Prevalence and Population Diversity of Listeria monocytogenes Isolated from Dairy Cattle Farms in the Cantabria Region of Spain
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Collection, Processing and L. monocytogenes Isolation
2.3. Decline of L. monocytogenes in Manure
2.4. Decline of L. monocytogenes in Pasture Crops
2.5. PCR Serogroup Determination
2.6. Pulse-Field Gel Electrophoresis (PFGE) Typing
2.7. Antimicrobial Susceptibility Test
2.8. DNA Extraction and Sequencing
2.9. Bioinformatics Analysis
2.10. Nucleotide Accession Numbers
2.11. Statistical Analyses
3. Results
3.1. Environmental Sampling, Incidence and L. monocytogenes Isolation
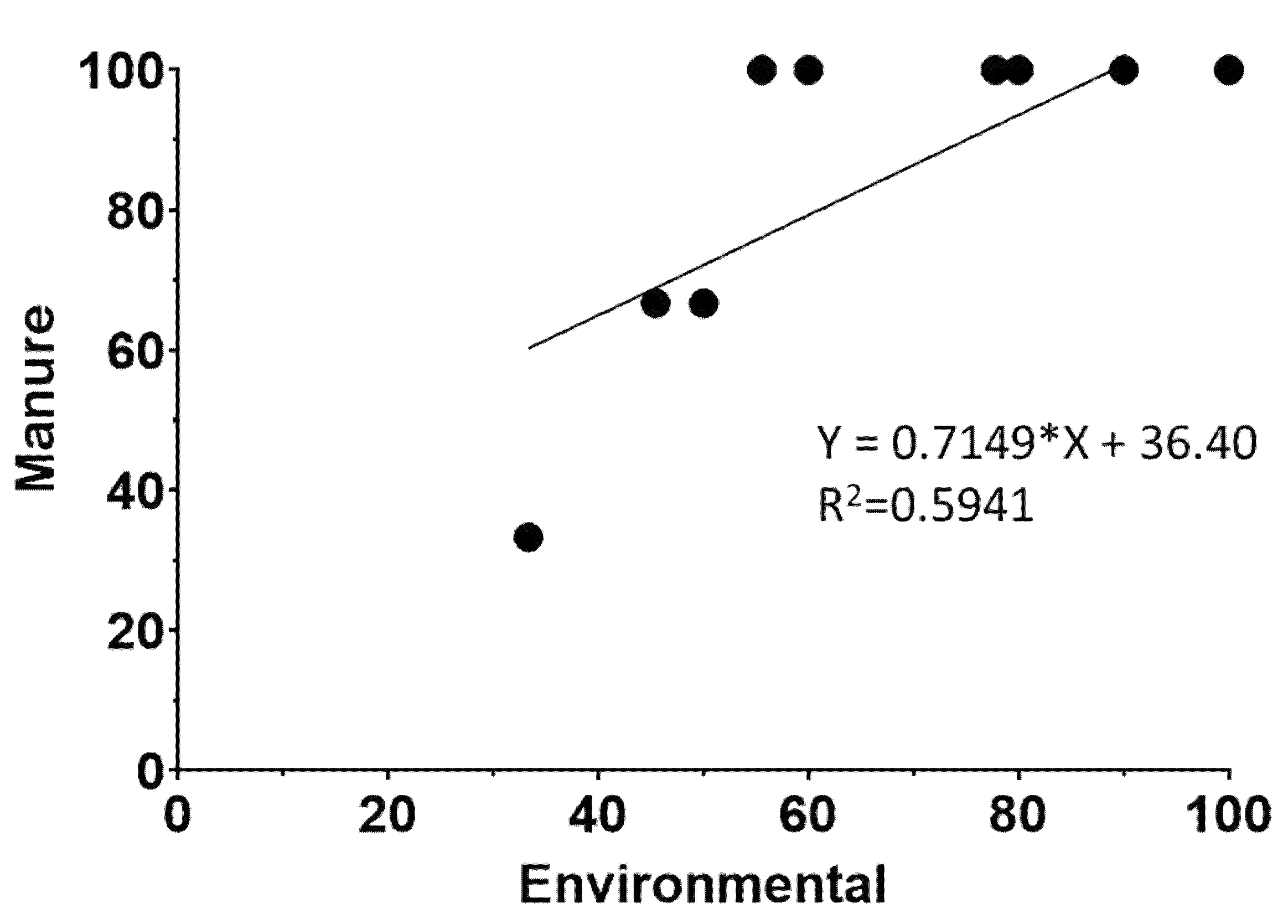
3.2. Transmission of L. monocytogenes
3.3. Decline in L. monocytogenes in Livestock Waste
3.4. Decline in L. monocytogenes in Pasture Crops
3.5. PCR Serogroups
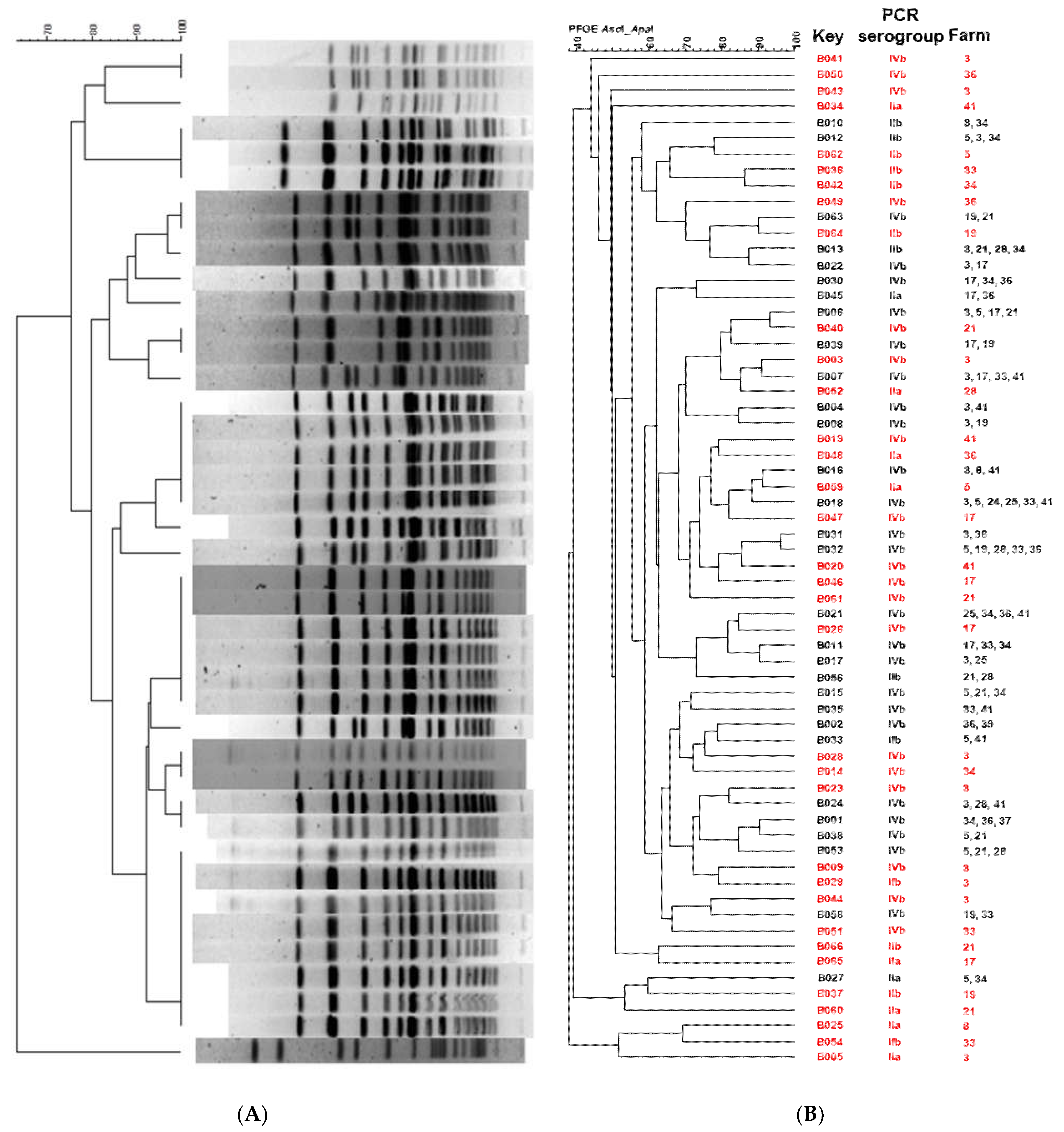
3.6. Pulsed-Field Gel Electrophoresis (PFGE)
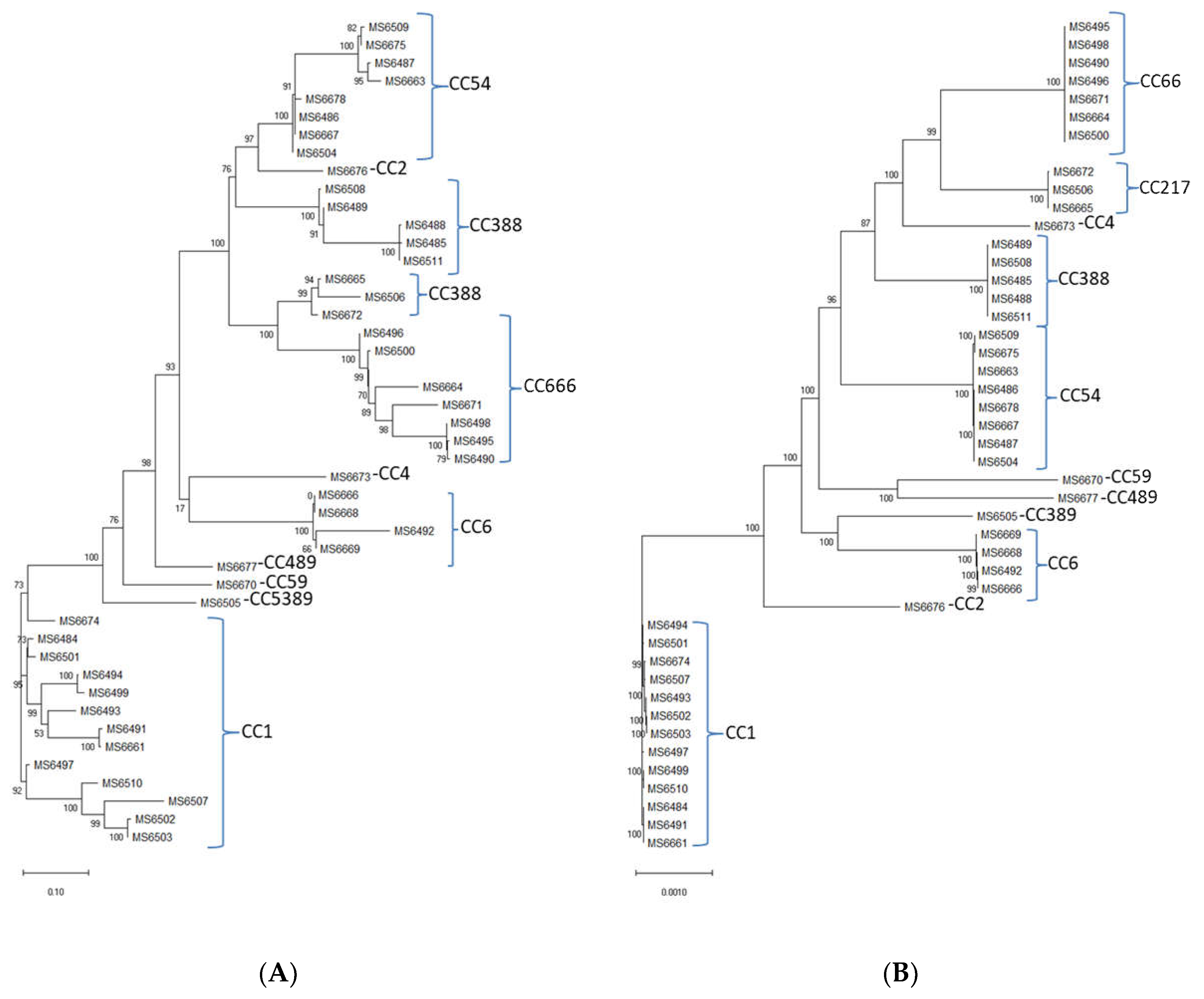
3.7. Genome Analysis of the L. monocytogenes Isolates and Multilocus Sequence Typing (MLST)
3.8. In Silico Analysis of Antimicrobial Resistance Genes and In Vitro Antimicrobial Susceptibility Testing
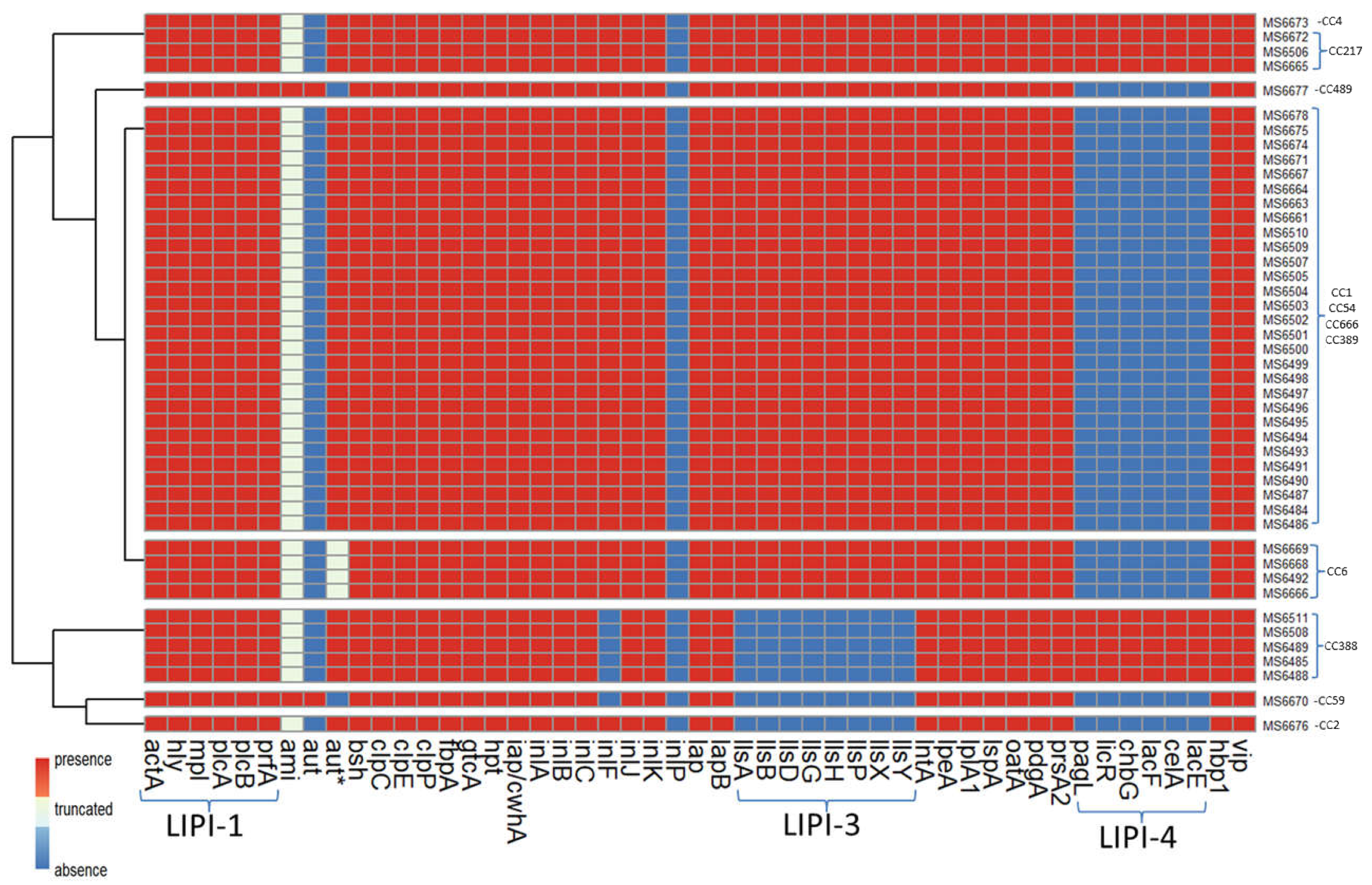
3.9. In Silico Analysis of Virulence Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roberts, A.J.; Wiedmann, M. Pathogen, Host and Environmental Factors Contributing to the Pathogenesis of Listeriosis. Cell. Mol. Life Sci. 2003, 60, 904–918. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Lanza, V.F.; Duval, M.; Coque, T.M. Ecogenetics of Antibiotic Resistance in Listeria monocytogenes. Mol. Microbiol. 2020, 113, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, E.; Courvalin, P. Antibiotic Resistance in Listeria spp. Antimicrob. Agents Chemother. 1999, 43, 2103. [Google Scholar] [CrossRef] [PubMed]
- Noll, M.; Kleta, S.; Al Dahouk, S. Antibiotic Susceptibility of 259 Listeria monocytogenes Strains Isolated from Food, Food-Processing Plants and Human Samples in Germany. J. Infect. Public Health 2018, 11, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Kayode, A.J.; Okoh, A.I. Assessment of Multidrug-Resistant Listeria monocytogenes in Milk and Milk Product and One Health Perspective. PLoS ONE 2022, 17, e0270993. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.A.C.; Carvalho, F.T.; Vallim, D.C.; Pereira, R.C.L.; Neto, A.C.; Vieira, B.S.; Carvalho, R.C.T.; Figueiredo, E.E.S. Listeria monocytogenes in Export-Approved Beef from Mato Grosso, Brazil: Prevalence, Molecular Characterization and Resistance to Antibiotics and Disinfectants. Microorganisms 2019, 8, 18. [Google Scholar] [CrossRef]
- Rabiey, S.; Hosseini, H.; Rezaei, M. The Hurdle Effect of Bunium Persicum Essential Oil, Smoke and NaCl for Controlling the Listeria monocytogenes Growth in Fish Model Systems. J. Food Saf. 2013, 33, 137–144. [Google Scholar] [CrossRef]
- Santorum, P.; Garcia, R.; Lopez, V.; Martinez-Suarez, J.V. Dairy Farm Management and Production Practices Associated with the Presence of Listeria monocytogenes in Raw Milk and Beef. Span. J. Agric. Res. 2012, 10, 360–371. [Google Scholar] [CrossRef]
- Oevermann, A.; Zurbriggen, A.; Vandevelde, M. Rhombencephalitis Caused by Listeria monocytogenes in Humans and Ruminants: A Zoonosis on the Rise? Interdiscip. Perspect. Infect. Dis. 2010, 2010, 632513. [Google Scholar] [CrossRef]
