Genome-Wide Association Study for Udder Conformation Traits in Chinese Holstein Cattle
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Phenotype and Genotype Data Collection
2.3. Phenotypic and Genotypic Parameters
2.4. Genotypiing Data and Quality Control
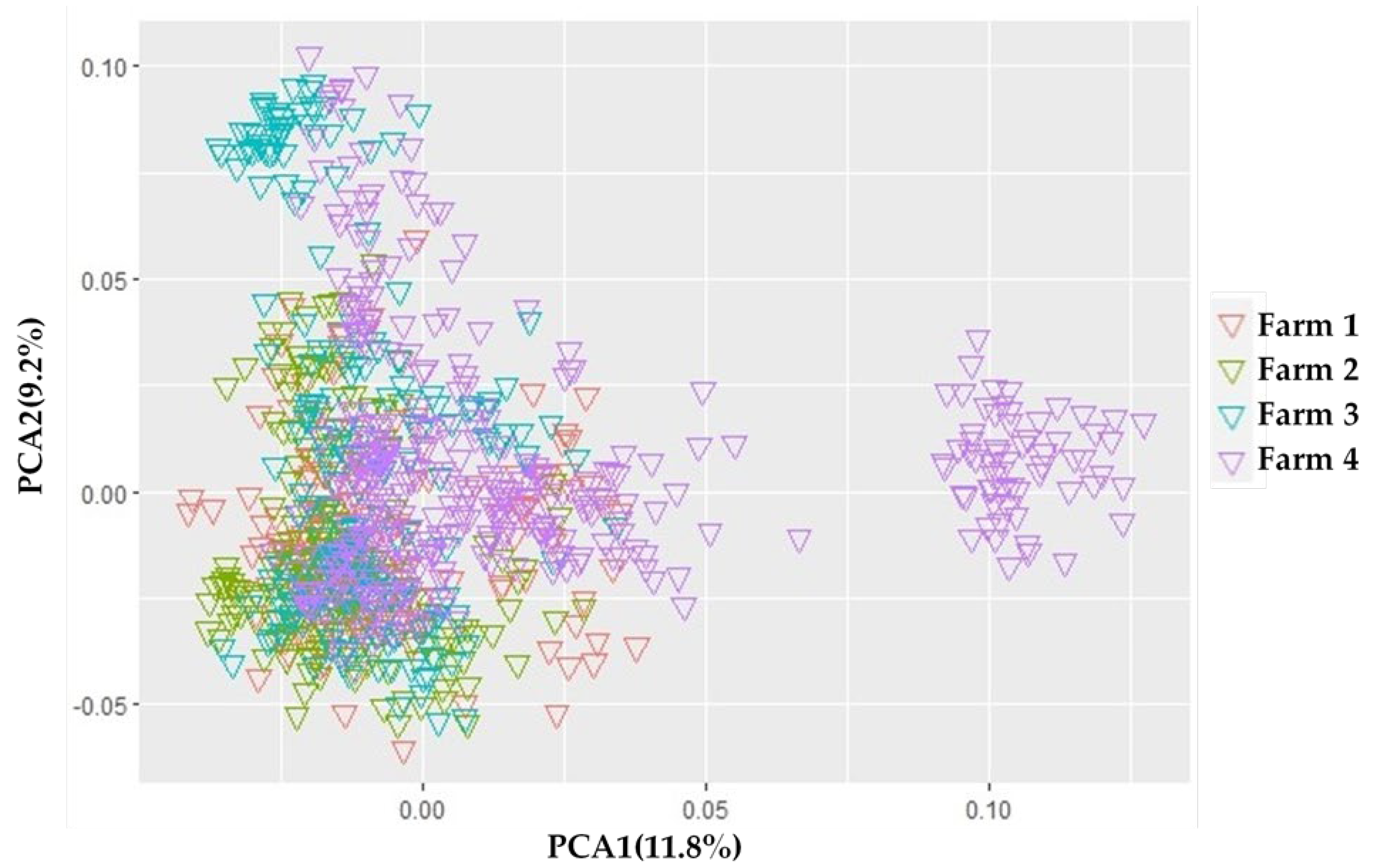
2.5. Principal Component Analysis
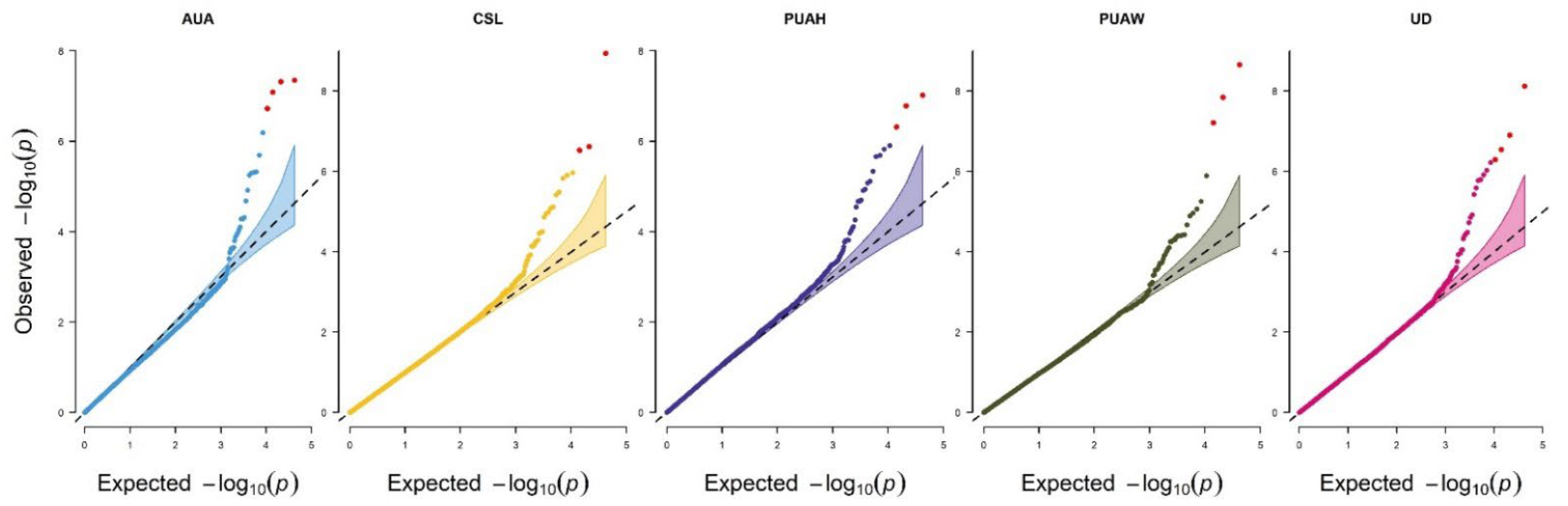
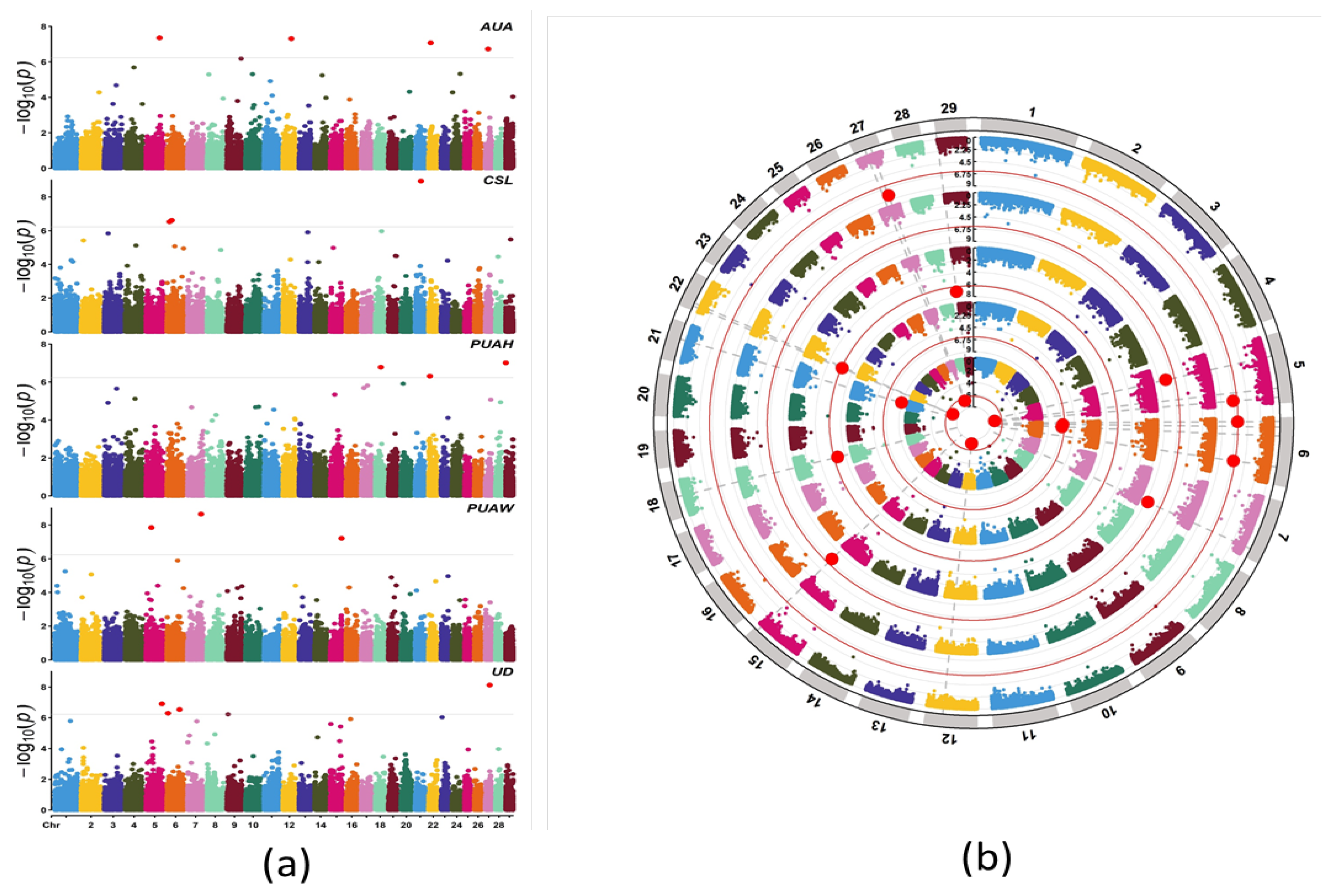
2.6. Association Analysis
2.7. Annotation of Candidate Genes
2.8. Functional Pathway-Enrichment and Gene Network Analysis of Candidate Gene
3. Results
3.1. Descriptive Statistical Data Analysis
3.2. Phenotypic and Genetic Correlations, and Heritability Estimation of Udder Traits
3.3. Population Structure
3.4. Genome-Wide Association Study
3.5. Annotation of the Candidate Genes
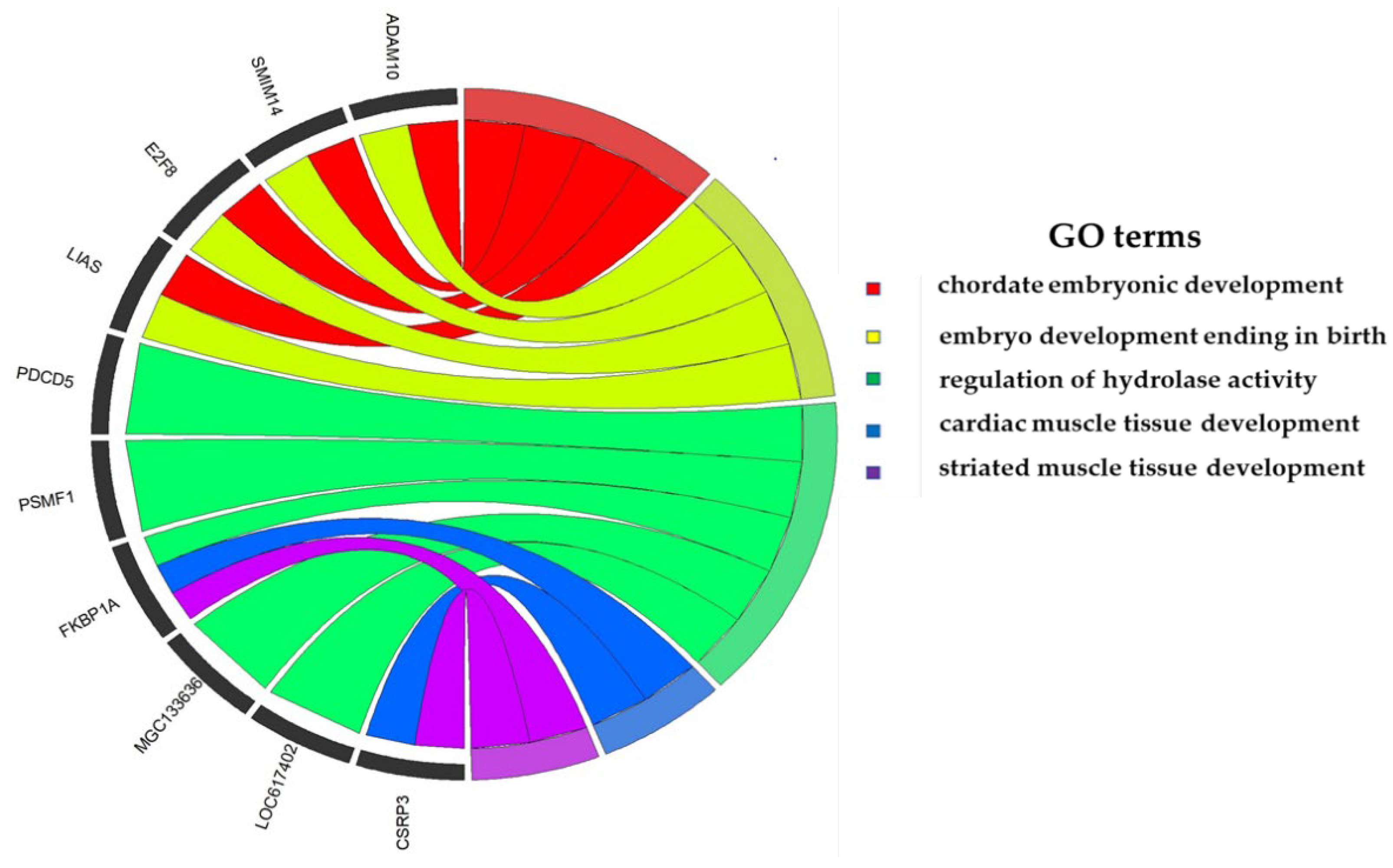
3.6. Enrichment Analysis
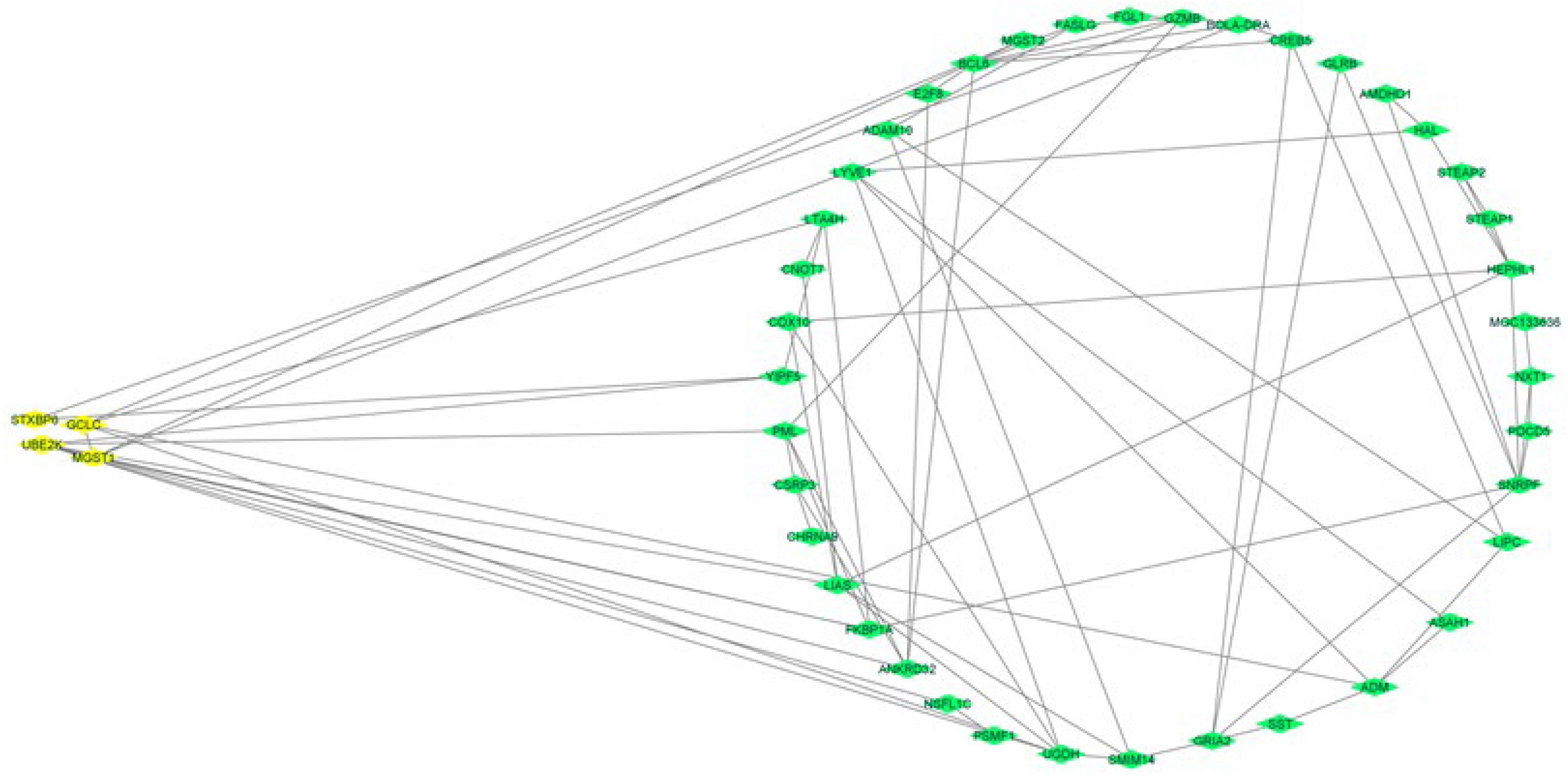
3.7. Gene Network Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Devani, K.; Plastow, G.; Orsel, K.; Valente, T.S. Genome-wide association study for mammary structure in Canadian Angus cows. PLoS ONE 2020, 15, e0237818. [Google Scholar] [CrossRef] [PubMed]
- Vukašinović, N.; Moll, J.; Künzi, N. Genetic relationships among longevity, milk production, and type traits in Swiss Brown cattle. Livest. Prod. Sci. 1995, 41, 11–18. [Google Scholar] [CrossRef]
- Wiggans, G.R.; VanRaden, P.M.; Cooper, T.A. The genomic evaluation system in the United States: Past, present, future. J. Dairy Sci. 2011, 94, 3202–3211. [Google Scholar] [CrossRef] [PubMed]
- Foster, M.K.; Caldwell, K.R.; Arnold, A.N.; Griffin, D.B.; Gehring, K.B.; Savell, J.W. Palatability assessments of beef strip loin steaks portioned by weight or by thickness sourced from various carcass weight/ribeye area size combinations. Meat Sci. 2021, 172, 108319. [Google Scholar] [CrossRef] [PubMed]
