Exosomes in Mastitis—Research Status, Opportunities, and Challenges
Abstract
Simple Summary
Abstract
1. Introduction
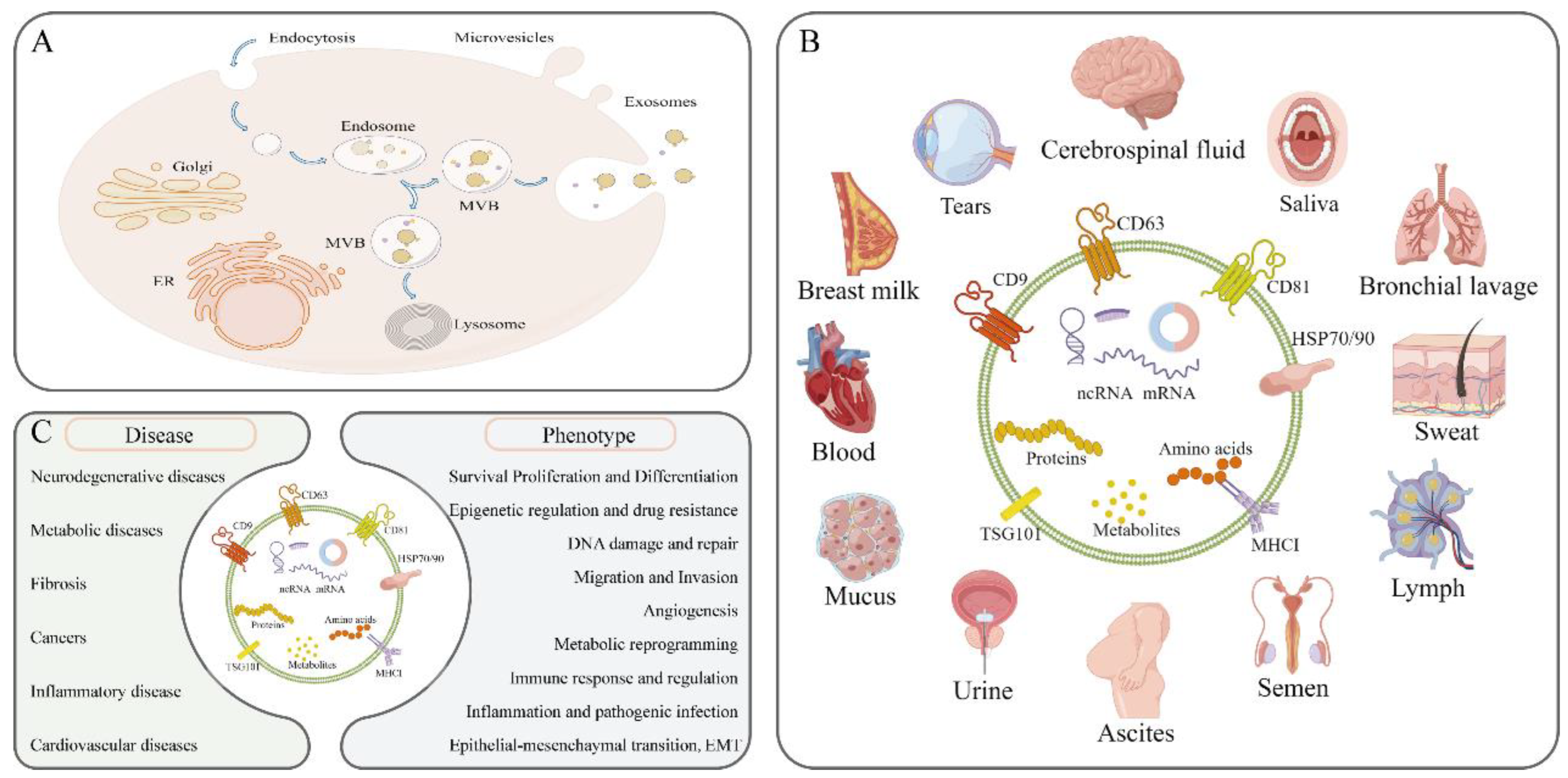
2. Introduction to Exosomes
2.1. The Biogenesis of Exosomes
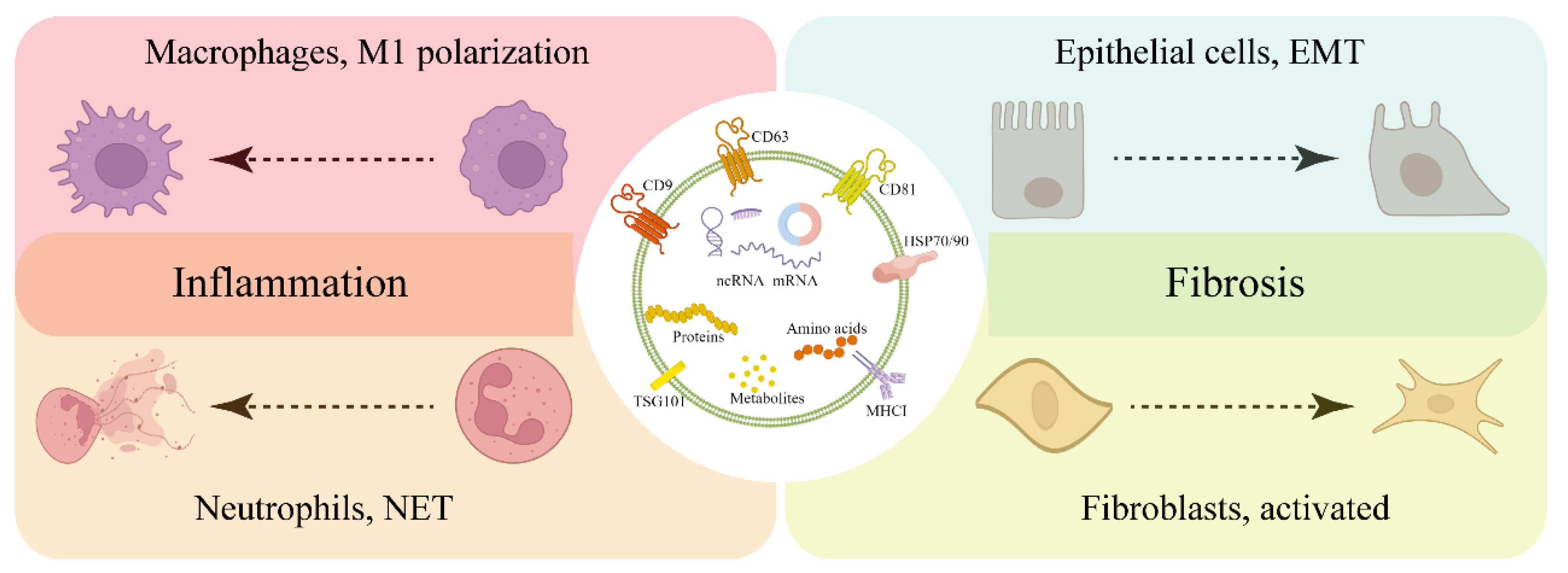
2.2. Biological Functions of Exosomes
2.3. Isolation and Identification of Exosomes
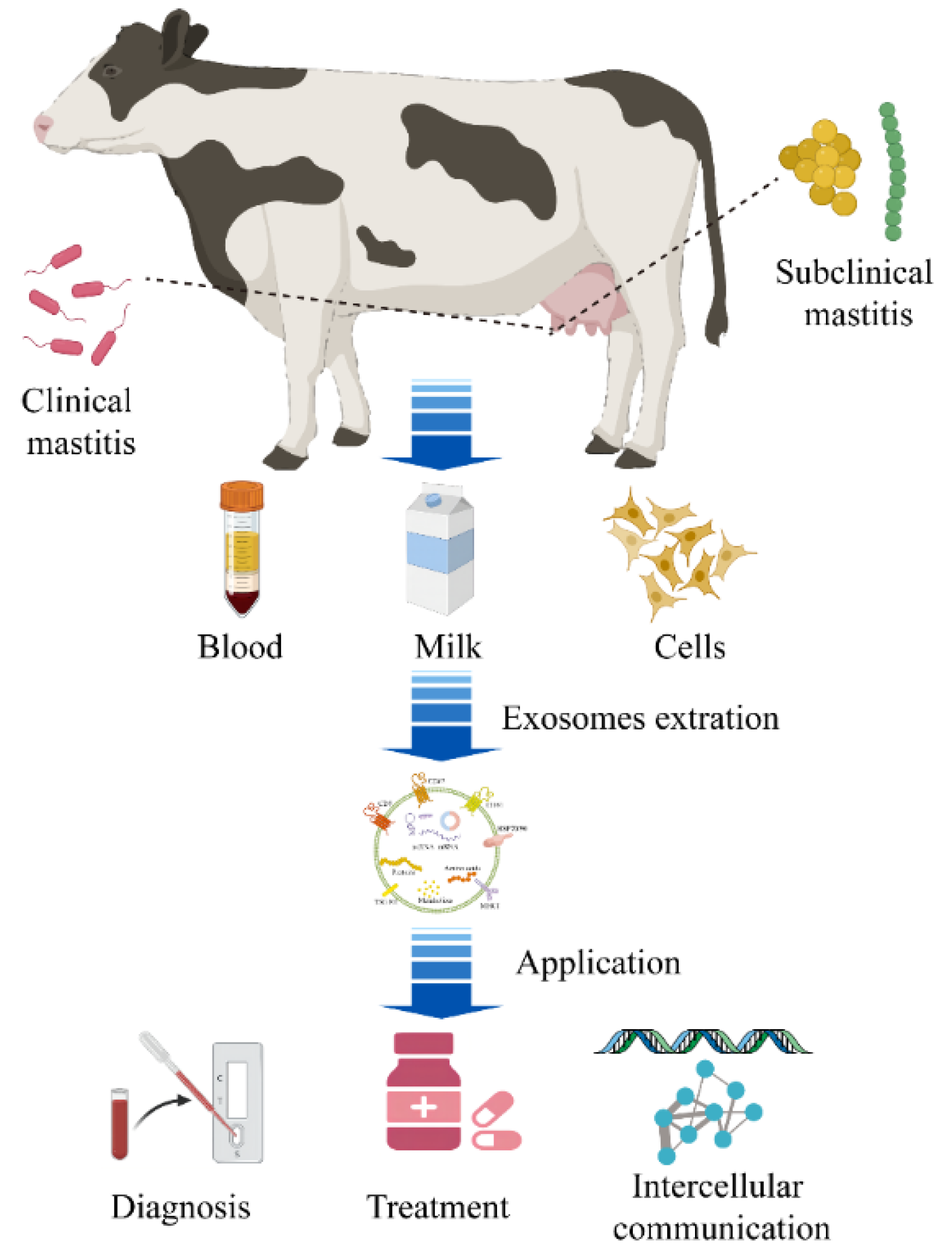
3. Exosomes Associated with Mastitis
3.1. Milk-Derived Exosomes
3.2. Mammary Epithelial Cell-Derived Exosomes
3.3. Peripheral Blood and Urine Exosomes
3.4. Mammary Tissue Exosomes
4. Research Status of Exosomes That Are Predictive and Diagnostic Markers in Mastitis
5. Application Prospects of Exosomes as Drug Delivery Vehicles in Mastitis Treatment
6. Exosome-Mediated Intercellular Communication Is Involved in the Occurrence and Development of Mastitis
7. Challenges of Using Exosomes in the Diagnosis and Treatment of Mastitis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Full name |
| AD | Alzheimer’s disease |
| AKI | Acute kidney injury |
| ALIX | Apoptosis-linked gene 2-interacting protein X |
| BMECs | Bovine mammary epithelial cells |
| circRNA | Circular RNA |
| E. coli | Escherichia coli |
| EMT | Epithelial-mesenchymal transition |
| ESCRT | Endosomal sorting complexes required for transport |
| FCM | Flow Cytometry |
| GC-MS | Gas Chromatography-Mass Spectrometer |
| HC11 | Mouse Mammary Epithelial Cell Lines-HC11 |
| HRS | Hepatocyte growth factor-regulated tyrosine kinase substrate |
| IBD | Inflammatory bowel disease |
| ISEV | International Society for Extracellular Vesicles |
| IVLs | Internal intraluminal vesicles |
| LC-MS | Liquid Chromatography-Mass Spectrometry |
| LncRNAs | Long noncoding RNAs |
| LPS | Lipopolysaccharide |
| LTA | Lipoteichoic acid |
| M1 | Macrophages type I |
| M2 | Macrophages type II |
| m6A | N6-Methyladenosine |
| MAC-T | Bovine Mammary Epithelial Cell Lines-MAC-T |
| MAPK | Mitogen-activated protein kinase |
| MVBs | Multivesicular bodies |
| NET | Neutrophil extracellular traps |
| NF-κB | Nuclear factor kappa B |
| nSMase2 | Neutral sphingomyelinase2 |
| NTA | Nanoparticle Tracking Analyzer |
| PD | Parkinson’s disease |
| PEG | Polymer polyethylene glycol |
| PON1 | Paraoxonase 1 |
| S. agalactiae | Streptococcus agalactiae |
| S. Aureus | Staphylococcus aureus |
| SCC | Somatic cell count |
| siRNA | Short interfering RNA |
| SNARE | Soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptor |
| STAM1 | Signal transducing adapter molecule 1 |
| TEM | Transmission Electron Microscopy |
| TSG101 | Tumor susceptibility gene 101 |
| WB | Western blot |
References
- Halasa, T.; Huijps, K.; Osteras, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q 2007, 29, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Ruegg, P.L. Investigation of mastitis problems on farms. Vet. Clin. North. Am. Food Anim. Pract. 2003, 19, 47–73. [Google Scholar] [CrossRef]
- Stevens, M.; Piepers, S.; De Vliegher, S. Mastitis prevention and control practices and mastitis treatment strategies associated with the consumption of (critically important) antimicrobials on dairy herds in Flanders, Belgium. J. Dairy Sci. 2016, 99, 2896–2903. [Google Scholar] [CrossRef] [PubMed]
- Ruegg, P.L. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef] [PubMed]
- Gruet, P.; Maincent, P.; Berthelot, X.; Kaltsatos, V. Bovine mastitis and intramammary drug delivery: Review and perspectives. Adv. Drug Deliv. Rev. 2001, 50, 245–259. [Google Scholar] [CrossRef]
- Saeed, S.I.; Mat Yazid, K.A.; Hashimy, H.A.; Dzulkifli, S.K.; Nordin, F.; Nik Him, N.A.; Omar, M.; Aklilu, E.; Mohamad, M.; Zalati, C.W.S.; et al. Prevalence, Antimicrobial Resistance, and Characterization of Staphylococcus aureus Isolated from Subclinical Bovine Mastitis in East Coast Malaysia. Animals 2022, 12, 1680. [Google Scholar] [CrossRef]
- Bari, M.S.; Rahman, M.M.; Persson, Y.; Derks, M.; Sayeed, M.A.; Hossain, D.; Singha, S.; Hoque, M.A.; Sivaraman, S.; Fernando, P.; et al. Subclinical mastitis in dairy cows in south-Asian countries: A review of risk factors and etiology to prioritize control measures. Vet. Res. Commun. 2022, 46, 621–640. [Google Scholar] [CrossRef]
- Tamba, M.; Rocca, R.; Prosperi, A.; Pupillo, G.; Bassi, P.; Galletti, G.; Martini, E.; Santi, A.; Casadei, G.; Arrigoni, N. Evaluation of Control Program Against Streptococcus agalactiae Infection in Dairy Herds During 2019–2021 in Emilia-Romagna Region, Northern Italy. Front. Vet. Sci. 2022, 9, 904527. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Tang, Y.; Liu, X.; Zhang, H.; Zhou, Y.; Wang, Y.; Xiao, W.; Yu, Y. Testing Two Somatic Cell Count Cutoff Values for Bovine Subclinical Mastitis Detection Based on Milk Microbiota and Peripheral Blood Leukocyte Transcriptome Profile. Animals 2022, 12, 1694. [Google Scholar] [CrossRef]
- Gunther, J.; Esch, K.; Poschadel, N.; Petzl, W.; Zerbe, H.; Mitterhuemer, S.; Blum, H.; Seyfert, H.M. Comparative kinetics of Escherichia coli- and Staphylococcus aureus-specific activation of key immune pathways in mammary epithelial cells demonstrates that S. aureus elicits a delayed response dominated by interleukin-6 (IL-6) but not by IL-1A or tumor necrosis factor alpha. Infect. Immun. 2011, 79, 695–707. [Google Scholar] [CrossRef]