- Van Kessel, J.S.; Santin-Duran, M.; Karns, J.S.; Schukken, Y. Tracing Zoonotic Pathogens in Dairy Production. Tracing zoonotic pathogens in dairy production. In Tracing Pathogens in the Food Chain; Brul, S., Fratamico, P.M., McMeekin, T.A., Eds.; Woodhead Publishing Limited: Philadelphia, PA, USA, 2011; pp. 503–526. [Google Scholar] [CrossRef]
- NicAogáin, K.; O’Byrne, C.P. The Role of Stress and Stress Adaptations in Determining the Fate of the Bacterial Pathogen Listeria monocytogenes in the Food Chain. Front. Microbiol. 2016, 7, 1865. [Google Scholar] [CrossRef]
- Fenlon, D.R. Wild Birds and Silage as Reservoirs of Listeria in the Agricultural Environment. J. Appl. Bacteriol. 1985, 59, 537–543. [Google Scholar] [CrossRef]
- Hasegawa, M.; Iwabuchi, E.; Yamamoto, S.; Muramatsu, M.; Takashima, I.; Hirai, K. Prevalence and Characteristics of Listeria monocytogenes in Feces of Black Beef Cattle Reared in Three Geographically Distant Areas in Japan. Foodborne Pathog. Dis. 2014, 11, 96–103. [Google Scholar] [CrossRef]
- Fox, E.; O’Mahony, T.; Clancy, M.; Dempsey, R.; O’Brien, M.; Jordan, K. Listeria monocytogenes in the Irish Dairy Farm Environment. J. Food Prot. 2009, 72, 1450–1456. [Google Scholar] [CrossRef]
- Haley, B.J.; Sonnier, J.; Schukken, Y.H.; Karns, J.S.; Van Kessel, J.A.S. Diversity of Listeria monocytogenes within a U.S. Dairy Herd, 2004–2010. Foodborne Pathog. Dis. 2015, 12, 844–850. [Google Scholar] [CrossRef]
- Latorre, A.A.; Van Kessel, J.S.; Karns, J.S.; Zurakowski, M.J.; Pradhan, A.K.; Boor, K.J.; Jayarao, B.M.; Houser, B.A.; Daugherty, C.S.; Schukken, Y.H. Biofilm in Milking Equipment on a Dairy Farm as a Potential Source of Bulk Tank Milk Contamination with Listeria monocytogenes. J. Dairy Sci. 2010, 93, 2792–2802. [Google Scholar] [CrossRef]
- Van Kessel, J.S.; Karns, J.S.; Gorski, L.; McCluskey, B.J.; Perdue, M.L. Prevalence of Salmonellae, Listeria monocytogenes, and Fecal Coliforms in Bulk Tank Milk on US Dairies. J. Dairy Sci. 2004, 87, 2822–2830. [Google Scholar] [CrossRef]
- Orsi, R.H.; de Bakker, H.C.; Wiedmann, M. Listeria monocytogenes Lineages: Genomics, Evolution, Ecology, and Phenotypic Characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef]
- Burall, L.S.; Grim, C.J.; Datta, A.R. A Clade of Listeria monocytogenes Serotype 4b Variant Strains Linked to Recent Listeriosis Outbreaks Associated with Produce from a Defined Geographic Region in the US. PLoS ONE 2017, 12, e0176912. [Google Scholar] [CrossRef]
- Jeršek, B.; Gilot, P.; Gubina, M.; Klun, N.; Mehle, J.; Tcherneva, E.; Rijpens, N.; Herman, L. Typing of Listeria Monocytogenes Strains by Repetitive Element Sequence-Based PCR. J. Clin. Microbiol. 1999, 37, 103–109. [Google Scholar] [CrossRef]
- Swaminathan, B.; Gerner-Smidt, P. The Epidemiology of Human Listeriosis. Microbes Infect. 2007, 9, 1236–1243. [Google Scholar] [CrossRef]
- Martin, P.; Jacquet, C.; Goulet, V.; Vaillant, V.; De Valk, H. Pulsed-Field Gel Electrophoresis of Listeria monocytogenes Strains: The PulseNet Europe Feasibility Study. Foodbourne Pathog. Dis. 2006, 3, 303–308. [Google Scholar] [CrossRef]