- Persson Waller, K.; Persson, Y.; Nyman, A.K.; Stengärde, L. Udder health in beef cows and its association with calf growth. Acta Vet. Scand. 2014, 56, 9. [Google Scholar] [CrossRef]
- Riley, D.G.; Sanders, J.O.; Knutson, R.E.; Lunt, D.K. Comparison of F1 Bos indicus x Hereford cows in central Texas: II. Udder, mouth, longevity, and lifetime productivity. J. Anim. Sci. 2001, 79, 1439–1449. [Google Scholar] [CrossRef]
- Lund, T.; Miglior, F.; Dekkers, J.C.M.; Burnside, E.B. Genetic relationships between clinical mastitis, somatic cell count, and udder conformation in Danish Holsteins. Livest. Prod. Sci. 1994, 39, 243–251. [Google Scholar] [CrossRef]
- Rupp, R.; Boichard, D. Genetic parameters for clinical mastitis, somatic cell score, production, udder type traits, and milking ease in first lactation Holsteins. J. Dairy Sci. 1999, 82, 2198–2204. [Google Scholar] [CrossRef]
- Carlström, C.; Pettersson, G.; Johansson, K.; Strandberg, E.; Stålhammar, H.; Philipsson, J. Feasibility of using automatic milking system data from commercial herds for genetic analysis of milkability. J. Dairy Sci. 2013, 96, 5324–5332. [Google Scholar] [CrossRef]
- Klein, D.; Flöck, M.; Khol, J.L.; Franz, S.; Stüger, H.P.; Baumgartner, W. Ultrasonographic measurement of the bovine teat: Breed differences, and the significance of the measurements for udder health. J. Dairy Res. 2005, 72, 296–302. [Google Scholar] [CrossRef]
- Vanraden, P.M.; Jensen, E.L.; Lawlor, T.J.; Funk, D.A. Prediction of Transmitting Abilities for Holstein Type Traits. J. Dairy Sci. 1990, 73, 191–197. [Google Scholar] [CrossRef]
- Dal Zotto, R.; De Marchi, M.; Dalvit, C.; Cassandro, M.; Gallo, L.; Carnier, P.; Bittante, G. Heritabilities and genetic correlations of body condition score and calving interval with yield, somatic cell score, and linear type traits in brown swiss cattle. J. Dairy Sci. 2007, 90, 5737–5743. [Google Scholar] [CrossRef]
