Genome-Wide Association Studies Revealed Significant QTLs and Candidate Genes Associated with Backfat and Loin Muscle Area in Pigs Using Imputation-Based Whole Genome Sequencing Data
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Phenotypes
2.2. Breeding Value Predictions
2.3. Genotype Imputation and Quality Control
2.4. Genome-Wide Association Study
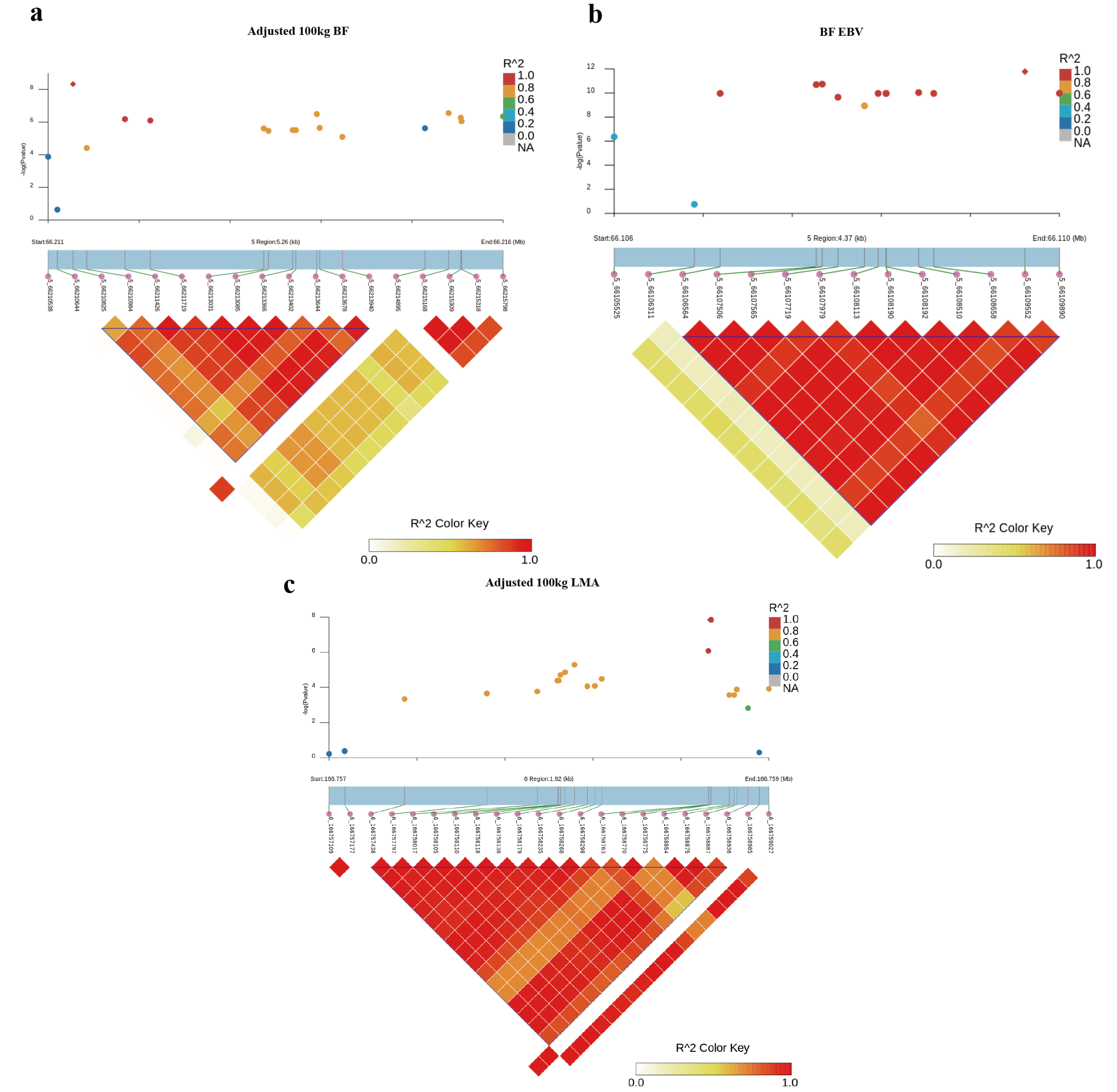
2.5. Haplotype Block Analysis and Candidate Gene Identification
3. Results
3.1. SNP Genotyping and Phenotypic Variation
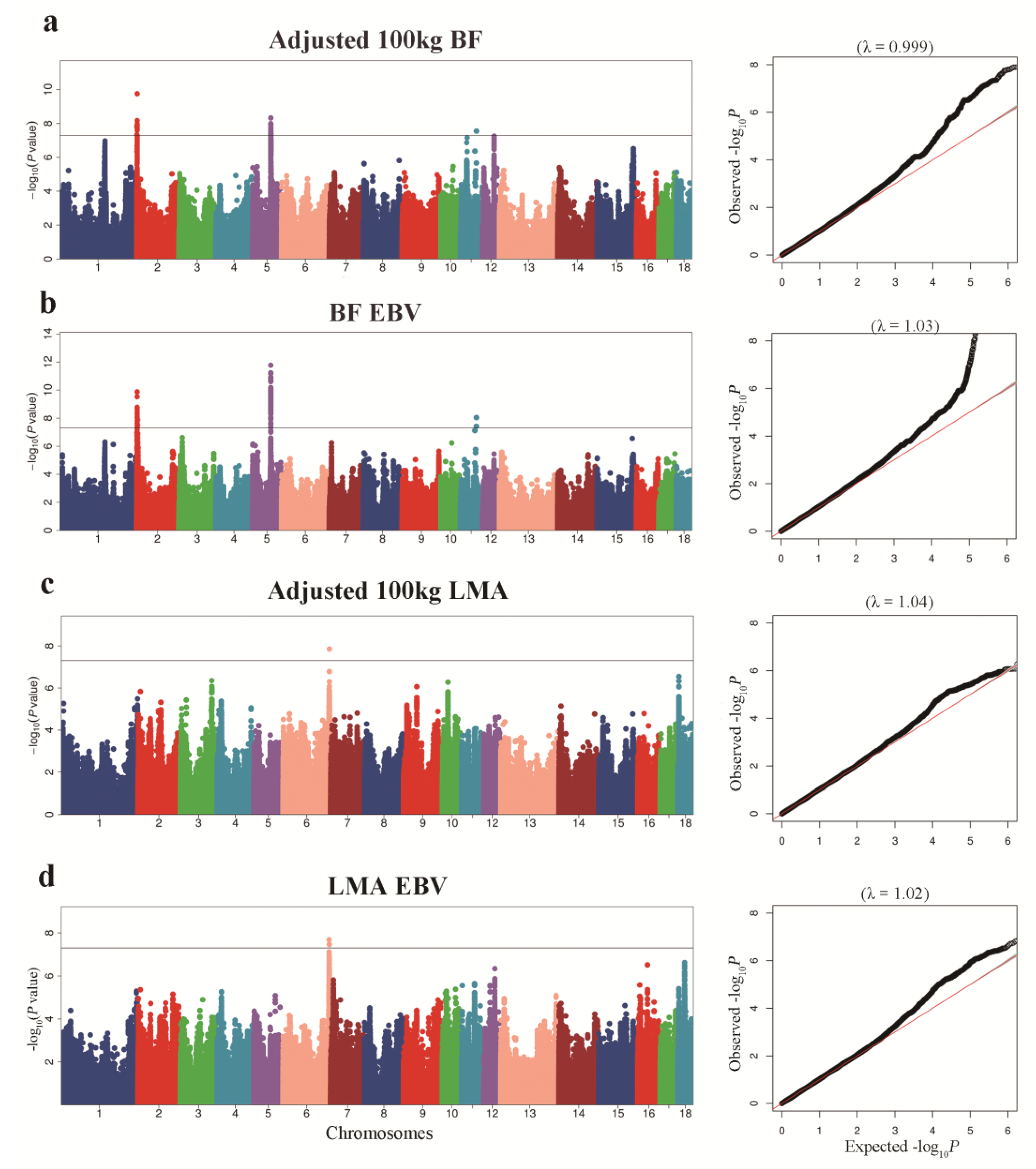
3.2. GWAS for BF Trait
3.3. GWAS for LMA Trait
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Do, D.N.; Ostersen, T.; Strathe, A.B.; Mark, T.; Jensen, J.; Kadarmideen, H.N. Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs. BMC Genet. 2014, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lavery, A.; Lawlor, P.G.; Magowan, E.; Miller, H.M.; O’driscoll, K.; Berry, D.P. An association analysis of sow parity, live-weight and back-fat depth as indicators of sow productivity. Animal 2019, 13, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Hoa, V.B.; Seo, H.W.; Seong, P.N.; Cho, S.H.; Kang, S.M.; Kim, Y.S.; Moon, S.S.; Choi, Y.M.; Kim, J.H.; Seol, K.H. Back-fat thickness as a primary index reflecting the yield and overall acceptance of pork meat. Anim. Sci. J. 2021, 92, e13515. [Google Scholar] [CrossRef]
- Hoque, M.A.; Suzuki, K.; Kadowaki, H.; Shibata, T.; Oikawa, T. Genetic parameters for feed efficiency traits and their relationships with growth and carcass traits in Duroc pigs. J. Anim. Breed. Genet. 2007, 124, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Iversen, M.W.; Bolhuis, J.E.; Camerlink, I.; Ursinus, W.W.; Reimert, I.; Duijvesteijn, N. Heritability of the backtest response in piglets and its genetic correlations with production traits. Animal 2017, 11, 556–563. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kline, E.A.; Hazel, L.N. Loin area at tenth and last rib as related to leanness of pork carcasses. J. Anim. Sci. 1955, 14, 659–663. [Google Scholar] [CrossRef]