- López, V.; Ortiz, S.; Corujo, A.; López, P.; Poza, D.; Navas, J.; Moreno, R.; Martínez-Suárez, J.V. Different Contamination Patterns of Lineage I and II Strains of Listeria monocytogenes in a Spanish Broiler Abattoir. Poult. Sci. 2008, 87, 1874–1882. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M. A New Perspective on Listeria monocytogenes Evolution. PLoS Pathog. 2008, 4, 1000146. [Google Scholar] [CrossRef]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole Genome-Based Population Biology and Epidemiological Surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef]
- Maiden, M.C.J.; Van Rensburg, M.J.J.; Bray, J.E.; Earle, S.G.; Ford, S.A.; Jolley, K.A.; McCarthy, N.D. MLST Revisited: The Gene-by-Gene Approach to Bacterial Genomics. Nat. Rev. Microbiol. 2013, 11, 728–736. [Google Scholar] [CrossRef]
- Van Walle, I.; Björkman, J.T.; Cormican, M.; Dallman, T.; Mossong, J.; Moura, A.; Pietzka, A.; Ruppitsch, W.; Takkinen, J.; Mattheus, W.; et al. Retrospective Validation of Whole Genome Sequencingenhanced Surveillance of Listeriosis in Europe, 2010 to 2015. Eurosurveillance 2018, 23, 1700798. [Google Scholar] [CrossRef] [PubMed]
- Varsaki, A.; Ortiz, S.; Santorum, P.; López, P.; López-Alonso, V.; Martínez-Suárez, J.V. Genetic Diversity, Antimicrobial Resistance and Survival upon Manure Storage of Campylobacter jejuni Isolated from Dairy Cattle Farms in the Cantabric Coast of Spain. Zoonotic Dis. 2022, 2, 82–94. [Google Scholar] [CrossRef]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the Major Listeria monocytogenes Serovars by Multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, A.; Chenal-Francisque, V.; Dieye, H.; Cantinelli, T.; Drali, R.; Brisse, S.; Lecuit, M. Characterization of the Novel Listeria monocytogenes PCR Serogrouping Profile IVb-V1. Int. J. Food Microbiol. 2011, 147, 74–77. [Google Scholar] [CrossRef]
- Halpin, J.L.; Garrett, N.M.; Ribot, E.M.; Graves, L.M.; Cooper, K.L. Re-Evaluation, Optimization, and Multilaboratory Validation of the PulseNet-Standardized Pulsed-Field Gel Electrophoresis Protocol for Listeria monocytogenes. Foodborne Pathog. Dis. 2010, 7, 293–298. [Google Scholar] [CrossRef]
- Ortiz, S.; López, V.; Villatoro, D.; López, P.; Dávila, J.C.; Martínez-Suárez, J.V. A 3-Year Surveillance of the Genetic Diversity and Persistence of Listeria monocytogenes in an Iberian Pig Slaughterhouse and Processing Plant. Foodborne Pathog. Dis. 2010, 7, 1177–1184. [Google Scholar] [CrossRef]
- Graves, L.M.; Swaminathan, B. PulseNet Standardized Protocol for Subtyping Listeria monocytogenes by Macrorestriction and Pulsed-Field Gel Electrophoresis. Int. J. Food Microbiol. 2001, 65, 55–62. [Google Scholar] [CrossRef]
- CLSI Clinical and Laboratory Standards Institude. Performance Standards for Antimicrobial Susceptibility Testing. Available online: https://clsi.org/standards/products/microbiology/documents/m100/ (accessed on 15 January 2022).
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 28 March 2022).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Hernández, M.; Iglesias, M.R.; Rodríguez-Lázaro, D.; Gallardo, A.; Quijada, N.M.; Miguela-Villoldo, P.; Campos, M.J.; Píriz, S.; López-Orozco, G.; de Frutos, C.; et al. Co-Occurrence of Colistin-Resistance Genes Mcr-1 and Mcr-3 among Multidrug-Resistant Escherichia coli Isolated from Cattle, Spain, September 2015. Eurosurveillance 2017, 22, 30586. [Google Scholar] [CrossRef]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M. TORMES: An Automated Pipeline for Whole Bacterial Genome Analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles Instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Cury, J.; Abby, S.S.; Doppelt-Azeroual, O.; Néron, B.; Rocha, E.P.C. Identifying Conjugative Plasmids and Integrative Conjugative Elements with CONJscan. Methods Mol. Biol. 2020, 2075, 265–283. [Google Scholar] [CrossRef]