- Campos, R.V.; Cobuci, J.A.; Kern, E.L.; Costa, C.N.; McManus, C.M. Genetic parameters for linear type traits and milk, fat, and protein production in Holstein cows in Brazil. Asian-Australas. J. Anim. Sci. 2015, 28, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Němcová, E.; Stípková, M.; Zavadilová, L. Genetic parameters for linear type traits in Czech Holstein cattle. Czech J. Anim. Sci. 2011, 56, 157–162. [Google Scholar] [CrossRef]
- Hogeveen, H.; Huijps, K.; Lam, T.J.G.M. Economic aspects of mastitis: New developments. N. Z. Vet. J. 2011, 59, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Nakov, D.; Hristov, S.; Andonov, S.; Trajchev, M. Udder-related risk factors for clinical mastitis in dairy cows. Vet. Arh. 2014, 84, 111–127. [Google Scholar]
- Jiang, J.; Ma, L.; Prakapenka, D.; VanRaden, P.M.; Cole, J.B.; Da, Y. A large-scale genome-wide association study in U.S. Holstein cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef]
- Kristensen, V.N.; Børresen-Dale, A.L. SNPs associated with molecular subtypes of breast cancer: On the usefulness of stratified Genome-wide Association Studies (GWAS) in the identification of novel susceptibility loci. Mol. Oncol. 2008, 2, 12–15. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, J.; Sun, D.; Ma, P.; Ding, X.; Yu, Y.; Zhang, Q. Genome wide association studies for milk production traits in Chinese Holstein population. PLoS ONE 2010, 5, e13661. [Google Scholar] [CrossRef]
- Li, C.; Sun, D.; Zhang, S.; Wang, S.; Wu, X.; Zhang, Q.; Liu, L.; Li, Y.; Qiao, L. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein. PLoS ONE 2014, 9, e0243343. [Google Scholar] [CrossRef]
- Li, X.; Buitenhuis, A.J.; Lund, M.S.; Li, C.; Sun, D.; Zhang, Q.; Poulsen, N.A.; Su, G. Joint genome-wide association study for milk fatty acid traits in Chinese and Danish Holstein populations. J. Dairy Sci. 2015, 98, 8152–8163. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Li, C.; Cai, W.; Liu, S.; Yin, H.; Shi, S.; Zhang, Q.; Zhang, S. Genome-wide association study for milk protein composition traits in a chiniese holstein population using a single-step approach. Front. Genet. 2019, 10, 72. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Sapi, E.; Brown, W.; Nilsen, J.; Tartaro, K.; Kacinski, B.M.; Craft, J.; Naftolin, F.; Mor, G. Roles of Fas and Fas ligand during mammary gland remodeling. J. Clin. Investig. 2000, 106, 1209–1220. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sahana, G.; Guldbrandtsen, B.; Lund, M.S. Genome-wide association study for calving traits in Danish and Swedish Holstein cattle. J. Dairy Sci. 2011, 94, 479–486. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, P.; Fu, W.; Dong, T.; Qi, C.; Liu, L.; Guo, G.; Li, C.; Cui, X.; Zhang, S.; et al. Genome-wide association study for pigmentation traits in Chinese Holstein population. Anim. Genet. 2014, 45, 740–744. [Google Scholar] [CrossRef]
- Liu, A.; Wang, Y.; Sahana, G.; Zhang, Q.; Liu, L.; Lund, M.S.; Su, G. Genome-wide Association Studies for Female Fertility Traits in Chinese and Nordic Holsteins. Sci. Rep. 2017, 7, 8487. [Google Scholar] [CrossRef]
- Cai, Z.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Prioritizing candidate genes post-GWAS using multiple sources of data for mastitis resistance in dairy cattle. BMC Genom. 2018, 19, 656. [Google Scholar] [CrossRef]
- Wang, X.; Ma, P.; Liu, J.; Zhang, Q.; Zhang, Y.; Ding, X.; Jiang, L.; Wang, Y.; Zhang, Y.; Sun, D.; et al. Genome-wide association study in Chinese Holstein cows reveal two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genet. 2015, 16, 111. [Google Scholar] [CrossRef]
- Vallée, A.; Daure, J.; Van Arendonk, J.A.M.; Bovenhuis, H. Genome-wide association study for behavior, type traits, and muscular development in Charolais beef cattle. J. Anim. Sci. 2016, 94, 2307–2316. [Google Scholar] [CrossRef]