- Gilbert, F.B.; Cunha, P.; Jensen, K.; Glass, E.J.; Foucras, G.; Robert-Granie, C.; Rupp, R.; Rainard, P. Differential response of bovine mammary epithelial cells to Staphylococcus aureus or Escherichia coli agonists of the innate immune system. Vet. Res. 2013, 44, 40. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Deng, R.; Li, X.; Zhang, Y.; Gao, M.Q. RNA-seq analysis of different inflammatory reactions induced by lipopolysaccharide and lipoteichoic acid in bovine mammary epithelial cells. Microb. Pathog. 2019, 130, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Chen, Z.; Tang, H.B.; Shan, L.Q.; Chen, Z.Y.; Wang, X.H.; Huang, D.G.; Liu, S.C.; Chen, X.; Yang, H.; et al. Exosomes derived from olfactory ensheathing cells provided neuroprotection for spinal cord injury by switching the phenotype of macrophages/microglia. Bioeng. Transl. Med. 2022, 7, e10287. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Cai, G.L.; Zhuang, Z.; Pei, S.Y.; Xu, S.N.; Wang, Y.N.; Wang, H.; Wang, X.; Cui, C.; Sun, M.C.; et al. Interleukin-1beta-Treated Mesenchymal Stem Cells Inhibit Inflammation in Hippocampal Astrocytes Through Exosome-Activated Nrf-2 Signaling. Int. J. Nanomed. 2021, 16, 1423–1434. [Google Scholar] [CrossRef]
- Cocucci, E.; Meldolesi, J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015, 25, 364–372. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Blanc, L.; Vidal, M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases 2018, 9, 95–106. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30, sup pp 11–13. [Google Scholar] [CrossRef]
- Song, L.; Tang, S.; Han, X.; Jiang, Z.; Dong, L.; Liu, C.; Liang, X.; Dong, J.; Qiu, C.; Wang, Y.; et al. KIBRA controls exosome secretion via inhibiting the proteasomal degradation of Rab27a. Nat. Commun. 2019, 10, 1639. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Bai, H.; Ren, L.; Zhang, L. The Role of Exosome and the ESCRT Pathway on Enveloped Virus Infection. Int. J. Mol. Sci. 2021, 22, 9060. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Thery, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed]
- Juan, T.; Furthauer, M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin. Cell Dev. Biol. 2018, 74, 66–77. [Google Scholar] [CrossRef]
- Raiborg, C.; Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009, 458, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020, 219, 1–22. [Google Scholar] [CrossRef]
- Wei, D.; Zhan, W.; Gao, Y.; Huang, L.; Gong, R.; Wang, W.; Zhang, R.; Wu, Y.; Gao, S.; Kang, T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021, 31, 157–177. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Dinkins, M.B.; Dasgupta, S.; Wang, G.; Zhu, G.; Bieberich, E. Exosome reduction in vivo is associated with lower amyloid plaque load in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging. 2014, 35, 1792–1800. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, R.; Liang, Y.; Fu, X.; Wang, D.; Wang, C. Blockade of lncRNA-ASLNCS5088-enriched exosome generation in M2 macrophages by GW4869 dampens the effect of M2 macrophages on orchestrating fibroblast activation. FASEB J. 2019, 33, 12200–12212. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, K.; Zhi, Y.; Wu, Y.; Chen, B.; Bai, J.; Wang, X. Tumor-derived exosomal miR-19b-3p facilitates M2 macrophage polarization and exosomal LINC00273 secretion to promote lung adenocarcinoma metastasis via Hippo pathway. Clin. Transl. Med. 2021, 11, e478. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, A.L.; Adams, A.; King, K.E.; Dunn, W.; Christenson, L.K.; Hung, W.T.; Weinman, S.A. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J. Cell Biol. 2020, 219, e201912074. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martin, R.; Wang, G.; Brandao, B.B.; Zanotto, T.M.; Shah, S.; Kumar Patel, S.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022, 601, 446–451. [Google Scholar] [CrossRef]
- Martins-Marques, T.; Costa, M.C.; Catarino, S.; Simoes, I.; Aasen, T.; Enguita, F.J.; Girao, H. Cx43-mediated sorting of miRNAs into extracellular vesicles. EMBO Rep. 2022, 23, e54312. [Google Scholar] [CrossRef]
- Buchet-Poyau, K.; Courchet, J.; Le Hir, H.; Seraphin, B.; Scoazec, J.Y.; Duret, L.; Domon-Dell, C.; Freund, J.N.; Billaud, M. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res. 2007, 35, 1289–1300. [Google Scholar] [CrossRef]
- Lin, F.; Zeng, Z.; Song, Y.; Li, L.; Wu, Z.; Zhang, X.; Li, Z.; Ke, X.; Hu, X. YBX-1 mediated sorting of miR-133 into hypoxia/reoxygenation-induced EPC-derived exosomes to increase fibroblast angiogenesis and MEndoT. Stem. Cell Res. Ther. 2019, 10, 263. [Google Scholar] [CrossRef]
- Temoche-Diaz, M.M.; Shurtleff, M.J.; Nottingham, R.M.; Yao, J.; Fadadu, R.P.; Lambowitz, A.M.; Schekman, R. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife 2019, 8, e47544. [Google Scholar] [CrossRef]
- Jia, L.; Qiu, Q.; Zhang, H.; Chu, L.; Du, Y.; Zhang, J.; Zhou, C.; Liang, F.; Shi, S.; Wang, S.; et al. Concordance between the assessment of Abeta42, T-tau, and P-T181-tau in peripheral blood neuronal-derived exosomes and cerebrospinal fluid. Alzheimers Dement. 2019, 15, 1071–1080. [Google Scholar] [CrossRef]
- Kim, K.U.; Kim, W.H.; Jeong, C.H.; Yi, D.Y.; Min, H. More than Nutrition: Therapeutic Potential of Breast Milk-Derived Exosomes in Cancer. Int. J. Mol. Sci. 2020, 21, 7327. [Google Scholar] [CrossRef]
- Zhang, D.; Lee, H.; Wang, X.; Groot, M.; Sharma, L.; Dela Cruz, C.S.; Jin, Y. A potential role of microvesicle-containing miR-223/142 in lung inflammation. Thorax 2019, 74, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Joyce, D.P.; Kerin, M.J.; Dwyer, R.M. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int. J. Cancer 2016, 139, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e418. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- Qi, Y.; Jin, C.; Qiu, W.; Zhao, R.; Wang, S.; Li, B.; Zhang, Z.; Guo, Q.; Zhang, S.; Gao, Z.; et al. The dual role of glioma exosomal microRNAs: Glioma eliminates tumor suppressor miR-1298-5p via exosomes to promote immunosuppressive effects of MDSCs. Cell Death Dis. 2022, 13, 426. [Google Scholar] [CrossRef]