- Nightingale, K.K.; Schukken, Y.H.; Nightingale, C.R.; Fortes, E.D.; Ho, A.J.; Her, Z.; Grohn, Y.T.; McDonough, P.L.; Wiedmann, M. Ecology and Transmission of Listeria monocytogenes Infecting Ruminants and in the Farm Environment. Appl. Environ. Microbiol. 2004, 70, 4458–4467. [Google Scholar] [CrossRef]
- Ireton, K.; Mortuza, R.; Gyanwali, G.C.; Gianfelice, A.; Hussain, M. Role of Internalin Proteins in the Pathogenesis of Listeria monocytogenes. Mol. Microbiol. 2021, 116, 1407–1419. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.A.; Domínguez-Bernal, G.; González-Zorn, B.; Kreft, J.; Goebel, W. Pathogenicity Islands and Virulence Evolution in Listeria. Microbes Infect. 2001, 3, 571–584. [Google Scholar] [CrossRef]
- Cotter, P.D.; Draper, L.A.; Lawton, E.M.; Daly, K.M.; Groeger, D.S.; Casey, P.G.; Ross, R.P.; Hill, C. Listeriolysin S, a Novel Peptide Haemolysin Associated with a Subset of Lineage I Listeria monocytogenes. PLoS Pathog. 2008, 4, e1000144. [Google Scholar] [CrossRef] [PubMed]
- Cahoon, L.A.; Freitag, N.E. Listeria monocytogenes Virulence Factor Secretion: Don’t Leave the Cell without a Chaperone. Front. Cell. Infect. Microbiol. 2014, 4, 13. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nair, S.; Milohanic, E.; Berche, P. ClpC ATPase Is Required for Cell Adhesion and Invasion of Listeria monocytogenes. Infect. Immun. 2000, 68, 7061–7068. [Google Scholar] [CrossRef]
- Nair, S.; Frehel, C.; Nguyen, L.; Escuyer, V.; Berche, P. ClpE, a Novel Member of the HSP100 Family, Is Involved in Cell Division and Virulence of Listeria monocytogenes. Mol. Microbiol. 1999, 31, 185–196. [Google Scholar] [CrossRef]
- Gaillot, O.; Bregenholt, S.; Jaubert, F.; Di Santo, J.P.; Berche, P. Stress-Induced ClpP Serine Protease of Listeria monocytogenes Is Essential for Induction of Listeriolysin O-Dependent Protective Immunity. Infect. Immun. 2001, 69, 4938. [Google Scholar] [CrossRef]
- Milohanic, E.; Jonquières, R.; Cossart, P.; Berche, P.; Gaillard, J.-L. The Autolysin Ami Contributes to the Adhesion of Listeria monocytogenes to Eukaryotic Cells via Its Cell Wall Anchor. Mol. Microbiol. 2001, 39, 1212–1224. [Google Scholar] [CrossRef]
- Cabanes, D.; Dussurget, O.; Dehoux, P.; Cossart, P. Auto, a Surface Associated Autolysin of Listeria Monocytogenes Required for Entry into Eukaryotic Cells and Virulence. Mol. Microbiol. 2004, 51, 1601–1614. [Google Scholar] [CrossRef]
- Painset, A.; Björkman, J.T.; Kiil, K.; Guillier, L.; Mariet, J.F.; Felix, B.; Amar, C.; Rotariu, O.; Roussel, S.; Perez-Reche, F.; et al. Liseq—Whole-Genome Sequencing of a Cross-Sectional Survey of Listeria monocytogenes in Ready-to-Eat Foods and Human Clinical Cases in Europe. Microb. Genomics 2019, 5, e000257. [Google Scholar] [CrossRef]
- Shenoy, A.G.; Oliver, H.F.; Deering, A.J. Listeria monocytogenes Internalizes in Romaine Lettuce Grown in Greenhouse Conditions. J. Food Prot. 2017, 80, 573–581. [Google Scholar] [CrossRef]
