Genetic Parameter Estimation and Genome-Wide Association Analysis of Social Genetic Effects on Average Daily Gain in Purebreds and Crossbreds
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Pedigree and Phenotype Data
2.2. Genotype Data
2.3. Genetic Parameters and Variance Components
2.4. Single-Step Genome-Wide Association Study
- D = I in the first step;
- Calculation of breeding values;
- Calculation of SNP effects, , where is the breeding value for genotyped individuals;
- Calculation of SNP weight for each SNP, , where i is the i-th SNP;
- Normalization of SNP weight for retaining constant total genetic variance;
- Then loop to step 2.
2.5. Candidate Genes and Gene Ontology (GO)
3. Results and Discussion
3.1. ADG Performance, Genetic Parameters and Variance Components
3.2. Genetic Correlations between Purebred and Crossbred Pigs
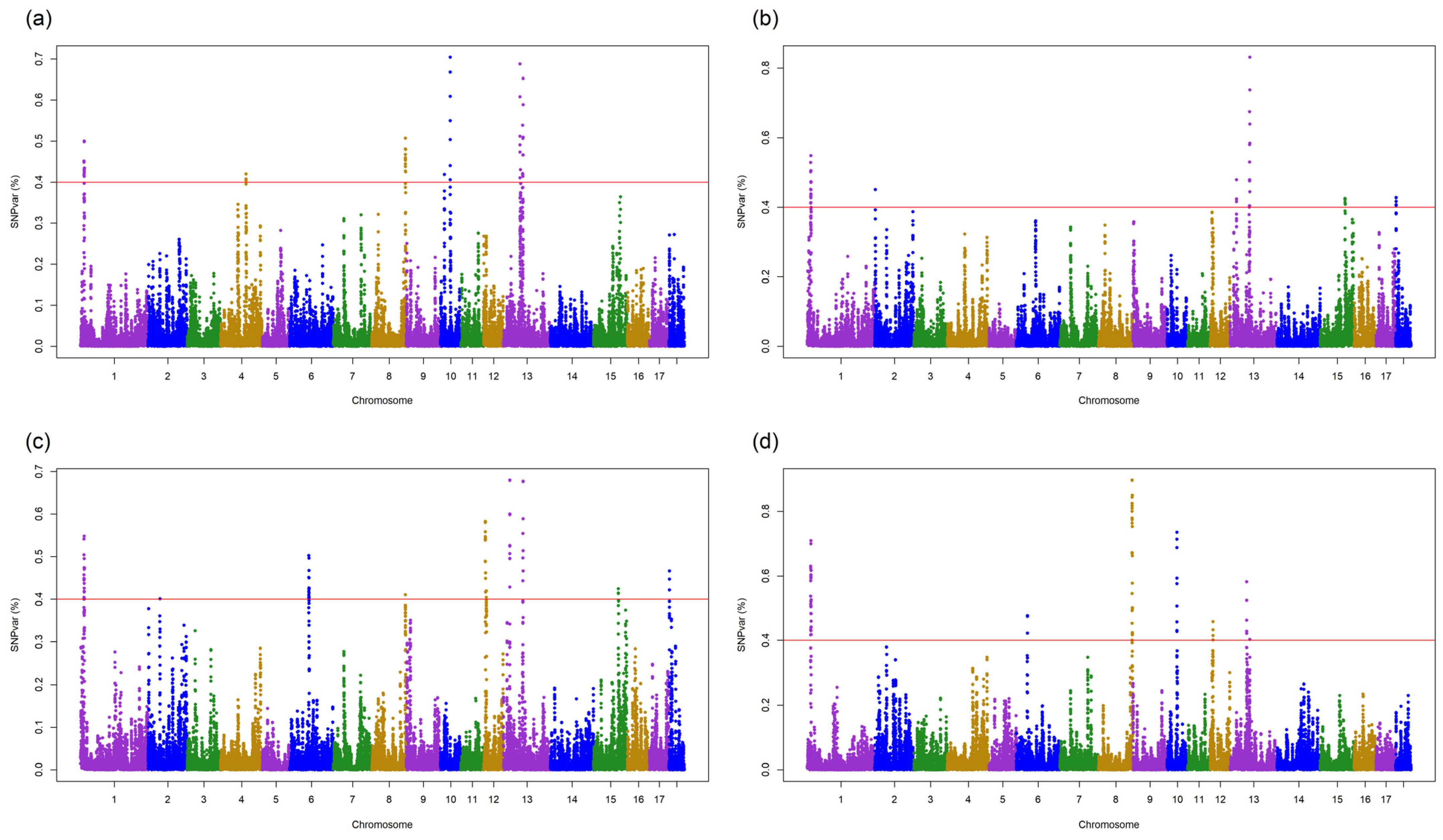
3.3. QTLs for DGE and SGE
3.4. GO and KEGG Analyses for SGE
3.5. GO and KEGG Analyses for DGE
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Canario, L.; Lundeheim, N.; Bijma, P. The early-life environment of a pig shapes the phenotypes of its social partners in adulthood. Heredity 2017, 118, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Van Nieuwamerongen, S. Development of Pigs Raised in a Group Housing System for Lactating Sows and Their Litters; Wageningen University and Research: Wageningen, The Netherlands, 2017. [Google Scholar]
- Angarita, B.K.; Cantet, R.J.C.; Wurtz, K.E.; O’Malley, C.I.O.; Siegford, J.M.; Ernst, C.W.; Turner, S.P.; Steibel, J.P. Estimation of indirect social genetic effects for skin lesion count in group-housed pigs by quantifying behavioral interactions1. J. Anim. Sci. 2019, 97, 3658–3668. [Google Scholar] [CrossRef] [PubMed]
- Griffing, B. Selection in reference to biological groups. I. Individual and group selection applied to populations of unordered groups. Aust. J. Biol. Sci. 1967, 20, 127–139. [Google Scholar] [PubMed]
- Hong, J.K.; Lee, J.B.; Ramayo-Caldas, Y.; Kim, S.D.; Cho, E.S.; Kim, Y.S.; Cho, K.H.; Lee, D.H.; Park, H.B. Single-step genome-wide association study for social genetic effects and direct genetic effects on growth in Landrace pigs. Sci. Rep. 2020, 10, 14958. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Xu, J.; Yin, L.; Yin, D.; Zhu, M.; Yu, M.; Li, X.; Zhao, S.; Liu, X. Genome-Wide Association Study Reveals Candidate Genes for Growth Relevant Traits in Pigs. Front. Genet. 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Ding, R.; Meng, F.; Wang, X.; Zhuang, Z.; Quan, J.; Geng, Q.; Wu, J.; Zheng, E.; Wu, Z.; et al. A meta-analysis of genome-wide association studies for average daily gain and lean meat percentage in two Duroc pig populations. BMC Genom. 2021, 22, 12. [Google Scholar] [CrossRef]