- Han, B.; Zhang, H.; Tian, R.; Liu, H.; Wang, Z.; Wang, Z.; Tian, J.; Cui, Y.; Ren, S.; Zuo, X.; et al. Exosomal EPHA2 derived from highly metastatic breast cancer cells promotes angiogenesis by activating the AMPK signaling pathway through Ephrin A1-EPHA2 forward signaling. Theranostics 2022, 12, 4127–4146. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Yu, H.; Cheng, Y.; Han, J.; Li, K.; Zhuang, J.; Lv, Q.; Yang, X.; Yang, H. Bladder cancer-derived exosomal KRT6B promotes invasion and metastasis by inducing EMT and regulating the immune microenvironment. J. Transl. Med. 2022, 20, 308. [Google Scholar] [CrossRef]
- Turiello, R.; Capone, M.; Morretta, E.; Monti, M.C.; Madonna, G.; Azzaro, R.; Del Gaudio, P.; Simeone, E.; Sorrentino, A.; Ascierto, P.A.; et al. Exosomal CD73 from serum of patients with melanoma suppresses lymphocyte functions and is associated with therapy resistance to anti-PD-1 agents. J. Immunother. Cancer 2022, 10, e004043. [Google Scholar] [CrossRef]
- Geng, X.; Zhang, Y.; Lin, X.; Zeng, Z.; Hu, J.; Hao, L.; Xu, J.; Wang, X.; Wang, H.; Li, Q. Exosomal circWDR62 promotes temozolomide resistance and malignant progression through regulation of the miR-370-3p/MGMT axis in glioma. Cell Death Dis. 2022, 13, 596. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Y.; Ma, L.; Yu, K.; Niu, Y.; Xu, X.; Shi, Y.; Guo, S.; Xue, X.; Wang, Y.; et al. Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma. Cancer Commun. 2022, 42, 287–313. [Google Scholar] [CrossRef]
- Wojda, U. The Perspective of Exosomal MicroRNAs as Biomarkers for Preclinical Alzheimer’s Disease. Biol. Psychiatry 2022, 92, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.T.; Liu, C.G.; Gao, S.C.; Zhang, Y.; Wang, P.C. The Serum Exosome Derived MicroRNA-135a, -193b, and -384 Were Potential Alzheimer’s Disease Biomarkers. Biomed. Environ. Sci. 2018, 31, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sun, Y.; Luo, Z.; Lin, J.; Qi, B.; Kang, X.; Ying, C.; Guo, C.; Yao, M.; Chen, X.; et al. Potential Mechanism Underlying Exercise Upregulated Circulating Blood Exosome miR-215-5p to Prevent Necroptosis of Neuronal Cells and a Model for Early Diagnosis of Alzheimer’s Disease. Front. Aging Neurosci. 2022, 14, 860364. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. [Google Scholar] [CrossRef]
- Sheykhhasan, M.; Amini, R.; Soleimani Asl, S.; Saidijam, M.; Hashemi, S.M.; Najafi, R. Neuroprotective effects of coenzyme Q10-loaded exosomes obtained from adipose-derived stem cells in a rat model of Alzheimer’s disease. Biomed. Pharmacother. 2022, 152, 113224. [Google Scholar] [CrossRef]
- Li, N.; Shu, J.; Yang, X.; Wei, W.; Yan, A. Exosomes Derived From M2 Microglia Cells Attenuates Neuronal Impairment and Mitochondrial Dysfunction in Alzheimer’s Disease Through the PINK1/Parkin Pathway. Front. Cell Neurosci. 2022, 16, 874102. [Google Scholar] [CrossRef]
- Zavatti, M.; Gatti, M.; Beretti, F.; Palumbo, C.; Maraldi, T. Exosomes Derived from Human Amniotic Fluid Mesenchymal Stem Cells Preserve Microglia and Neuron Cells from Abeta. Int. J. Mol. Sci. 2022, 23, 4967. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, M.M.; Wang, M.; Jiang, Z.H.; Tan, Z.G. Bone Marrow-Derived Mesenchymal Stem Cell-Derived Exosomes Containing Gli1 Alleviate Microglial Activation and Neuronal Apoptosis In Vitro and in a Mouse Parkinson Disease Model by Direct Inhibition of Sp1 Signaling. J. Neuropathol. Exp. Neurol. 2022, 81, 522–534. [Google Scholar] [CrossRef]
- Xiong, Y.; Tang, R.; Xu, J.; Jiang, W.; Gong, Z.; Zhang, L.; Ning, Y.; Huang, P.; Xu, J.; Chen, G.; et al. Tongxinluo-pretreated mesenchymal stem cells facilitate cardiac repair via exosomal transfer of miR-146a-5p targeting IRAK1/NF-kappaB p65 pathway. Stem Cell Res. Ther. 2022, 13, 289. [Google Scholar] [CrossRef]
- Wang, D.; Xue, H.; Tan, J.; Liu, P.; Qiao, C.; Pang, C.; Zhang, L. Bone marrow mesenchymal stem cells-derived exosomes containing miR-539-5p inhibit pyroptosis through NLRP3/caspase-1 signalling to alleviate inflammatory bowel disease. Inflamm. Res. 2022, 71, 833–846. [Google Scholar] [CrossRef]
- Lv, L.L.; Feng, Y.; Wu, M.; Wang, B.; Li, Z.L.; Zhong, X.; Wu, W.J.; Chen, J.; Ni, H.F.; Tang, T.T.; et al. Exosomal miRNA-19b-3p of tubular epithelial cells promotes M1 macrophage activation in kidney injury. Cell Death Differ. 2020, 27, 210–226. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Huo, P.; Cui, K.; Wei, H.; Cao, J.; Wang, J.; Liu, Q.; Lei, X.; Zhang, S. Follicular fluid-derived exosomal miR-143-3p/miR-155-5p regulate follicular dysplasia by modulating glycolysis in granulosa cells in polycystic ovary syndrome. Cell Commun. Signal 2022, 20, 61. [Google Scholar] [CrossRef] [PubMed]
- Wan, Z.; Yang, X.; Liu, X.; Sun, Y.; Yu, P.; Xu, F.; Deng, H. M2 macrophage-derived exosomal microRNA-411-5p impedes the activation of hepatic stellate cells by targeting CAMSAP1 in NASH model. iScience 2022, 25, 104597. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Jin, Z.; Bandyopadhyay, G.; Cunha, E.R.K.; Liu, X.; Zhao, H.; Zhang, D.; Jouihan, H.; Pourshahian, S.; Kisseleva, T.; et al. MiR-690 treatment causes decreased fibrosis and steatosis and restores specific Kupffer cell functions in NASH. Cell Metab. 2022, 34, 978–990.e974. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.M.; Chen, S.; Liu, N.; Zhao, Q.; et al. Progress, opportunity, and perspective on exosome isolation—Efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Soares Martins, T.; Catita, J.; Martins Rosa, I.; da Cruz e Silva, O.A.B.; Henriques, A.G. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE 2018, 13, e0198820. [Google Scholar] [CrossRef]