- Fenlon, D.R.; Wilson, J.; Donachie, W. The Incidence and Level of Listeria monocytogenes Contamination of Food Sources at Primary Production and Initial Processing. J. Appl. Bacteriol. 1996, 81, 641–650. [Google Scholar] [CrossRef]
- Bardsley, C.A.; Boyer, R.R.; Rideout, S.L.; Strawn, L.K. Survival of Listeria monocytogenes on the Surface of Basil, Cilantro, Dill, and Parsley Plants. Food Control 2019, 95, 90–94. [Google Scholar] [CrossRef]
- Kljujev, I.; Raicevic, V.; Jovicic-Petrovic, J.; Vujovic, B.; Mirkovic, M.; Rothballer, M. Listeria monocytogenes—Danger for Health Safety Vegetable Production. Microb. Pathog. 2018, 120, 23–31. [Google Scholar] [CrossRef]
- Truong, H.N.; Garmyn, D.; Gal, L.; Fournier, C.; Sevellec, Y.; Jeandroz, S.; Piveteau, P. Plants as a Realized Niche for Listeria monocytogenes. Microbiologyopen 2021, 10, e1255. [Google Scholar] [CrossRef]
- Kõiv, V.; Arbo, K.; Maiväli, Ü.; Kisand, V.; Roosaare, M.; Remm, M.; Tenson, T. Endophytic Bacterial Communities in Peels and Pulp of Five Root Vegetables. PLoS ONE 2019, 14, e0210542. [Google Scholar] [CrossRef]
- Kutter, S.; Hartmann, A.; Schmid, M. Colonization of Barley (Hordeum Vulgare) with Salmonella enterica and Listeria spp. FEMS Microbiol. Ecol. 2006, 56, 262–271. [Google Scholar] [CrossRef]
- Lyautey, E.; Lapen, D.R.; Wilkes, G.; McCleary, K.; Pagotto, F.; Tyler, K.; Hartmann, A.; Piveteau, P.; Rieu, A.; Robertson, W.J.; et al. Distribution and Characteristics of Listeria monocytogenes Isolates from Surface Waters of the South Nation River Watershed, Ontario, Canada. Appl. Environ. Microbiol. 2007, 73, 5401–5410. [Google Scholar] [CrossRef]
- Hingston, P.; Chen, J.; Dhillon, B.K.; Laing, C.; Bertelli, C.; Gannon, V.; Tasara, T.; Allen, K.; Brinkman, F.S.L.; Hansen, L.T.; et al. Genotypes Associated with Listeria monocytogenes Isolates Displaying Impaired or Enhanced Tolerances to Cold, Salt, Acid, or Desiccation Stress. Front. Microbiol. 2017, 8, 369. [Google Scholar] [CrossRef]
- Lukinmaa, S.; Aarnisalo, K.; Suihko, M.L.; Siitonen, A. Diversity of Listeria monocytogenes Isolates of Human and Food Origin Studied by Serotyping, Automated Ribotyping and Pulsed-Field Gel Electrophoresis. Clin. Microbiol. Infect. 2004, 10, 562–568. [Google Scholar] [CrossRef][Green Version]
- Esteban, J.I.; Oporto, B.; Aduriz, G.; Juste, R.A.; Hurtado, A. Faecal Shedding and Strain Diversity of Listeria monocytogenes in Healthy Ruminants and Swine in Northern Spain. BMC Vet. Res. 2009, 5, 2. [Google Scholar] [CrossRef]
- Muchaamba, F.; Eshwar, A.K.; Stevens, M.J.A.; Stephan, R.; Tasara, T. Different Shades of Listeria monocytogenes: Strain, Serotype, and Lineage-Based Variability in Virulence and Stress Tolerance Profiles. Front. Microbiol. 2022, 12, 792162. [Google Scholar] [CrossRef]
- Lee, B.H.; Garmyn, D.; Gal, L.; Guérin, C.; Guillier, L.; Rico, A.; Rotter, B.; Nicolas, P.; Piveteau, P. Exploring Listeria monocytogenes Transcriptomes in Correlation with Divergence of Lineages and Virulence as Measured in Galleria mellonella. Appl. Environ. Microbiol. 2019, 85, e01370-19. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes Hypervirulence by Harnessing Its Biodiversity. Nat. Genet. 2016, 48, 308. [Google Scholar] [CrossRef]