- Hu, Z.-L.; Park, C.A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the future: Meeting new challenges and providing updated services. Nucleic Acids Res. 2022, 50, D956–D961. [Google Scholar] [CrossRef]
- Fan, B.; Onteru, S.K.; Du, Z.-Q.; Garrick, D.J.; Stalder, K.J.; Rothschild, M.F. Genome-wide association study identifies loci for body composition and structural soundness traits in pigs. PLoS ONE 2011, 6, e14726. [Google Scholar] [CrossRef]
- Guo, Y.-Y.; Zhang, L.-C.; Wang, L.-X.; Liu, W.-Z. Genome-wide association study for rib eye muscle area in a Large White×Minzhu F2 pig resource population. J. Integr. Agric. 2015, 14, 2590–2597. [Google Scholar] [CrossRef]
- Fontanesi, L.; Davoli, R.; Costa, L.N.; Beretti, F.; Scotti, E.; Tazzoli, M.; Tassone, F.; Colombo, M.; Buttazzoni, L.; Russo, V. Investigation of candidate genes for glycolytic potential of porcine skeletal muscle: Association with meat quality and production traits in Italian Large White pigs. Meat Sci. 2008, 80, 780–787. [Google Scholar] [CrossRef]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.K. That BLUP is a good thing: The estimation of random effects. Stat. Sci. 1991, 6, 15–32. [Google Scholar]
- Ding, R.; Quan, J.; Yang, M.; Wang, X.; Zheng, E.; Yang, H.; Fu, D.; Yang, Y.; Yang, L.; Li, Z.; et al. Genome-wide association analysis reveals genetic loci and candidate genes for feeding behavior and eating efficiency in Duroc boars. PLoS ONE 2017, 12, e0183244. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Savegnago, R.; Liu, J.; Long, N.; Tan, C.; Cai, G.; Zhuang, Z.; Wu, J.; Yang, M.; Qiu, Y.; et al. Nucleotide resolution genetic mapping in pigs by publicly accessible whole genome imputation. bioRxiv 2022. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Yan, G.; Liu, X.; Xiao, S.; Xin, W.; Xu, W.; Li, Y.; Huang, T.; Qin, J.; Xie, L.; Ma, J.; et al. An imputed whole-genome sequence-based GWAS approach pinpoints causal mutations for complex traits in a specific swine population. Sci. China Life Sci. 2022, 65, 781–794. [Google Scholar] [CrossRef]
- Ma, J.; Yang, J.; Zhou, L.; Ren, J.; Liu, X.; Zhang, H.; Yang, B.; Zhang, Z.; Ma, H.; Xie, X.; et al. A splice mutation in the PHKG1 gene causes high glycogen content and low meat quality in pig skeletal muscle. PLoS Genet. 2014, 10, e1004710. [Google Scholar] [CrossRef]
- Dong, S.-S.; He, W.-M.; Ji, J.-J.; Zhang, C.; Guo, Y.; Yang, T.-L. LDBlockShow: A fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief. Bioinform. 2021, 22. [Google Scholar] [CrossRef]
- Ding, R.; Zhuang, Z.; Qiu, Y.; Ruan, D.; Wu, J.; Ye, J.; Cao, L.; Zhou, S.; Zheng, E.; Huang, W.; et al. Identify known and novel candidate genes associated with backfat thickness in Duroc pigs by large-scale genome-wide association analysis. J. Anim. Sci. 2022, 100, skac012. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Li, S.; Ding, R.; Yang, M.; Zheng, E.; Yang, H.; Gu, T.; Xu, Z.; Cai, G.; Wu, Z.; et al. Meta-analysis of genome-wide association studies for loin muscle area and loin muscle depth in two Duroc pig populations. PLoS ONE 2019, 14, e0218263. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Xu, J.; Yin, L.; Yin, D.; Zhu, M.; Yu, M.; Li, X.; Zhao, S.; Liu, X. Genome-wide association study reveals candidate genes for growth relevant traits in pigs. Front. Genet. 