- Garcia-Romero, N.; Madurga, R.; Rackov, G.; Palacin-Aliana, I.; Nunez-Torres, R.; Asensi-Puig, A.; Carrion-Navarro, J.; Esteban-Rubio, S.; Peinado, H.; Gonzalez-Neira, A.; et al. Polyethylene glycol improves current methods for circulating extracellular vesicle-derived DNA isolation. J. Transl. Med. 2019, 17, 75. [Google Scholar] [CrossRef]
- Kanchi Ravi, R.; Khosroheidari, M.; DiStefano, J.K. A modified precipitation method to isolate urinary exosomes. J. Vis. Exp. 2015, 95, e51158. [Google Scholar] [CrossRef]
- Wan, Z.; Zhao, L.; Lu, F.; Gao, X.; Dong, Y.; Zhao, Y.; Wei, M.; Yang, G.; Xing, C.; Liu, L. Mononuclear phagocyte system blockade improves therapeutic exosome delivery to the myocardium. Theranostics 2020, 10, 218–230. [Google Scholar] [CrossRef]
- Peng, C.; Wang, J.; Bao, Q.; Wang, J.; Liu, Z.; Wen, J.; Zhang, W.; Shen, Y. Isolation of extracellular vesicle with different precipitation-based methods exerts a tremendous impact on the biomarker analysis for clinical plasma samples. Cancer Biomark 2020, 29, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, E.; Boing, A.N.; Gool, E.L.; Nieuwland, R. Recent developments in the nomenclature, presence, isolation, detection and clinical impact of extracellular vesicles. J. Thromb. Haemost. 2016, 14, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef]
- Sun, J.; Aswath, K.; Schroeder, S.G.; Lippolis, J.D.; Reinhardt, T.A.; Sonstegard, T.S. MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection. BMC Genom. 2015, 16, 806. [Google Scholar] [CrossRef]
- Cai, M.; He, H.; Jia, X.; Chen, S.; Wang, J.; Shi, Y.; Liu, B.; Xiao, W.; Lai, S. Genome-wide microRNA profiling of bovine milk-derived exosomes infected with Staphylococcus aureus. Cell Stress Chaperones 2018, 23, 663–672. [Google Scholar] [CrossRef]
- Ma, S.; Tong, C.; Ibeagha-Awemu, E.M.; Zhao, X. Identification and characterization of differentially expressed exosomal microRNAs in bovine milk infected with Staphylococcus aureus. BMC Genom. 2019, 20, 934. [Google Scholar] [CrossRef]
- Saenz-de-Juano, M.D.; Silvestrelli, G.; Bauersachs, S.; Ulbrich, S.E. Determining extracellular vesicles properties and miRNA cargo variability in bovine milk from healthy cows and cows undergoing subclinical mastitis. BMC Genom. 2022, 23, 189. [Google Scholar] [CrossRef]
- Chen, W.; Wang, R.; Li, D.; Zuo, C.; Wen, P.; Liu, H.; Chen, Y.; Fujita, M.; Wu, Z.; Yang, G. Comprehensive Analysis of the Glycome and Glycoproteome of Bovine Milk-Derived Exosomes. J. Agric. Food Chem. 2020, 68, 12692–12701. [Google Scholar] [CrossRef]
- Vaswani, K.M.; Peiris, H.; Qin Koh, Y.; Hill, R.J.; Harb, T.; Arachchige, B.J.; Logan, J.; Reed, S.; Davies, P.S.W.; Mitchell, M.D. A complete proteomic profile of human and bovine milk exosomes by liquid chromatography mass spectrometry. Expert Rev. Proteom. 2021, 18, 719–735. [Google Scholar] [CrossRef]
- Yang, M.; Song, D.; Cao, X.; Wu, R.; Liu, B.; Ye, W.; Wu, J.; Yue, X. Comparative proteomic analysis of milk-derived exosomes in human and bovine colostrum and mature milk samples by iTRAQ-coupled LC-MS/MS. Food Res. Int. 2017, 92, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Benmoussa, A.; Laugier, J.; Beauparlant, C.J.; Lambert, M.; Droit, A.; Provost, P. Complexity of the microRNA transcriptome of cow milk and milk-derived extracellular vesicles isolated via differential ultracentrifugation. J. Dairy Sci. 2020, 103, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Chen, T.; Xie, M.Y.; Luo, J.Y.; He, J.J.; Xi, Q.Y.; Sun, J.J.; Zhang, Y.L. Exploration of long noncoding RNA in bovine milk exosomes and their stability during digestion in vitro. J. Dairy Sci. 2019, 102, 6726–6737. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, J.; Zeng, H.F.; Zhong, J.F.; Li, H.X.; Chen, K.L. Identification and bioinformatics analysis of differentially expressed milk exosomal microRNAs in milk exosomes of heat-stressed Holstein cows. Funct. Integr. Genom. 2022, 22, 77–87. [Google Scholar] [CrossRef]
- Colitti, M.; Sgorlon, S.; Licastro, D.; Stefanon, B. Differential expression of miRNAs in milk exosomes of cows subjected to group relocation. Res. Vet. Sci. 2019, 122, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, S. Identification and comparison of exosomal microRNAs in the milk and colostrum of two different cow breeds. Gene 2020, 743, 144609. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Pei, Y.; Wang, X.; Feng, J.; Zhang, Y.; Gao, M.Q. LncRNA XIST mediates bovine mammary epithelial cell inflammatory response via NF-kappaB/NLRP3 inflammasome pathway. Cell Prolif. 2019, 52, e12525. [Google Scholar] [CrossRef]
- Wang, M.Q.; Zhou, C.H.; Cong, S.; Han, D.X.; Wang, C.J.; Tian, Y.; Zhang, J.B.; Jiang, H.; Yuan, B. Lipopolysaccharide inhibits triglyceride synthesis in dairy cow mammary epithelial cells by upregulating miR-27a-3p, which targets the PPARG gene. J. Dairy Sci. 2021, 104, 989–1001. [Google Scholar] [CrossRef]
- Ogunnaike, M.; Wang, H.; Zempleni, J. Bovine mammary alveolar MAC-T cells afford a tool for studies of bovine milk exosomes in drug delivery. Int. J. Pharm. 2021, 610, 121263. [Google Scholar] [CrossRef]
- Zhang, M.; Ma, Z.; Li, R.; Guo, S.; Qiu, Y.; Gao, X. Proteomic Analysis Reveals Proteins and Pathways Associated with Lactation in Bovine Mammary Epithelial Cell-Derived Exosomes. J. Proteome Res. 2020, 19, 3211–3219. [Google Scholar] [CrossRef]
- Chen, Y.; Jing, H.; Chen, M.; Liang, W.; Yang, J.; Deng, G.; Guo, M. Transcriptional Profiling of Exosomes Derived from Staphylococcus aureus-Infected Bovine Mammary Epithelial Cell Line MAC-T by RNA-Seq Analysis. Oxid. Med. Cell Longev. 2021, 2021, 8460355. [Google Scholar] [CrossRef] [PubMed]