- Cabal, A.; Pietzka, A.; Huhulescu, S.; Allerberger, F.; Ruppitsch, W.; Schmid, D. Isolate-Based Surveillance of Listeria monocytogenes by Whole Genome Sequencing in Austria. Front. Microbiol. 2019, 10, 2282. [Google Scholar] [CrossRef]
- Halbedel, S.; Wilking, H.; Holzer, A.; Kleta, S.; Fischer, M.A.; Lüth, S.; Pietzka, A.; Huhulescu, S.; Lachmann, R.; Krings, A.; et al. Large Nationwide Outbreak of Invasive Listeriosis Associated with Blood Sausage, Germany, 2018–2019. Emerg. Infect. Dis. 2020, 26, 1456–1464. [Google Scholar] [CrossRef]
- Palacios-Gorba, C.; Moura, A.; Gomis, J.; Leclercq, A.; Gómez-Martín, Á.; Bracq-Dieye, H.; Mocé, M.L.; Tessaud-Rita, N.; Jiménez-Trigos, E.; Vales, G.; et al. Ruminant-Associated Listeria monocytogenes Isolates Belong Preferentially to Dairy-Associated Hypervirulent Clones: A Longitudinal Study in 19 Farms. Environ. Microbiol. 2021, 23, 7617–7631. [Google Scholar] [CrossRef]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria Monocytogenes Clones’ Adaption to Mammalian Gut Accounts for Their Association with Dairy Products. Nat. Commun. 2019, 10, 2488. [Google Scholar] [CrossRef]
- Huemer, M.; Mairpady Shambat, S.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic resistance and persistence-Implications for human health and treatment perspectives. EMBO Rep. 2020, 21, e51034. [Google Scholar] [CrossRef]
- Haubert, L.; Mendonça, M.; Lopes, G.V.; de Itapema Cardoso, M.R.; da Silva, W.P. Listeria monocytogenes Isolates from Food and Food Environment Harbouring TetM and ErmB Resistance Genes. Lett. Appl. Microbiol. 2016, 62, 23–29. [Google Scholar] [CrossRef]
- Neil, K.; Allard, N.; Rodrigue, S. Molecular Mechanisms Influencing Bacterial Conjugation in the Intestinal Microbiota. Front. Microbiol. 2021, 12, 1415. [Google Scholar] [CrossRef]
- Alvarez-Molina, A.; Cobo-Díaz, J.F.; López, M.; Prieto, M.; de Toro, M.; Alvarez-Ordóñez, A. Unraveling the Emergence and Population Diversity of Listeria monocytogenes in a Newly Built Meat Facility through Whole Genome Sequencing. Int. J. Food Microbiol. 2021, 340, 109043. [Google Scholar] [CrossRef] [PubMed]
- Garcillán-Barcia, M.P.; Redondo-Salvo, S.; Vielva, L.; de la Cruz, F. MOBscan: Automated annotation of MOB relaxases. Methods Mol. Biol. 2020, 2075, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Garcillán-Barcia, M.P.; Francia, M.V.; de la Cruz, F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009, 33, 657–687. [Google Scholar] [CrossRef] [PubMed]
- Coluzzi, C.; Garcillán-Barcia, M.D.P.; de la Cruz, F.; Rocha, E.P.C. Evolution of Plasmid Mobility: Origin and Fate of Non-Conjugative Plasmids. bioRxiv 2021. [Google Scholar] [CrossRef]
- Disson, O.; Moura, A.; Lecuit, M. Making sense of the biodiversity and virulence of Listeria monocytogenes. Trends Microbiol. 2021, 29, 811–822. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Paramithiotis, S.; Drosinos, E.H. Genetic Analysis of the Listeria Pathogenicity Island 1 of Listeria monocytogenes 1/2a and 4b Isolates. Curr. Microbiol. 2018, 75, 857–865. [Google Scholar] [CrossRef]
- Poimenidou, S.V.; Dalmasso, M.; Papadimitriou, K.; Fox, E.M.; Skandamis, P.N.; Jordan, K. Virulence Gene Sequencing Highlights Similarities and Differences in Sequences in Listeria monocytogenes Serotype 1/2a and 4b Strains of Clinical and Food Origin from 3 Different Geographic Locations. Front. Microbiol. 2018, 9, 1103. [Google Scholar] [CrossRef]