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Wang, K.; Zhou, J.; Chen, D.; Jiang, A.; Jiang, Y.; Zhu, L.; Qiu, X.; Li, X.; Tang, G. A combined GWAS approach reveals key loci for socially-affected traits in Yorkshire pigs. Commun. Biol. 2021, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Le, T.H.; Christensen, O.F.; Nielsen, B.; Sahana, G. Genome-wide association study for conformation traits in three Danish pig breeds. Genet. Sel. Evol. 2017, 49, 1–12. [Google Scholar] [CrossRef]
- Brown, S.D.M.; Moore, M.W. The International Mouse Phenotyping Consortium: Past and future perspectives on mouse phenotyping. Mamm. Genome 2012, 23, 632–640. [Google Scholar] [CrossRef]
- Lo, C.-L.; Lossie, A.C.; Liang, T.; Liu, Y.; Xuei, X.; Lumeng, L.; Zhou, F.C.; Muir, W.M. High resolution genomic scans reveal genetic architecture controlling alcohol preference in bidirectionally selected rat model. PLoS Genet. 2016, 12, e1006178. [Google Scholar] [CrossRef]
- Comuzzie, A.G.; Cole, S.A.; Laston, S.L.; Voruganti, V.S.; Haack, K.; Gibbs, R.A.; Butte, N.F. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS ONE 2012, 7, e51954. [Google Scholar] [CrossRef]
- Falker-Gieske, C.; Blaj, I.; Preuß, S.; Bennewitz, J.; Thaller, G.; Tetens, J. GWAS for meat and carcass traits using imputed sequence level genotypes in pooled F2-designs in pigs. G3 Genes Genomes Genet. 2019, 9, 2823–2834. [Google Scholar] [CrossRef]
- Ma, P.; Li, Y.; Wang, H.; Mao, B. Haploinsufficiency of the TDP43 ubiquitin E3 ligase RNF220 leads to ALS-like motor neuron defects in the mouse. J. Mol. Cell Biol. 2021, 13, 374–382. [Google Scholar] [CrossRef]
- Ballester, M.; Ramayo-Caldas, Y.; González-Rodríguez, O.; Pascual, M.; Reixach, J.; Díaz, M.; Blanc, F.; López-Serrano, S.; Tibau, J.; Quintanilla, R. Genetic parameters and associated genomic regions for global immunocompetence and other health-related traits in pigs. Sci. Rep. 2020, 10, 18462. [Google Scholar] [CrossRef] [PubMed]
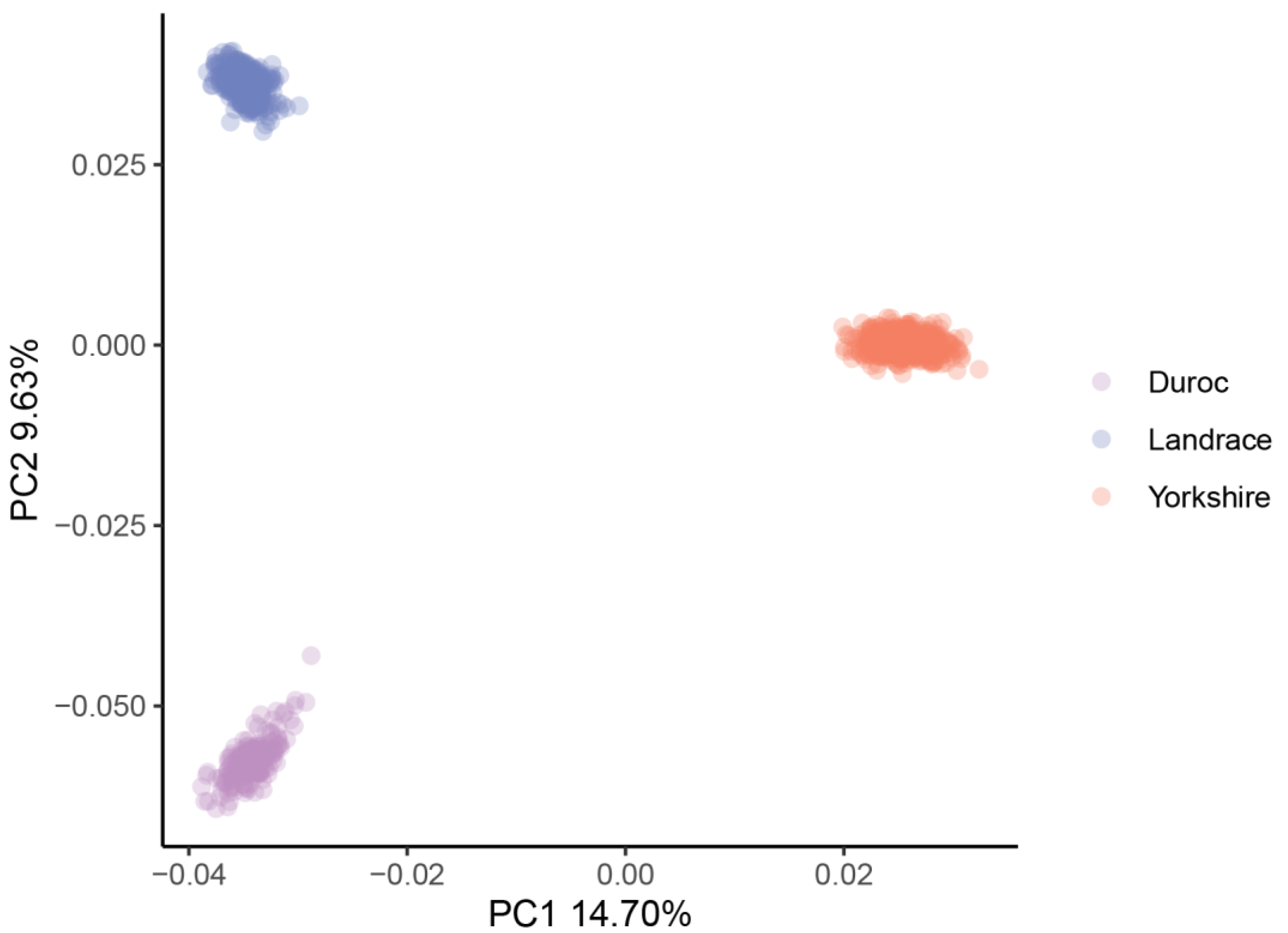