- Bates, A.J.; Wells, M.; Laven, R.A. The effect of pre-calving injection of trace mineral supplements on periparturient disease incidence in pasture based dairy cows. Vet. J. 2022, 286, 105867. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Xu, S.; Li, J.; Cui, L.; Dong, J.; Meng, X.; Zhu, G.; Wang, H. Selenomethionine protected BMECs from inflammatory injury and oxidative damage induced by Klebsiella pneumoniae by inhibiting the NF-kappaB and activating the Nrf2 signaling pathway. Int. Immunopharmacol. 2022, 110, 109027. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Z.; Ma, Y.; Xiao, J.; Chen, T.; Ma, J.; Liu, S.; Wang, Y.; Khan, A.; Alugongo, G.M.; Cao, Z. Role of Selenium and Vitamins E and B9 in the Alleviation of Bovine Mastitis during the Periparturient Period. Antioxidants 2022, 11, 657. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Lin, J.; He, W.; Huang, K. Selenium and Taurine Combination Is Better Than Alone in Protecting Lipopolysaccharide-Induced Mammary Inflammatory Lesions via Activating PI3K/Akt/mTOR Signaling Pathway by Scavenging Intracellular ROS. Oxid. Med. Cell Longev. 2021, 2021, 5048375. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.; Chen, B.; Zhao, B.; Gao, X.J. Selenium Deficiency Promotes Oxidative Stress-Induced Mastitis via Activating the NF-kappaB and MAPK Pathways in Dairy Cow. Biol. Trace Elem. Res. 2022, 200, 2716–2726. [Google Scholar] [CrossRef]
- Jing, H.; Chen, Y.; Liang, W.; Chen, M.; Qiu, C.; Guo, M.Y. Effects of Selenium on MAC-T Cells in Bovine Mastitis: Transcriptome Analysis of Exosomal mRNA Interactions. Biol. Trace Elem. Res. 2021, 199, 2904–2912. [Google Scholar] [CrossRef]
- Cai, M.; Shi, Y.; Zheng, T.; Hu, S.; Du, K.; Ren, A.; Jia, X.; Chen, S.; Wang, J.; Lai, S. Mammary epithelial cell derived exosomal MiR-221 mediates M1 macrophage polarization via SOCS1/STATs to promote inflammatory response. Int. Immunopharmacol. 2020, 83, 106493. [Google Scholar] [CrossRef]
- Cai, M.; Fan, W.; Li, X.; Sun, H.; Dai, L.; Lei, D.; Dai, Y.; Liao, Y. The Regulation of Staphylococcus aureus-Induced Inflammatory Responses in Bovine Mammary Epithelial Cells. Front. Vet. Sci. 2021, 8, 683886. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, J.; Huang, Z.; Jing, H.; Yin, B.; Guo, S.; Deng, G.; Guo, M. Exosomal lnc-AFTR as a novel translation regulator of FAS ameliorates Staphylococcus aureus-induced mastitis. Biofactors 2022, 48, 148–163. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, T.; Hu, X.; Xie, Y.; Wu, R.; Lian, S.; Wang, J. Exosomal miR-193b-5p as a regulator of LPS-induced inflammation in dairy cow mammary epithelial cells. In Vitro Cell Dev. Biol. Anim. 2021, 57, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, H.; Chen, L.; Wang, L.; Liu, X.; Ru, C.; Song, A. Identification and characterization of novel and differentially expressed microRNAs in peripheral blood from healthy and mastitis Holstein cattle by deep sequencing. Anim. Genet. 2014, 45, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.C.; Habiby, G.H.; Jasing Pathiranage, C.C.; Rahman, M.M.; Chen, H.W.; Husna, A.A.; Kubota, C.; Miura, N. Bovine serum miR-21 expression affected by mastitis. Res. Vet. Sci. 2021, 135, 290–292. [Google Scholar] [CrossRef] [PubMed]
- Luoreng, Z.M.; Yang, J.; Wang, X.P.; Wei, D.W.; Zan, L.S. Expression Profiling of microRNA From Peripheral Blood of Dairy Cows in Response to Staphylococcus aureus-Infected Mastitis. Front. Vet. Sci. 2021, 8, 691196. [Google Scholar] [CrossRef]
- Luoreng, Z.M.; Wang, X.P.; Mei, C.G.; Zan, L.S. Expression profiling of peripheral blood miRNA using RNAseq technology in dairy cows with Escherichia coli-induced mastitis. Sci. Rep. 2018, 8, 12693. [Google Scholar] [CrossRef]
- Dervishi, E.; Zhang, G.; Dunn, S.M.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. GC-MS Metabolomics Identifies Metabolite Alterations That Precede Subclinical Mastitis in the Blood of Transition Dairy Cows. J. Proteome. Res. 2017, 16, 433–446. [Google Scholar] [CrossRef]
- Zhang, G.; Tobolski, D.; Zwierzchowski, G.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. Identification of Serum-Predictive Biomarkers for Subclinical Mastitis in Dairy Cows and New Insights into the Pathobiology of the Disease. J. Agric. Food Chem. 2022, 70, 1724–1746. [Google Scholar] [CrossRef]
- Zwierzchowski, G.; Zhang, G.; Mandal, R.; Wishart, D.S.; Ametaj, B.N. Mass-spec-based urinary metabotyping around parturition identifies screening biomarkers for subclinical mastitis in dairy cows. Res. Vet. Sci. 2020, 129, 39–52. [Google Scholar] [CrossRef]
- Li, R.; Zhang, C.L.; Liao, X.X.; Chen, D.; Wang, W.Q.; Zhu, Y.H.; Geng, X.H.; Ji, D.J.; Mao, Y.J.; Gong, Y.C.; et al. Transcriptome microRNA profiling of bovine mammary glands infected with Staphylococcus aureus. Int. J. Mol. Sci. 2015, 16, 4997–5013. [Google Scholar] [CrossRef]
- Wang, X.; Fan, Y.; He, Y.; Han, Z.; Gong, Z.; Peng, Y.; Meng, Y.; Mao, Y.; Yang, Z.; Yang, Y. Integrative Analysis of miRNA and mRNA Expression Profiles in Mammary Glands of Holstein Cows Artificially Infected with Staphylococcus aureus. Pathogens 2021, 10, 506. [Google Scholar] [CrossRef]
- Luoreng, Z.M.; Wang, X.P.; Mei, C.G.; Zan, L.S. Comparison of microRNA Profiles between Bovine Mammary Glands Infected with Staphylococcus aureus and Escherichia coli. Int. J. Biol. Sci. 2018, 14, 87–99. [Google Scholar] [CrossRef]
- Pu, J.; Li, R.; Zhang, C.; Chen, D.; Liao, X.; Zhu, Y.; Geng, X.; Ji, D.; Mao, Y.; Gong, Y.; et al. Expression profiles of miRNAs from bovine mammary glands in response to Streptococcus agalactiae-induced mastitis. J. Dairy Res. 2017, 84, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci. Rep. 2016, 6, 20680. [Google Scholar] [CrossRef] [PubMed]