- Tolleson, M.W.; Gill, C.A.; Herring, A.D.; Riggs, P.K.; Sawyer, J.E.; Sanders, J.O.; Riley, D.G. Association of udder traits with single nucleotide polymorphisms in crossbred bos indicus–bos taurus cows. J. Anim. Sci. 2017, 95, 2399–2407. [Google Scholar] [CrossRef]
- Pausch, H.; Emmerling, R.; Schwarzenbacher, H.; Fries, R. A multi-trait meta-analysis with imputed sequence variants reveals twelve QTL for mammary gland morphology in Fleckvieh cattle. Genet. Sel. Evol. 2016, 48, 47. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.C.C.; Guimarães, S.E.F.; Kelly, M.J.; Fortes, M.R.S.; e Silva, F.F.; Verardo, L.L.; Mota, R.; Moore, S. Multiple-trait genomewide mapping and gene network analysis for scrotal circumference growth curves in Brahman cattle. J. Anim. Sci. 2017, 95, 3331. [Google Scholar] [CrossRef] [PubMed]
- Marete, A.; Lund, M.S.; Boichard, D.; Ramayo-Caldas, Y. A system-based analysis of the genetic determinism of udder conformation and health phenotypes across three French dairy cattle breeds. PLoS ONE 2018, 13, e0199931. [Google Scholar] [CrossRef] [PubMed]
- Madsen, P.; Jensen, J.A. A User’S Guide to DMU. A Package for Analysing Multivariate Mixed Models. Available online: https://dmu.ghpc.au.dk/dmu/index.html (accessed on 25 June 2022).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Reich, D.; Price, A.L.; Patterson, N. Principal component analysis of genetic data. Nat. Genet. 2008, 40, 491–492. [Google Scholar] [CrossRef]
- Wang, Q.; Tian, F.; Pan, Y.; Buckler, E.S.; Zhang, Z. A SUPER powerful method for genome wide association study. PLoS ONE 2014, 9, e107684. [Google Scholar] [CrossRef]
- Duggal, P.; Gillanders, E.M.; Holmes, T.N.; Bailey-Wilson, J.E. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genom. 2008, 9, 516. [Google Scholar] [CrossRef] [PubMed]
- Lilin-yin, A. Package ‘CMplot: Circle Manhattan Plot. 2022. Available online: https://github.com/YinLiLin/CMplot (accessed on 30 June 2022).