| Traits | Mean (±sd) | C.V. | h2 | Phenotypic Correlation | Genetic Correlation |
|---|---|---|---|---|---|
| Adjusted 100 kg BF | 11.38 ± 2.82 | 24.78 | 0.56 ± 0.05 | −0.23 | −0.33 ± 0.09 |
| Adjusted 100 kg LMA | 37.22 ± 4.75 | 12.76 | 0.50 ± 0.05 | ||
| BF EBV | 0.05 ± 0.74 | 14.80 | |||
| LMA EBV | 0.04 ± 1.90 | 47.50 |
| Traits | SSC | Positoin | p-Value | Consequence | Candidate Gene |
|---|---|---|---|---|---|
| Adjusted 100 kg BF | 5 | 66210825 | 4.73 × 10−9 | intergenic_variant | CCND2, TSPAN11 |
| 5 | 66109552 | 1.02 × 10−8 | splice polypyrimidine tract variant, splice region variant, intron variant | ||
| 5 | 66225671 | 1.23 × 10−8 | intergenic_variant | ||
| 5 | 66231984 | 1.78 × 10−8 | intergenic_variant | ||
| 5 | 66231994 | 2.31 × 10−8 | intergenic_variant | ||
| 5 | 66231774 | 2.61 × 10−8 | intergenic_variant | ||
| 5 | 66231974 | 3.28 × 10−8 | intergenic_variant | ||
| 5 | 66232303 | 3.38 × 10−8 | intergenic_variant | ||
| 5 | 66223267 | 3.72 × 10−8 | intergenic_variant | ||
| 5 | 66224507 | 4.03 × 10−8 | intergenic_variant | ||
| 2 | 2915752 | 1.76 × 10−10 | intron_variant | SHANK2 | |
| 2 | 2927340 | 6.84 × 10−9 | intron_variant | ||
| 2 | 2924189 | 1.08 × 10−8 | intron_variant | ||
| 2 | 2913910 | 1.27 × 10−8 | intron_variant | ||
| 2 | 2913928 | 1.27 × 10−8 | intron_variant | ||
| 2 | 2913943 | 1.27 × 10−8 | intron_variant | ||
| 2 | 2914889 | 1.27 × 10−8 | intron_variant | ||
| 2 | 2915433 | 1.27 × 10−8 | intron_variant | ||
| 2 | 1521939 | 1.37 × 10−8 | intergenic_variant | ||
| 2 | 2919066 | 1.50 × 10−8 | intron_variant | ||
| BF EBV | 5 | 66109552 | 1.70 × 10−12 | splice polypyrimidine tract variant, splice region variant, intron variant | CCND2 |
| 5 | 66230535 | 5.98 × 10−12 | intergenic_variant | ||
| 5 | 66227438 | 6.17 × 10−12 | intergenic_variant | ||
| 5 | 66103958 | 6.77 × 10−12 | intron_variant | ||