- Ruan, D.; Zhuang, Z.; Ding, R.; Qiu, Y.; Zhou, S.; Wu, J.; Xu, C.; Hong, L.; Huang, S.; Zheng, E.; et al. Weighted Single-Step GWAS Identified Candidate Genes Associated with Growth Traits in a Duroc Pig Population. Genes 2021, 12, 117. [Google Scholar] [CrossRef]
- Wu, P.; Wang, K.; Zhou, J.; Chen, D.; Jiang, A.; Jiang, Y.; Zhu, L.; Qiu, X.; Li, X.; Tang, G. A combined GWAS approach reveals key loci for socially-affected traits in Yorkshire pigs. Commun. Biol. 2021, 4, 891. [Google Scholar] [CrossRef]
- Wu, P.; Wang, K.; Yang, Q.; Zhou, J.; Chen, D.; Liu, Y.; Ma, J.; Tang, Q.; Jin, L.; Xiao, W.; et al. Whole-genome re-sequencing association study for direct genetic effects and social genetic effects of six growth traits in Large White pigs. Sci. Rep. 2019, 9, 9667. [Google Scholar] [CrossRef]
- Ask, B.; Pedersen, L.V.; Christensen, O.F.; Nielsen, H.M.; Turner, S.P.; Nielsen, B. Selection for social genetic effects in purebreds increases growth in crossbreds. Genet. Sel. Evol. 2021, 53, 15. [Google Scholar] [CrossRef]
- Christensen, O.F.; Lund, M.S. Genomic prediction when some animals are not genotyped. Genet. Sel. Evol. 2010, 42, 2. [Google Scholar]
- Legarra, A.; Aguilar, I.; Misztal, I. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar]
- Salek Ardestani, S.; Jafarikia, M.; Sargolzaei, M.; Sullivan, B.; Miar, Y. Genomic prediction of average daily gain, back-fat thickness, and loin muscle depth using different genomic tools in Canadian swine populations. Front. Genet. 2021, 12, 665344. [Google Scholar] [PubMed]
- Long, Y.; Ruan, G.R.; Su, Y.; Xiao, S.J.; Zhang, Z.Y.; Ren, J.; Ding, N.S.; Huang, L.S. Genome-wide association study identifies QTLs for EBV of backfat thickness and average daily gain in Duroc pigs. Genetika 2015, 51, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Ostersen, T.; Christensen, O.F.; Xiang, T. Single-step genomic evaluation with metafounders for feed conversion ratio and average daily gain in Danish Landrace and Yorkshire pigs. Genet. Sel. Evol. 2021, 53, 79. [Google Scholar] [CrossRef]
- Bergamaschi, M.; Maltecca, C.; Fix, J.; Schwab, C.; Tiezzi, F. Genome-wide association study for carcass quality traits and growth in purebred and crossbred pigs. J. Anim. Sci. 2020, 98, skz360. [Google Scholar]
- Ibanez-Escriche, N.; Fernando, R.L.; Toosi, A.; Dekkers, J.C. Genomic selection of purebreds for crossbred performance. Genet. Sel. Evol. 2009, 41, 12. [Google Scholar] [CrossRef]