- Sanchez, M.P.; Govignon-Gion, A.; Croiseau, P.; Fritz, S.; Hozé, C.; Miranda, G.; Martin, P.; Barbat-Leterrier, A.; Letaïef, R.; Rocha, D.; et al. Within-breed and multi-breed GWAS on imputed whole-genome sequence variants reveal candidate mutations affecting milk protein composition in dairy cattle. Genet. Sel. Evol. 2017, 49, 68. [Google Scholar] [CrossRef]
- Mota, L.F.M.; Lopes, F.B.; Fernandes Júnior, G.A.; Rosa, G.J.M.; Magalhães, A.F.B.; Carvalheiro, R.; Albuquerque, L.G. Genome-wide scan highlights the role of candidate genes on phenotypic plasticity for age at first calving in Nellore heifers. Sci. Rep. 2020, 10, 6481. [Google Scholar] [CrossRef]
- Navarro Gonzalez, J.; Zweig, A.S.; Speir, M.L.; Schmelter, D.; Rosenbloom, K.R.; Raney, B.J.; Powell, C.C.; Nassar, L.R.; Maulding, N.D.; Lee, C.M.; et al. The UCSC genome browser database: 2021 update. Nucleic Acids Res. 2021, 49, D1046–D1057. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Dietze, H.; Lewis, S.E.; Mungall, C.J.; Munoz-Torres, M.C.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; Fey, P.; Thomas, P.D.; et al. Expansion of the gene ontology knowledgebase and resources: The gene ontology consortium. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Wu, X.; Fang, M.; Liu, L.; Wang, S.; Liu, J.; Ding, X.; Zhang, S.; Zhang, Q.; Zhang, Y.; Qiao, L.; et al. Genome wide association studies for body conformation traits in the Chinese Holstein cattle population. BMC Genom. 2013, 14, 897. [Google Scholar] [CrossRef]
- DeGroot, B.J.; Keown, J.F.; Van Vleck, L.D.; Marotz, E.L. Genetic parameters and responses of linear type, yield traits, and somatic cell scores to divergent selection for predicted transmitting ability for type in Holsteins. J. Dairy Sci. 2002, 85, 1578–1585. [Google Scholar] [CrossRef]
- Royal, M.D.; Pryce, J.E.; Woolliams, J.A.; Flint, A.P.F. The genetic relationship between commencement of luteal activity and calving interval, body condition score, production, and linear type traits in holstein-friesian dairy cattle. J. Dairy Sci. 2002, 85, 3071–3080. [Google Scholar] [CrossRef]
- Wang, D.; Sun, Y.; Stang, P.; Berlin, J.A.; Wilcox, M.A.; Li, Q. Comparison of methods for correcting population stratification in a genome-wide association study of rheumatoid arthritis: Principal-component analysis versus multidimensional scaling. BMC Proc. 2009, 3, S109. [Google Scholar] [CrossRef]
- Lin, S.; Ke, C.; Liu, L.; Gao, Y.; Xu, L.; Han, B.; Zhao, Y.; Zhang, S.; Sun, D. Genome-wide association studies for immunoglobulin concentrations in colostrum and serum in Chinese Holstein. BMC Genom. 2022, 23, 840. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Zaitlen, N.A.; Reich, D.; Patterson, N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 2010, 11, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Devlin, B.; Roeder, K. Genomic control for association studies. Biometrics 1999, 55, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Haldar, T.; Ghosh, S. Effect of Population Stratification on False Positive Rates of Population-Based Association Analyses of Quantitative Traits. Ann. Hum. Genet. 2012, 76, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Sharmaa, A.; Lee, J.S.; Dang, C.G.; Sudrajad, P.; Kim, H.C.; Yeon, S.H.; Kang, H.S.; Lee, S.H. Stories and challenges of genome wide association studies in livestock—A review. Asian-Australas. J. Anim. Sci. 2015, 28, 1371–1379. [Google Scholar] [CrossRef]
- Scales, S.J.; Hesser, B.A.; Masuda, E.S.; Scheller, R.H. Amisyn, a novel syntaxin-binding protein that may regulate SNARE complex assembly. J. Biol. Chem. 2002, 277, 28271–28279. [Google Scholar] [CrossRef]
- Krintel, S.B.; Essioux, L.; Wool, A.; Johansen, J.S.; Schreiber, E.; Zekharya, T.; Akiva, P.; Østergaard, M.; Hetland, M.L. CD6 and syntaxin binding protein 6 variants and response to tumor necrosis factor alpha inhibitors in danish patients with rheumatoid arthritis. PLoS ONE 2012, 7, e38539. [Google Scholar] [CrossRef]
- Lenka, G.; Shan, J.; Halabi, N.; Abuaqel, S.W.J.; Goswami, N.; Schmidt, F.; Zaghlool, S.; Romero, A.R.; Subramanian, M.; Boujassoum, S.; et al. STXBP6, reciprocally regulated with autophagy, reduces triple negative breast cancer aggressiveness. Clin. Transl. Med. 2020, 10, e147. [Google Scholar] [CrossRef]
- Räschle, M.; Smeenk, G.; Hansen, R.K.; Temu, T.; Oka, Y.; Hein, M.Y.; Nagaraj, N.; Long, D.T.; Walter, J.C.; Hofmann, K.; et al. Proteomics reveals dynamic assembly of Repair complexes during bypass of DNA cross-links. Science 2015, 348, 1253671. [Google Scholar] [CrossRef]