| 5 | 66210825 | 1.24 × 10−11 | intergenic_variant | ||
| 5 | 66220545 | 1.27 × 10−11 | intergenic_variant | ||
| 5 | 66224773 | 1.64 × 10−11 | intergenic_variant | ||
| 5 | 66224775 | 1.64 × 10−11 | intergenic_variant | ||
| 5 | 66224782 | 1.64 × 10−11 | intergenic_variant | ||
| 5 | 66230608 | 1.70 × 10−11 | intergenic_variant | ||
| 2 | 2915752 | 1.35 × 10−10 | intron_variant | SHANK2 | |
| 2 | 2927340 | 2.99 × 10−10 | intron_variant | ||
| 2 | 2919084 | 1.71 × 10−9 | intron_variant | ||
| 2 | 2919066 | 2.07 × 10−9 | intron_variant | ||
| 2 | 2928547 | 2.13 × 10−9 | intron_variant | ||
| 2 | 2927984 | 3.34 × 10−9 | intron_variant | ||
| 2 | 2924104 | 5.16 × 10−9 | intron_variant | ||
| 2 | 2924106 | 5.16 × 10−9 | intron_variant | ||
| 2 | 2927278 | 7.83 × 10−9 | intron_variant | ||
| 2 | 2928297 | 9.32 × 10−9 | intron_variant |
| Traits | SSC | Position | p-Value | Consequence | Candidate Gene |
|---|---|---|---|---|---|
| Adjusted 100 kg LMA | 6 | 166758770 | 1.41−8 | intron_variant | GPBP1L1, IPP, TMEM69, NASP, PRDX1, TESK2, MMACHC, ARMH1, ENSSSCG00000021448, ENSSSCG00000046843 |
| 6 | 166758775 | 1.41−8 | intron_variant | ||
| 6 | 166603876 | 1.67−7 | intron_variant | ||
| 6 | 166786086 | 5.03−7 | intron_variant | ||
| 6 | 166758763 | 8.36−7 | intron_variant | ||
| 6 | 165650769 | 1.13−6 | intergenic_variant | ||
| 6 | 165953286 | 1.24−6 | intron_variant | ||
| 6 | 166744870 | 1.34−6 | intron_variant | ||
| LMA EBV | 6 | 165987408 | 2.06−8 | intron_variant | MAST2, PLK3, AKR1A1, UROD, EIF2B3, HECTD3, BEST4, GPBP1L1, IPP, TMEM69, NASP, PRDX1, TESK2, ZSWIM5, DYNLT4, BTBD19, RPS8, MMACHC, ARMH1, ENSSSCG00000021448, ENSSSCG00000046843, ENSSSCG0000003909, ENSSSCG00000035907 |
| 6 | 166603876 | 3.45−8 | intron_variant | ||
| 6 | 165924110 | 7.65−8 | intron_variant | ||
| 6 | 165924144 | 7.65−8 | intron_variant | ||