- Hong, J.K.; Cho, K.H.; Kim, Y.S.; Chung, H.J.; Baek, S.Y.; Cho, E.S.; Sa, S.J. Genetic relationship between purebred and synthetic pigs for growth performance using single step method. Anim. Biosci. 2021, 34, 967. [Google Scholar]
- Kim, Y.M.; Seong, H.S.; Kim, Y.S.; Hong, J.K.; Sa, S.J.; Lee, J.; Lee, J.H.; Cho, K.H.; Chung, W.H.; Choi, J.W.; et al. Genome-Wide Assessment of a Korean Composite Pig Breed, Woori-Heukdon. Front. Genet. 2022, 13, 779152. [Google Scholar] [CrossRef]
- Aguilar, I.; Tsuruta, S.; Masuda, Y.; Lourenco, D.; Legarra, A.; Misztal, I. BLUPF90 suite of programs for animal breeding with focus on genomics. In Proceedings of the World Congress on Genetics Applied to Livestock Production, Auckland, New Zealand, 11–16 February 2018; Volume 11, p. 751. [Google Scholar]
- Bijma, P. A general definition of the heritable variation that determines the potential of a population to respond to selection. Genetics 2011, 189, 1347–1359. [Google Scholar] [PubMed]
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Fernando, R.L.; Vitezica, Z.; Okimoto, R.; Wing, T.; Hawken, R.; Muir, W.M. Genome-wide association mapping including phenotypes from relatives without genotypes in a single-step (ssGWAS) for 6-week body weight in broiler chickens. Front. Genet. 2014, 5, 134. [Google Scholar]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Cameron, N.D.; Enser, M.B. Fatty acid composition of lipid in Longissimus dorsi muscle of Duroc and British Landrace pigs and its relationship with eating quality. Meat Sci. 1991, 29, 295–307. [Google Scholar] [CrossRef]
- Alam, M.; Chang, H.K.; Lee, S.S.; Choi, T.J. Genetic Analysis of Major Production and Reproduction Traits of Korean Duroc, Landrace and Yorkshire Pigs. Animals 2021, 11, 1321. [Google Scholar] [CrossRef]
- Willson, H.E.; Rojas de Oliveira, H.; Schinckel, A.P.; Grossi, D.; Brito, L.F. Estimation of Genetic Parameters for Pork Quality, Novel Carcass, Primal-Cut and Growth Traits in Duroc Pigs. Animals 2020, 10, 779. [Google Scholar] [CrossRef]
- Hoque, M.; Kadowaki, H.; Shibata, T.; Oikawa, T.; Suzuki, K. Genetic parameters for measures of residual feed intake and growth traits in seven generations of Duroc pigs. Livest. Sci. 2009, 121, 45–49. [Google Scholar]
- Suzuki, K.; Irie, M.; Kadowaki, H.; Shibata, T.; Kumagai, M.; Nishida, A. Genetic parameter estimates of meat quality traits in Duroc pigs selected for average daily gain, longissimus muscle area, backfat thickness, and intramuscular fat content. J. Anim. Sci. 2005, 83, 2058–2065. [Google Scholar] [CrossRef] [PubMed]