- Sanchez, M.P.; Ramayo-Caldas, Y.; Wolf, V.; Laithier, C.; El Jabri, M.; Michenet, A.; Boussaha, M.; Taussat, S.; Fritz, S.; Delacroix-Buchet, A.; et al. Sequence-based GWAS, network and pathway analyses reveal genes co-associated with milk cheese-making properties and milk composition in Montbéliarde cows. Genet. Sel. Evol. 2019, 51, 34. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, E.; Komolka, K.; Ponsuksili, S.; Gotoh, T.; Wimmers, K.; Maak, S. Transcriptome profiling of Musculus longissimus dorsi in two cattle breeds with different intramuscular fat deposition. Genomics Data 2016, 7, 109–111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sun, T.; Huang, G.Y.; Wang, Z.H.; Teng, S.H.; Cao, Y.H.; Sun, J.L.; Hanif, Q.; Chen, N.B.; Lei, C.Z.; Liao, Y.Y. Selection signatures of Fuzhong Buffalo based on whole-genome sequences. BMC Genom. 2020, 21, 674. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, H.; Xu, L.; Zhu, B.; Liu, Y.; Bordbar, F.; Chen, Y.; Zhang, L.; Gao, X.; Gao, H.; et al. Genome-wide scan identifies selection signatures in chinese wagyu cattle using a high-density SNP array. Animals 2019, 9, 296. [Google Scholar] [CrossRef] [PubMed]
- Braz, C.U.; Rowan, T.N.; Schnabel, R.D.; Decker, J.E. Genome-wide association analyses identify genotype-by-environment interactions of growth traits in Simmental cattle. Sci. Rep. 2021, 11, 13335. [Google Scholar] [CrossRef]
- Christensen, J.; Cloos, P.; Toftegaard, U.; Klinkenberg, D.; Bracken, A.P.; Trinh, E.; Heeran, M.; Di Stefano, L.; Helin, K. Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res. 2005, 33, 5458–5470. [Google Scholar] [CrossRef] [PubMed]
- Morrell, B.C.; Zhang, L.; Schütz, L.F.; Perego, M.C.; Maylem, E.R.S.; Spicer, L.J. Regulation of the transcription factor E2F8 gene expression in bovine ovarian cells. Mol. Cell. Endocrinol. 2019, 498, 110572. [Google Scholar] [CrossRef]
- Yang, B.; Jiao, B.; Ge, W.; Zhang, X.; Wang, S.; Zhao, H.; Wang, X. Transcriptome sequencing to detect the potential role of long non-coding RNAs in bovine mammary gland during the dry and lactation period. BMC Genom. 2018, 19, 605. [Google Scholar] [CrossRef]
- Bakhtiarizadeh, M.R.; Mirzaei, S.; Norouzi, M.; Sheybani, N.; Vafaei Sadi, M.S. Identification of Gene Modules and Hub Genes Involved in Mastitis Development Using a Systems Biology Approach. Front. Genet. 2020, 11, 722. [Google Scholar] [CrossRef]
- Zhu, B.; Niu, H.; Zhang, W.; Wang, Z.; Liang, Y.; Guan, L.; Guo, P.; Chen, Y.; Zhang, L.; Guo, Y.; et al. Genome wide association study and genomic prediction for fatty acid composition in Chinese Simmental beef cattle using high density SNP array. BMC Genom. 2017, 18, 464. [Google Scholar] [CrossRef]
- Buaban, S.; Lengnudum, K.; Boonkum, W.; Phakdeedindan, P. Genome-wide association study on milk production and somatic cell score for Thai dairy cattle using weighted single-step approach with random regression test-day model. J. Dairy Sci. 2022, 105, 468–494. [Google Scholar] [CrossRef]
- Cai, Z.; Dusza, M.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Distinguishing pleiotropy from linked QTL between milk production traits and mastitis resistance in Nordic Holstein cattle. Genet. Sel. Evol. 2020, 52, 19. [Google Scholar] [CrossRef] [PubMed]
- Freebern, E.; Santos, D.J.A.; Fang, L.; Jiang, J.; Parker Gaddis, K.L.; Liu, G.E.; Vanraden, P.M.; Maltecca, C.; Cole, J.B.; Ma, L. GWAS and fine-mapping of livability and six disease traits in holstein cattle. BMC Genom. 2022, 21, 41. [Google Scholar] [CrossRef] [PubMed]
- de Souza Iung, L.H.; Mulder, H.A.; de Rezende Neves, H.H.; Carvalheiro, R. Genomic regions underlying uniformity of yearling weight in Nellore cattle evaluated under different response variables. BMC Genom. 2018, 19, 619. [Google Scholar] [CrossRef] [PubMed]
- Han, L.Q.; Zhou, Z.; Ma, Y.; Batistel, F.; Osorio, J.S.; Loor, J.J. Phosphorylation of nuclear factor erythroid 2-like 2 (NFE2L2) in mammary tissue of Holstein cows during the periparturient period is associated with mRNA abundance of antioxidant gene networks. J. Dairy Sci. 2018, 101, 6511–6522. [Google Scholar] [CrossRef]
- Li, P.; Fei, H.; Wang, L.; Xu, H.; Zhang, H.; Zheng, L. Pdcd5 regulates cell proliferation, cell cycle progression and apoptosis. Oncol. Lett. 2018, 15, 1177–1183. [Google Scholar] [CrossRef]
- Casey, T.; Patel, O.V.; Plaut, K. Transcriptomes reveal alterations in gravity impact circadian clocks and activate mechanotransduction pathways with adaptation through epigenetic change. Physiol. Genom. 2015, 47, 113–128. [Google Scholar] [CrossRef]
- Xu, Q.; Mei, G.; Sun, D.; Zhang, Q.; Zhang, Y.; Yin, C.; Chen, H.; Ding, X.; Liu, J. Detection of genetic association and functional polymorphisms of UGDH affecting milk production trait in Chinese Holstein cattle. BMC Genom. 2012, 13, 661. [Google Scholar] [CrossRef]
- Libera, K.; Konieczny, K.; Witkowska, K.; Żurek, K.; Szumacher-Strabel, M.; Cieslak, A.; Smulski, S. The association between selected dietary minerals and mastitis in dairy cows—A review. Animals 2021, 11, 2330. [Google Scholar] [CrossRef]