- Quereda, J.J.; Dussurget, O.; Nahori, M.A.; Ghozlane, A.; Volant, S.; Dillies, M.A.; Regnault, B.; Kennedy, S.; Mondot, S.; Villoing, B.; et al. Bacteriocin from Epidemic Listeria Strains Alters the Host Intestinal Microbiota to Favor Infection. Proc. Natl. Acad. Sci. USA 2016, 113, 5706–5711. [Google Scholar] [CrossRef]
- Chen, M.; Cheng, J.; Wu, Q.; Zhang, J.; Chen, Y.; Xue, L.; Lei, T.; Zeng, H.; Wu, S.; Ye, Q.; et al. Occurrence, Antibiotic Resistance, and Population Diversity of Listeria monocytogenes isolated from Fresh Aquatic Products in China. Front. Microbiol. 2018, 9, 2215. [Google Scholar] [CrossRef]
- Lee, S.; Chen, Y.; Gorski, L.; Ward, T.J.; Osborne, J.; Kathariou, S. Listeria monocytogenes Source Distribution Analysis Indicates Regional Heterogeneity and Ecological Niche Preference among Serotype 4b Clones. mBio 2018, 9, e00396-18. [Google Scholar] [CrossRef]
- Quereda, J.J.; Rodríguez-Gómez, I.M.; Meza-Torres, J.; Gómez-Laguna, J.; Nahori, M.A.; Dussurget, O.; Carrasco, L.; Cossart, P.; Pizarro-Cerdá, J. Reassessing the Role of Internalin B in Listeria monocytogenes Virulence Using the Epidemic Strain F2365. Clin. Microbiol. Infect. 2019, 25, 252.e1–252.e4. [Google Scholar] [CrossRef]
- Dortet, L.; Mostowy, S.; Louaka, A.S.; Gouin, E.; Nahori, M.A.; Wiemer, E.A.C.; Dussurget, O.; Cossart, P. Recruitment of the Major Vault Protein by InlK: A Listeria monocytogenes Strategy to Avoid Autophagy. PLoS Pathog. 2011, 7, e1002168. [Google Scholar] [CrossRef]
- Ghosh, P.; Halvorsen, E.M.; Ammendolia, D.A.; Mor-Vaknin, N.; O’Riordan, M.X.D.; Brumell, J.H.; Markovitz, D.M.; Higgins, D.E. Invasion of the Brain by Listeria monocytogenes Is Mediated by InlF and Host Cell Vimentin. mBio 2018, 9, e00160-18. [Google Scholar] [CrossRef]
- Asano, K.; Sashinami, H.; Osanai, A.; Asano, Y.; Nakane, A. Autolysin Amidase of Listeria monocytogenes Promotes Efficient Colonization of Mouse Hepatocytes and Enhances Host Immune Response. Int. J. Med. Microbiol. 2011, 301, 480–487. [Google Scholar] [CrossRef]


| Sample Type | No of Samples Tested | No (%) of Samples Negative for Listeria | No (%) of Samples Positive for Listeria | No (%) of Samples Positive for L. monocytogenes |
|---|---|---|---|---|
| Slurry tanker | 19 | 1 (5%) | 18 (95%) | 13 (68%) |
| Feeding through/corridor | 36 | 1 (3%) | 35 (97%) | 29 (80%) |
| Stable floor | 40 | 1 (3%) | 39 (97%) | 32 (80%) |
| Slurry drain | 19 | 0 (0%) | 19 (100%) | 16 (84%) |
| Total environmental samples | 114 | 3 (3%) | 111 (97%) | 90 (79%) |
| Dry forage | 42 | 3 (7%) | 39 (93%) | 10 (24%) |
| Fresh grass crops | 24 | 3 (12%) | 21 (88%) | 11 (46%) |
| Maize cured forage | 11 | 0 (0%) | 11 (100%) | 3 (27%) |
| Grass cured forage | 8 | 0 (0%) | 8 (100%) | 0 (0%) |
| Concentrate for dairy cows | 41 | 2 (5%) | 39 (95%) | 12 (29%) |
| Total feed samples | 126 | 8 (6%) | 118 (94%) | 36 (29%) |
| Filter from the milk tank | 28 | 9 (32%) | 19 (68%) | 4 (14%) |
| Raw milk from the tank | 34 | 10 (29%) | 24 (71%) | 2 (6%) |
| Total raw dairy samples | 62 | 19 (31%) | 43 (69%) | 6 (10%) |
| Chicken manure | 9 | 1 (11%) | 8 (89%) | 7 (78%) |
| Stored dairy manure | 39 | 0 (0%) | 39 (100%) | 36 (92%) |