- Le Guillou, S.; Leduc, A.; Laubier, J.; Barbey, S.; Rossignol, M.N.; Lefebvre, R.; Marthey, S.; Laloe, D.; Le Provost, F. Characterization of Holstein and Normande whole milk miRNomes highlights breed specificities. Sci. Rep. 2019, 9, 20345. [Google Scholar] [CrossRef] [PubMed]
- Nedic, S.; Vakanjac, S.; Samardzija, M.; Borozan, S. Paraoxonase 1 in bovine milk and blood as marker of subclinical mastitis caused by Staphylococcus aureus. Res. Vet. Sci. 2019, 125, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.C.; Fujikawa, T.; Maemura, T.; Ando, T.; Kitahara, G.; Endo, Y.; Yamato, O.; Koiwa, M.; Kubota, C.; Miura, N. Inflammation-related microRNA expression level in the bovine milk is affected by mastitis. PLoS ONE 2017, 12, e0177182. [Google Scholar] [CrossRef]
- Tzelos, T.; Ho, W.; Charmana, V.I.; Lee, S.; Donadeu, F.X. MiRNAs in milk can be used towards early prediction of mammary gland inflammation in cattle. Sci. Rep. 2022, 12, 5131. [Google Scholar] [CrossRef]
- Moyes, K.M.; Larsen, T.; Friggens, N.C.; Drackley, J.K.; Ingvartsen, K.L. Identification of potential markers in blood for the development of subclinical and clinical mastitis in dairy cattle at parturition and during early lactation. J. Dairy Sci. 2009, 92, 5419–5428. [Google Scholar] [CrossRef]
- Izumi, H.; Tsuda, M.; Sato, Y.; Kosaka, N.; Ochiya, T.; Iwamoto, H.; Namba, K.; Takeda, Y. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J. Dairy Sci. 2015, 98, 2920–2933. [Google Scholar] [CrossRef]
- Del Pozo-Acebo, L.; Hazas, M.L.L.; Tome-Carneiro, J.; Gil-Cabrerizo, P.; San-Cristobal, R.; Busto, R.; Garcia-Ruiz, A.; Davalos, A. Bovine Milk-Derived Exosomes as a Drug Delivery Vehicle for miRNA-Based Therapy. Int. J. Mol. Sci. 2021, 22, 1105. [Google Scholar] [CrossRef]
- Luo, S.; Sun, X.; Huang, M.; Ma, Q.; Du, L.; Cui, Y. Enhanced Neuroprotective Effects of Epicatechin Gallate Encapsulated by Bovine Milk-Derived Exosomes against Parkinson’s Disease through Antiapoptosis and Antimitophagy. J. Agric. Food Chem. 2021, 69, 5134–5143. [Google Scholar] [CrossRef] [PubMed]
- Yenuganti, V.R.; Afroz, S.; Khan, R.A.; Bharadwaj, C.; Nabariya, D.K.; Nayak, N.; Subbiah, M.; Chintala, K.; Banerjee, S.; Reddanna, P.; et al. Milk exosomes elicit a potent anti-viral activity against dengue virus. J. Nanobiotechnology 2022, 20, 317. [Google Scholar] [CrossRef]
- Arntz, O.J.; Pieters, B.C.; Oliveira, M.C.; Broeren, M.G.; Bennink, M.B.; de Vries, M.; van Lent, P.L.; Koenders, M.I.; van den Berg, W.B.; van der Kraan, P.M.; et al. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol. Nutr. Food Res. 2015, 59, 1701–1712. [Google Scholar] [CrossRef] [PubMed]
- Aarts, J.; Boleij, A.; Pieters, B.C.H.; Feitsma, A.L.; van Neerven, R.J.J.; Ten Klooster, J.P.; M’Rabet, L.; Arntz, O.J.; Koenders, M.I.; van de Loo, F.A.J. Flood Control: How Milk-Derived Extracellular Vesicles Can Help to Improve the Intestinal Barrier Function and Break the Gut-Joint Axis in Rheumatoid Arthritis. Front. Immunol. 2021, 12, 703277. [Google Scholar] [CrossRef] [PubMed]
- Maghraby, M.K.; Li, B.; Chi, L.; Ling, C.; Benmoussa, A.; Provost, P.; Postmus, A.C.; Abdi, A.; Pierro, A.; Bourdon, C.; et al. Extracellular vesicles isolated from milk can improve gut barrier dysfunction induced by malnutrition. Sci. Rep. 2021, 11, 7635. [Google Scholar] [CrossRef]
- Babaker, M.A.; Aljoud, F.A.; Alkhilaiwi, F.; Algarni, A.; Ahmed, A.; Khan, M.I.; Saadeldin, I.M.; Alzahrani, F.A. The Therapeutic Potential of Milk Extracellular Vesicles on Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 6812. [Google Scholar] [CrossRef]
- Gonzalez-Sarrias, A.; Iglesias-Aguirre, C.E.; Cortes-Martin, A.; Vallejo, F.; Cattivelli, A.; Del Pozo-Acebo, L.; Del Saz, A.; Lopez de Las Hazas, M.C.; Davalos, A.; Espin, J.C. Milk-Derived Exosomes as Nanocarriers to Deliver Curcumin and Resveratrol in Breast Tissue and Enhance Their Anticancer Activity. Int. J. Mol. Sci. 2022, 23, 2860. [Google Scholar] [CrossRef]
- Ahmed, F.; Tamma, M.; Pathigadapa, U.; Reddanna, P.; Yenuganti, V.R. Drug Loading and Functional Efficacy of Cow, Buffalo, and Goat Milk-Derived Exosomes: A Comparative Study. Mol. Pharm. 2022, 19, 763–774. [Google Scholar] [CrossRef]
- Gao, H.N.; Hu, H.; Wen, P.C.; Lian, S.; Xie, X.L.; Song, H.L.; Yang, Z.N.; Ren, F.Z. Yak milk-derived exosomes alleviate lipopolysaccharide-induced intestinal inflammation by inhibiting PI3K/AKT/C3 pathway activation. J. Dairy Sci. 2021, 104, 8411–8424. [Google Scholar] [CrossRef]
- Gao, H.N.; Guo, H.Y.; Zhang, H.; Xie, X.L.; Wen, P.C.; Ren, F.Z. Yak-milk-derived exosomes promote proliferation of intestinal epithelial cells in an hypoxic environment. J. Dairy Sci. 2019, 102, 985–996. [Google Scholar] [CrossRef]
- Gao, H.N.; Ren, F.Z.; Wen, P.C.; Xie, L.X.; Wang, R.; Yang, Z.N.; Li, Y.X. Yak milk-derived exosomal microRNAs regulate intestinal epithelial cells on proliferation in hypoxic environment. J. Dairy Sci. 2021, 104, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.; Atalla, H.; Karrow, N.; Mallard, B.A. The bioactivity of colostrum and milk exosomes of high, average, and low immune responder cows on human intestinal epithelial cells. J. Dairy Sci. 2021, 104, 2499–2510. [Google Scholar] [CrossRef] [PubMed]
- Somiya, M.; Yoshioka, Y.; Ochiya, T. Biocompatibility of highly purified bovine milk-derived extracellular vesicles. J. Extracell Vesicles 2018, 7, 1440132. [Google Scholar] [CrossRef] [PubMed]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016, 371, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Manca, S.; Upadhyaya, B.; Mutai, E.; Desaulniers, A.T.; Cederberg, R.A.; White, B.R.; Zempleni, J. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci. Rep. 2018, 8, 11321. [Google Scholar] [CrossRef]