| 6 | 165927020 | 7.83−8 | intron_variant | ||
| 6 | 166000906 | 1.06−7 | missense variant, upstream gene variant, downstream gene variant | ||
| 6 | 166017041 | 1.35−7 | upstream gene variant, downstream gene variant | ||
| 6 | 166629589 | 1.40−7 | intron_variant | ||
| 6 | 166634843 | 1.40−7 | intron_variant | ||
| 6 | 166637452 | 1.54−7 | intron_variant |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Wu, J.; Jian, Y.; Zhuang, Z.; Qiu, Y.; Huang, R.; Lu, P.; Guan, X.; Huang, X.; Li, S.; et al. Genome-Wide Association Studies Revealed Significant QTLs and Candidate Genes Associated with Backfat and Loin Muscle Area in Pigs Using Imputation-Based Whole Genome Sequencing Data. Animals 2022, 12, 2911. https://doi.org/10.3390/ani12212911
Li J, Wu J, Jian Y, Zhuang Z, Qiu Y, Huang R, Lu P, Guan X, Huang X, Li S, et al. Genome-Wide Association Studies Revealed Significant QTLs and Candidate Genes Associated with Backfat and Loin Muscle Area in Pigs Using Imputation-Based Whole Genome Sequencing Data. Animals. 2022; 12(21):2911. https://doi.org/10.3390/ani12212911
Chicago/Turabian StyleLi, Jie, Jie Wu, Yunhua Jian, Zhanwei Zhuang, Yibin Qiu, Ruqu Huang, Pengyun Lu, Xiang Guan, Xiaoling Huang, Shaoyun Li, and et al. 2022. "Genome-Wide Association Studies Revealed Significant QTLs and Candidate Genes Associated with Backfat and Loin Muscle Area in Pigs Using Imputation-Based Whole Genome Sequencing Data" Animals 12, no. 21: 2911. https://doi.org/10.3390/ani12212911
APA StyleLi, J., Wu, J., Jian, Y., Zhuang, Z., Qiu, Y., Huang, R., Lu, P., Guan, X., Huang, X., Li, S., Min, L., & Ye, Y. (2022). Genome-Wide Association Studies Revealed Significant QTLs and Candidate Genes Associated with Backfat and Loin Muscle Area in Pigs Using Imputation-Based Whole Genome Sequencing Data. Animals, 12(21), 2911. https://doi.org/10.3390/ani12212911