- Necasova, A.; Pechova, A.; Bodor, R.; Masar, M. Evaluation of the glutathione concentration in whole blood of dairy Holstein cows. Vet. Med. 2021, 66, 179–188. [Google Scholar] [CrossRef]
- Moro, J.; Tome, D.; Schmidely, P.; Demersay, T.; Azzout-Marniche, D. Histidine: A Systematic Review on Metabolism and. Nutrients 2020, 12, 1414. [Google Scholar] [CrossRef] [PubMed]
- Akers, R.M. Major advances associated with hormone and growth factor regulation of mammary growth and lactation in dairy cows. J. Dairy Sci. 2006, 89, 1222–1234. [Google Scholar] [CrossRef]

| Traits | Mean | SE | Min | Max | SD | CV% | Skewness | Kurtosis | p Value |
|---|---|---|---|---|---|---|---|---|---|
| AUA | 0.17 | 0.09 | −10.15 | 8.04 | 2.81 | 7.94 | 0.22 | 0.32 | 2.53 × 10−6 |
| CSL | 0.45 | 0.04 | −1.88 | 8.85 | 2.67 | 3.50 | 0.33 | 0.50 | 2.20 × 10−7 |
| PUAH | 1.73 | 0.05 | −2.2 | 7.32 | 1.55 | 2.41 | 0.34 | 0.53 | 5.70 × 10−8 |
| PUAW | 1.34 | 0.05 | −5.9 | 5.59 | 1.68 | 2.83 | 0.26 | 0.23 | 1.00 × 10−4 |
| UD | 0.18 | 0.05 | −5.29 | 4.75 | 1.56 | 2.43 | 0.24 | 0.34 | 3.206 × 10−5 |
| Traits | AUA | CSL | PUAH | PUAW | UD |
|---|---|---|---|---|---|
| AUA | 0.24 (0.02) | −0.38 | 0.22 | −0.62 | 0.10 |
| CSL | 0.02 | 0.34 (0.03) | −0.44 | 0.52 | −0.45 |
| PUAH | −0.11 ** | −0.37 ** | 0.04 (0.00) | 0.14 | 0.09 |
| PUAW | 0.17 ** | 0.14 ** | 0.002 | 0.13 (0.01) | −0.49 |
| UD | 0.26 ** | 0.14 ** | −0.07 * | 0.07 * | 0.49 (0.03) |
| Traits | SNPs | CHR | Position (kb) | MAF | Nearest Gene | Distance (kb) | p-Value | Effect |
|---|---|---|---|---|---|---|---|---|
| AUA | DB-340-seq-rs208014256 | 5 | 93520616 | 0.46 | MGST1 | Within | 4.48 × 10−8 | 0.330783 |
| Hapmap58214-rs29015775 | 22 | 13159539 | 0.49 | LOC101903734 | Within | 8.34 × 10−8 | −0.35043 | |
| BovineHD2700005329 | 27 | 19594311 | 0.16 | MTUS1 | Within | 1.90 × 10−7 | −0.47118 | |
| BovineHD0900028603 | 9 | 97665052 | 0.25 | PRKN | Within | 6.48 × 10−7 | 0.371653 | |
| CSL | ARS-BFGL-BAC-29174 | 21 | 40773446 | 0.43 | STXBP6 | 100 kb | 1.16 × 10−9 | 0.36344 |
| Hapmap32447-BTC-033214 | 6 | 32254947 | 0.42 | GRID2 | Within | 2.45 × 10−7 | −0.31119 | |
| BovineHD0600005127 | 6 | 17417238 | 0.39 | LOC112447148 | Within | 3.02 × 10−7 | −0.31736 | |
| PUAH | BovineHD2900000083 | 29 | 702083 | 0.44 | E2F8 | 100 kb | 9.70 × 10−8 | 0.298361 |
| BovineHD1800011193 | 18 | 37485453 | 0.47 | CDH11 | 100 kb | 1.66 × 10−7 | 0.26997 | |
| BovineHD2200002408 | 22 | 8008314 | 0.12 | FOXP1 | Within | 4.89 × 10−7 | −0.42465 | |
| PUAW | BovineHD0700028083 | 7 | 93970405 | 0.38 | SLF1 | Within | 2.26 × 10−9 | −0.33939 |
| BovineHD0500010522 | 5 | 36446050 | 0.50 | TMEM117 | Within | 1.45 × 10−8 | −0.33019 | |
| BovineHD1500023322 | 15 | 78715609 | 0.40 | SBF2 | Within | 6.19 × 10−8 | −0.30795 | |
| UD | BTA-75047-no-rs | 5 | 109433376 | 0.05 | LGALS2 | Within | 1.26 × 10−7 | −0.66283 |
| BovineHD0600024277 | 6 | 86964714 | 0.21 | GC | Within | 2.92 × 10−7 | 0.321971 | |
| BovineHD0600001885 | 6 | 7087395 | 0.35 | UBE2K | 50 kb | 5.16 × 10−7 | −0.27781 | |
| BovineHD0900001933 | 9 | 8043006 | 0.28 | ADGRB3 | Within | 5.98 × 10−7 | −0.28799 | |
| BovineHD2300001734 | 23 | 6986268 | 0.26 | GCLC | Within | 9.36 × 10−7 | −0.32781 |
| KEGG ID | Description | Count | Gene Name |
|---|---|---|---|
| bta04978 | Mineral absorption | 3 | STEAP1, STEAP2, HEPHL1 |
| bta00480 | Glutathione metabolism | 3 | MGST1, MGST2, GCLC |
| bta00340 | Histidine metabolism | 2 | HAL, AMDHD1 |
| bta04935 | Growth hormone synthesis, secretion and action | 3 | CREB5, SST, CACNA1D |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazar, M.; Abdalla, I.M.; Chen, Z.; Ullah, N.; Liang, Y.; Chu, S.; Xu, T.; Mao, Y.; Yang, Z.; Lu, X. Genome-Wide Association Study for Udder Conformation Traits in Chinese Holstein Cattle. Animals 2022, 12, 2542. https://doi.org/10.3390/ani12192542
Nazar M, Abdalla IM, Chen Z, Ullah N, Liang Y, Chu S, Xu T, Mao Y, Yang Z, Lu X. Genome-Wide Association Study for Udder Conformation Traits in Chinese Holstein Cattle. Animals. 2022; 12(19):2542. https://doi.org/10.3390/ani12192542
Chicago/Turabian StyleNazar, Mudasir, Ismail Mohamed Abdalla, Zhi Chen, Numan Ullah, Yan Liang, Shuangfeng Chu, Tianle Xu, Yongjiang Mao, Zhangping Yang, and Xubin Lu. 2022. "Genome-Wide Association Study for Udder Conformation Traits in Chinese Holstein Cattle" Animals 12, no. 19: 2542. https://doi.org/10.3390/ani12192542
APA StyleNazar, M., Abdalla, I. M., Chen, Z., Ullah, N., Liang, Y., Chu, S., Xu, T., Mao, Y., Yang, Z., & Lu, X. (2022). Genome-Wide Association Study for Udder Conformation Traits in Chinese Holstein Cattle. Animals, 12(19), 2542. https://doi.org/10.3390/ani12192542