- Zempleni, J.; Sukreet, S.; Zhou, F.; Wu, D.; Mutai, E. Milk-Derived Exosomes and Metabolic Regulation. Annu. Rev. Anim. Biosci. 2019, 7, 245–262. [Google Scholar] [CrossRef]
- Betker, J.L.; Angle, B.M.; Graner, M.W.; Anchordoquy, T.J. The Potential of Exosomes From Cow Milk for Oral Delivery. J. Pharm. Sci. 2019, 108, 1496–1505. [Google Scholar] [CrossRef]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef]
- Ngu, A.; Wang, S.; Wang, H.; Khanam, A.; Zempleni, J. Milk exosomes in nutrition and drug delivery. Am. J. Physiol. Cell Physiol. 2022, 322, C865–C874. [Google Scholar] [CrossRef]
- Sedykh, S.; Kuleshova, A.; Nevinsky, G. Milk Exosomes: Perspective Agents for Anticancer Drug Delivery. Int. J. Mol. Sci. 2020, 21, 6646. [Google Scholar] [CrossRef]
- Li, X.; Su, L.; Zhang, X.; Chen, Q.; Wang, Y.; Shen, Z.; Zhong, T.; Wang, L.; Xiao, Y.; Feng, X.; et al. Recent Advances on the Function and Purification of Milk Exosomes: A Review. Front. Nutr. 2022, 9, 871346. [Google Scholar] [CrossRef] [PubMed]
- Ghai, S.; Saini, S.; Ansari, S.; Verma, V.; Chopra, S.; Sharma, V.; Devi, P.; Malakar, D. Allogenic umbilical cord blood-mesenchymal stem cells are more effective than antibiotics in alleviating subclinical mastitis in dairy cows. Theriogenology 2022, 187, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, J.; Huang, Z.; Yin, B.; Umar, T.; Yang, C.; Zhang, X.; Jing, H.; Guo, S.; Guo, M.; et al. Vitexin Mitigates Staphylococcus aureus-Induced Mastitis via Regulation of ROS/ER Stress/NF-kappaB/MAPK Pathway. Oxid. Med. Cell Longev. 2022, 2022, 7977433. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Jin, G.; Liu, W.; Dou, M.; Wang, X.; Shi, W.; Bao, Y. Salvia miltiorrhiza polysaccharides ameliorates Staphylococcus aureus-induced mastitis in rats by inhibiting activation of the NF-kappaB and MAPK signaling pathways. BMC Vet. Res. 2022, 18, 201. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, Y.; Yang, Y.; Wang, H.; Ye, J.; Liu, D.; Chen, Y.; Lian, C.; Wang, R.; Gao, Y.; et al. Houttuynia Essential Oil and its Self-Microemulsion Preparation Protect Against LPS-Induced Murine Mastitis by Restoring the Blood-Milk Barrier and Inhibiting Inflammation. Front. Immunol. 2022, 13, 842189. [Google Scholar] [CrossRef]
- Hassel, C.; Gausseres, B.; Guzylack-Piriou, L.; Foucras, G. Ductal Macrophages Predominate in the Immune Landscape of the Lactating Mammary Gland. Front. Immunol. 2021, 12, 754661. [Google Scholar] [CrossRef]
- Dawson, C.A.; Pal, B.; Vaillant, F.; Gandolfo, L.C.; Liu, Z.; Bleriot, C.; Ginhoux, F.; Smyth, G.K.; Lindeman, G.J.; Mueller, S.N.; et al. Tissue-resident ductal macrophages survey the mammary epithelium and facilitate tissue remodelling. Nat. Cell. Biol. 2020, 22, 546–558. [Google Scholar] [CrossRef]
- Huang, T.; Zhou, C.; Che, Y.; Zhang, M.; Ren, W.; Lei, L. Exosomes Derived from Bovine Mammary Epithelial Cells Treated with Transforming Growth Factor-beta1 Inhibit the Proliferation of Bovine Macrophages. J. Interferon. Cytokine Res. 2019, 39, 752–759. [Google Scholar] [CrossRef]
- Kudo, T.; Nakazawa, D.; Watanabe-Kusunoki, K.; Kanda, M.; Shiratori-Aso, S.; Abe, N.; Nishio, S.; Koga, J.I.; Iwasaki, S.; Tsuji, T.; et al. Cyclophilin D regulates NETosis and inflammation in myeloperoxidase-antineutrophil cytoplasmic antibody-associated vasculitis. Arthritis Rheumatol. 2022. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, W.; Wang, W.; Tong, X.; Xia, R.; Fan, J.; Du, J.; Zhang, C.; Shi, X. Platelet-derived exosomes promote neutrophil extracellular trap formation during septic shock. Crit. Care 2020, 24, 380. [Google Scholar] [CrossRef]
- Jiang, L.Y.; Sun, H.Z.; Guan, R.W.; Shi, F.; Zhao, F.Q.; Liu, J.X. Formation of Blood Neutrophil Extracellular Traps Increases the Mastitis Risk of Dairy Cows During the Transition Period. Front. Immunol. 2022, 13, 880578. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Yang, S.; Zhou, M.; Lu, Y.; Deng, B.; Zhang, J.; Fan, H.; Wang, G. NADPH oxidase-derived reactive oxygen species production activates the ERK1/2 pathway in neutrophil extracellular traps formation by Streptococcus agalactiae isolated from clinical mastitis bovine. Vet. Microbiol. 2022, 268, 109427. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Ma, M.; Yang, W.; Wang, H.; Zhang, Y.; Gao, M.Q. SDF-1 in Mammary Fibroblasts of Bovine with Mastitis Induces EMT and Inflammatory Response of Epithelial Cells. Int. J. Biol. Sci. 2017, 13, 604–614. [Google Scholar] [CrossRef]
- Wijenayake, S.; Eisha, S.; Tawhidi, Z.; Pitino, M.A.; Steele, M.A.; Fleming, A.S.; McGowan, P.O. Comparison of methods for pre-processing, exosome isolation, and RNA extraction in unpasteurized bovine and human milk. PLoS ONE 2021, 16, e0257633. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.F.; Zhao, Y.; Dong, C.; Liu, L.; Pan, Y.; Lai, J.; Liu, Z.; Yu, G.T.; Chen, X.; Rao, L. Genetically Programmable Fusion Cellular Vesicles for Cancer Immunotherapy. Angew. Chem. Int. Ed. Engl. 2021, 60, 26320–26326. [Google Scholar] [CrossRef]
- Li, T.; Zhu, Y.; Lin, C.; Chen, J.; Yin, Y.; Tang, X.; Chen, Y.; Guo, A.; Hu, C. N (6)-Methyladenosine Modification Profile in Bovine Mammary Epithelial Cells Treated with Heat-Inactivated Staphylococcus aureus. Oxid. Med. Cell Longev. 2022, 2022, 1704172. [Google Scholar] [CrossRef]