| Fresh feces from unhealthy dairy cows (diarrhea/infection/miscarriage) | 33 | 3 (9%) | 30 (91%) | 28 (93%) |
| Fresh feces from healthy dairy cows (without symptoms) | 41 | 3 (7%) | 38 (93%) | 8 (19%) |
| Total fecal samples | 122 | 7 (6%) | 115 (94%) | 79 (65%) |
| Total samples | 424 | 37 (9%) | 387 (91%) | 211 (50%) |
| Farm Type | No of Samples Tested | No (%) of Samples Negative for Listeria | No (%) of Samples Positive for Listeria | No (%) of Samples Positive for L. monocytogenes |
|---|---|---|---|---|
| Organic | 147 | 9 (6%) | 138 (94%) | 83 (56%) |
| Conventional | 277 | 28 (10%) | 249 (90%) | 128 (46%) |
| Total samples | 424 | 37 (8%) | 387 (91%) | 211 (50%) |
| Isolates | ST | CC | Lineage |
|---|---|---|---|
| MS6484, MS6491, MS6493, MS6494, MS6497, MS6499, MS6501, MS6502, MS6503, MS6507, MS6510, MS6661, MS6674 | 1 | 1 | I |
| MS6676 | 2 | 2 | I |
| MS6673 | 4 | 4 | I |
| MS6492, MS6666, MS6668, MS6669 | 6 | 6 | I |
| MS6486, MS6487, MS6504, MS6509, MS6667, MS6675, MS6678 | 54 | 54 | I |
| MS6670 | 59 | 59 | I |
| MS6506, MS6665, MS6672 | 217 | 217 | I |
| MS6485, MS6488, MS6489, MS6508, MS6511 | 388 | 388 | I |
| MS6505 | 389 | 389 | I |
| MS6677 | 489 | 489 | I |
| MS6490, MS6495, MS6496, MS6498, MS6500, MS6664, MS6671 | 666 | 666 | I |
| MS6663 | 2921 (novel) | 54 | I |
| Isolate | AMP | CIP | ERY | GEN | TET | VAN | MEM | FOX |
|---|---|---|---|---|---|---|---|---|
| MS6484 | 2(R) | 4(R) | 2(R) | 0.5(S) | 4(R) | 4(R) | 1(R) | 16(R) |
| MS6488 | 1(S) | 2(R) | 2(R) | 0.5(S) | 2(S) | 1(S) | 0.25(S) | 32(R) |
| MS6490 | 2(R) | 0.5(S) | 1(S) | 0.5(S) | 4(R) | 4(R) | 0.25(S) | 64(R) |
| MS6485 | 0.25(S) | 2(R) | 1(S) | 0.5(S) | 4(R) | 1(S) | 0.25(S) | 32(R) |
| MS6499 | 2(R) | 2(R) | 1(S) | 0.25(S) | 4(R) | 1(S) | 0.25(S) | 64(R) |
| MS6501 | 4(R) | 0.5(S) | 0.25(S) | 0.5(S) | 2(S) | 1(S) | 0.5(R) | 16(R) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varsaki, A.; Ortiz, S.; Santorum, P.; López, P.; López-Alonso, V.; Hernández, M.; Abad, D.; Rodríguez-Grande, J.; Ocampo-Sosa, A.A.; Martínez-Suárez, J.V. Prevalence and Population Diversity of Listeria monocytogenes Isolated from Dairy Cattle Farms in the Cantabria Region of Spain. Animals 2022, 12, 2477. https://doi.org/10.3390/ani12182477
Varsaki A, Ortiz S, Santorum P, López P, López-Alonso V, Hernández M, Abad D, Rodríguez-Grande J, Ocampo-Sosa AA, Martínez-Suárez JV. Prevalence and Population Diversity of Listeria monocytogenes Isolated from Dairy Cattle Farms in the Cantabria Region of Spain. Animals. 2022; 12(18):2477. https://doi.org/10.3390/ani12182477
Chicago/Turabian StyleVarsaki, Athanasia, Sagrario Ortiz, Patricia Santorum, Pilar López, Victoria López-Alonso, Marta Hernández, David Abad, Jorge Rodríguez-Grande, Alain A. Ocampo-Sosa, and Joaquín V. Martínez-Suárez. 2022. "Prevalence and Population Diversity of Listeria monocytogenes Isolated from Dairy Cattle Farms in the Cantabria Region of Spain" Animals 12, no. 18: 2477. https://doi.org/10.3390/ani12182477
APA StyleVarsaki, A., Ortiz, S., Santorum, P., López, P., López-Alonso, V., Hernández, M., Abad, D., Rodríguez-Grande, J., Ocampo-Sosa, A. A., & Martínez-Suárez, J. V. (2022). Prevalence and Population Diversity of Listeria monocytogenes Isolated from Dairy Cattle Farms in the Cantabria Region of Spain. Animals, 12(18), 2477. https://doi.org/10.3390/ani12182477






