimethoprim n = 108 | Sulfonamides n = 171 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amoxicillin n = 507 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | |
| Amoxicillin/ Clavulanic acid n = 54 | p = 0.0002 | p < 0.001 | p = 0.007 | p = 0.6032 | p < 0.001 | p = 0.0001 | p = 0.1295 | p < 0.001 | p < 0.001 | p < 0.001 | ||
| Colistin n = 23 | p < 0.001 | p < 0.001 | p = 0.0015 | p < 0.001 | p < 0.001 | p = 0.0270 | p < 0.001 | p < 0.001 | p < 0.001 | |||
| Doxycycline n = 175 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p = 0.3594 | p < 0.001 | p = 0.7911 | ||||
| Enrofloxacin n = 83 | p = 0.0015 | p < 0.001 | p = 0.1651 | p < 0.001 | p < 0.001 | p = 0.0447 | p < 0.001 | |||||
| Florfenicol n = 49 | p < 0.001 | p < 0.001 | p = 0.3179 | p < 0.001 | p < 0.001 | p < 0.001 | ||||||
| Flumequine n = 138 | p = 0.0049 | p < 0.001 | p = 0.0006 | p = 0.0280 | p = 0.0244 | |||||||
| Lincomycin/ Spectinomycin n = 100 | p < 0.001 | p < 0.001 | p = 0.5338 | p < 0.001 | ||||||||
| Neomycin n = 40 | p < 0.001 | p < 0.001 | p < 0.001 | |||||||||
| Oxytetracycline n = 189 | p < 0.001 | p = 0.2375 | ||||||||||
| Sulfamethoxazole/ Trimethoprim n = 108 | p < 0.001 | |||||||||||
| Sulfonamides n = 171 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kowalczyk, J.; Czokajło, I.; Gańko, M.; Śmiałek, M.; Koncicki, A. Identification and Antimicrobial Resistance in Klebsiella spp. Isolates from Turkeys in Poland between 2019 and 2022. Animals 2022, 12, 3157. https://doi.org/10.3390/ani12223157
Kowalczyk J, Czokajło I, Gańko M, Śmiałek M, Koncicki A. Identification and Antimicrobial Resistance in Klebsiella spp. Isolates from Turkeys in Poland between 2019 and 2022. Animals. 2022; 12(22):3157. https://doi.org/10.3390/ani12223157
Chicago/Turabian StyleKowalczyk, Joanna, Ilona Czokajło, Marta Gańko, Marcin Śmiałek, and Andrzej Koncicki. 2022. "Identification and Antimicrobial Resistance in Klebsiella spp. Isolates from Turkeys in Poland between 2019 and 2022" Animals 12, no. 22: 3157. https://doi.org/10.3390/ani12223157
APA StyleKowalczyk, J., Czokajło, I., Gańko, M., Śmiałek, M., & Koncicki, A. (2022). Identification and Antimicrobial Resistance in Klebsiella spp. Isolates from Turkeys in Poland between 2019 and 2022. Animals, 12(22), 3157. https://doi.org/10.3390/ani12223157