- Li, T.; Lin, C.; Zhu, Y.; Xu, H.; Yin, Y.; Wang, C.; Tang, X.; Song, T.; Guo, A.; Chen, Y.; et al. Transcriptome Profiling of m (6)A mRNA Modification in Bovine Mammary Epithelial Cells Treated with Escherichia coli. Int. J. Mol. Sci. 2021, 22, 6254. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Lin, C.; Li, T.; Zhu, Y.; Yang, J.; Chen, S.; Chen, J.; Chen, X.; Chen, Y.; Guo, A.; et al. N (6)-Methyladenosine-Modified circRNA in the Bovine Mammary Epithelial Cells Injured by Staphylococcus aureus and Escherichia coli. Front. Immunol. 2022, 13, 873330. [Google Scholar] [CrossRef]
- Zhang, R.; Qu, Y.; Ji, Z.; Hao, C.; Su, Y.; Yao, Y.; Zuo, W.; Chen, X.; Yang, M.; Ma, G. METTL3 mediates Ang-II-induced cardiac hypertrophy through accelerating pri-miR-221/222 maturation in an m6A-dependent manner. Cell Mol. Biol. Lett. 2022, 27, 55. [Google Scholar] [CrossRef] [PubMed]
| Source of Exosomes | Isolation Method | Identification Method | Key Results | Ref |
|---|---|---|---|---|
| BMEC | Ultracentrifugation | TEM + WB | Identified protein species in normal exosomes | [90] |
| MAC-T | Ultracentrifugation | TEM + NTA + FCM | Constructed mRNA and LncRNA expression profiles of normal and S. aureus-infected MAC-T cell-derived exosomes | [91] |
| MAC-T | Ultracentrifugation | TEM + NTA | Constructed mRNA expression profiles of selenium-pretreated normal and S. aureus-infected MAC-T cell-derived exosomes | [97] |
| MAC-T and milk | Ultracentrifugation | TEM + NTA + WB | MAC-T cell-derived and milk-derived exosomes showed similar miRNA and protein expression profiles | [89] |
| HC11 | Ultracentrifugation | TEM + NTA + WB | HC11 cell-derived exosome miR-211 induces macrophages M1-type polarization by targeting SOCS1 | [98] |
| MAC-T | Ultracentrifugation | TEM + NTA + WB | MAC-T cell-derived exosomes induce macrophages M1-type polarization | [99] |
| MAC-T | Ultracentrifugation | TEM + NTA + FCM | MAC-T cell-derived exosomal lnc-AFTR plays a regulatory role in apoptosis and inflammation induced by S. aureus by mediating FAS degradation | [100] |
| MAC-T | Ultracentrifugation | TEM + WB | MAC-T cell-derived exosome miR-193b-5p promotes inflammatory response | [101] |
| Milk | Sucrose Gradient Centrifugation + Filtration Purification | - | Constructed miRNA expression profiles of milk exosomes after Staphylococcus spp. infection, and bta-miR-142-5p and bta-miR-223 were initially identified as potential early detection markers | [75] |
| Milk | Ultracentrifugation | TEM + NTA + WB | The results showed that 18 miRNAs were differentially expressed between the control and S. aureus-infected groups and revealed that bta-miR-142-5p and bta-miR-223 may play a role in mastitis | [76] |
| Milk | Ultracentrifugation | TEM + NTA + WB | Analysis of the results of 3 consecutive days of sampling revealed the stability of miRNAs within the exosomes, and bta-miR-223-3p was found to be significantly upregulated in subclinical mastitis | [78] |
| Milk | Ultracentrifugation | TEM + NTA + FCM | Constructed exosome miRNA expression profiles for normal and S. aureus-infected groups | [77] |
| Sample | Types of Mastitis | Key Molecule | Expression | Ref |
|---|---|---|---|---|
| Peripheral Blood | Subclinical mastitis | bta-miR-21/bta-miR-146a/bta-miR-155/bta-miR-222/bta- miR-383 | UP | [116] |
| Peripheral Blood | Subclinical mastitis | bta-miR-223/bta-miR-142-5p | UP | [117] |
| Peripheral Blood and Milk | Subclinical mastitis (S. aureus) | PON1 | DOWN | [115] |
| Peripheral Blood | Subclinical mastitis (S. aureus) | bta-miR-21 | UP | [103] |
| Peripheral Blood | Mastitis | bta-miR-1301 | UP | [104] |
| Peripheral Blood | Subclinical mastitis (S. aureus) | bta-miR-2284r | DOWN | [104] |
| Peripheral Blood | Subclinical mastitis | Valine (Val)/serine (Ser)/tyrosine (Tyr)/phenylalanine (Phe)/isoleucine (Ile) | - | [106] |
| Peripheral Blood | Mastitis | Nonesterified fatty acid/aspartate aminotransferase | UP | [118] |
| Peripheral Blood | Subclinical mastitis | Lysine/leucine/isoleucine/kynurenine/sphingomyelin (SM) C26:0/Ornithine/lysoPC A C17:0/SM C26:1 | - | [107] |
| Urine | Subclinical mastitis | Acylcarnitines (ACs)/phosphatidylcholines (PCs)/amino acids (AAs)/biogenic amines (BAs) | - | [108] |
| Milk | Subclinical mastitis (S. aureus) | bta-miR-142-5p/bta-miR-223 | UP | [75,76] |
| Milk | Subclinical mastitis | bta-miR-223-3p | UP | [78] |
| Milk | Subclinical mastitis (S. aureus) | bta-miR-378/bta-miR-185 | UP | [77] |
| Bovine Mammary Gland Tissue | Clinical mastitis (E. coli) | bta-miR-202/bta-miR-2357 | UP | [111] |
| Bovine Mammary Gland Tissue | Subclinical mastitis (S. aureus) | bta-miR-7863 | UP | [111] |
| Bovine Mammary Gland Tissue | Subclinical mastitis (S. agalactiae) | bta-miR-223 | UP | [112] |
| Bovine Mammary Gland Tissue | Subclinical mastitis (S. agalactiae) | bta-miR-26a | DOWN | [112] |
| Bovine Mammary Gland Tissue | Subclinical mastitis (S. aureus) | bta-miR-223/bta-miR-132/bta-miR-1246 | UP | [109] |
| HC11 cells | LTA | mmu-miR-211 | UP | [98] |
| MAC-T cells | S. aureus | Lnc-AFTR | DOWN | [100] |
| MAC-T cells | LPS | bta-miR-193b-5p | UP | [101] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Z.-H.; Ren, W.-Z.; Wu, H.-Y.; Zhang, J.-B.; Yuan, B. Exosomes in Mastitis—Research Status, Opportunities, and Challenges. Animals 2022, 12, 2881. https://doi.org/10.3390/ani12202881
Ji Z-H, Ren W-Z, Wu H-Y, Zhang J-B, Yuan B. Exosomes in Mastitis—Research Status, Opportunities, and Challenges. Animals. 2022; 12(20):2881. https://doi.org/10.3390/ani12202881
Chicago/Turabian StyleJi, Zhong-Hao, Wen-Zhi Ren, Hong-Yu Wu, Jia-Bao Zhang, and Bao Yuan. 2022. "Exosomes in Mastitis—Research Status, Opportunities, and Challenges" Animals 12, no. 20: 2881. https://doi.org/10.3390/ani12202881
APA StyleJi, Z.-H., Ren, W.-Z., Wu, H.-Y., Zhang, J.-B., & Yuan, B. (2022). Exosomes in Mastitis—Research Status, Opportunities, and Challenges. Animals, 12(20), 2881. https://doi.org/10.3390/ani12202881