- Bergsma, R.; Kanis, E.; Knol, E.F.; Bijma, P. The contribution of social effects to heritable variation in finishing traits of domestic pigs (Sus scrofa). Genetics 2008, 178, 1559–1570. [Google Scholar] [CrossRef] [PubMed]
- Esfandyari, H.; Sørensen, A.C.; Bijma, P. Maximizing crossbred performance through purebred genomic selection. Genet. Sel. Evol. 2015, 47, 16. [Google Scholar] [CrossRef]
- Liu, G.; Jennen, D.G.; Tholen, E.; Juengst, H.; Kleinwachter, T.; Holker, M.; Tesfaye, D.; Un, G.; Schreinemachers, H.J.; Murani, E.; et al. A genome scan reveals QTL for growth, fatness, leanness and meat quality in a Duroc-Pietrain resource population. Anim. Genet. 2007, 38, 241–252. [Google Scholar] [CrossRef]
- Knott, S.A.; Marklund, L.; Haley, C.S.; Andersson, K.; Davies, W.; Ellegren, H.; Fredholm, M.; Hansson, I.; Hoyheim, B.; Lundström, K. Multiple marker mapping of quantitative trait loci in a cross between outbred wild boar and large white pigs. Genetics 1998, 149, 1069–1080. [Google Scholar] [CrossRef]
- Wang, K.; Liu, D.; Hernandez-Sanchez, J.; Chen, J.; Liu, C.; Wu, Z.; Fang, M.; Li, N. Genome wide association analysis reveals new production trait genes in a male Duroc population. PLoS ONE 2015, 10, e0139207. [Google Scholar]
- Guo, Y.M.; Lee, G.J.; Archibald, A.L.; Haley, C.S. Quantitative trait loci for production traits in pigs: A combined analysis of two Meishan x Large White populations. Anim. Genet. 2008, 39, 486–495. [Google Scholar] [CrossRef]
- Vidal, O.; Noguera, J.L.; Amills, M.; Varona, L.; Gil, M.; Jimenez, N.; Davalos, G.; Folch, J.M.; Sanchez, A. Identification of carcass and meat quality quantitative trait loci in a Landrace pig population selected for growth and leanness. J. Anim. Sci. 2005, 83, 293–300. [Google Scholar] [CrossRef]
- Duthie, C.; Simm, G.; Doeschl-Wilson, A.; Kalm, E.; Knap, P.; Roehe, R. Quantitative trait loci for chemical body composition traits in pigs and their positional associations with body tissues, growth and feed intake. Anim. Genet. 2008, 39, 130–140. [Google Scholar] [CrossRef]
- Cho, I.-C.; Yoo, C.-K.; Lee, J.-B.; Jung, E.-J.; Han, S.-H.; Lee, S.-S.; Ko, M.-S.; Lim, H.-T.; Park, H.-B. Genome-wide QTL analysis of meat quality-related traits in a large F2 intercross between Landrace and Korean native pigs. Genet. Sel. Evol. 2015, 47, 7. [Google Scholar] [CrossRef]
- Reiner, G.; Kohler, F.; Berge, T.; Fischer, R.; Hubner-Weitz, K.; Scholl, J.; Willems, H. Mapping of quantitative trait loci affecting behaviour in swine. Anim. Genet. 2009, 40, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Xiong, Y.Z.; Zuo, B.; Lei, M.G.; Jiang, S.W.; Li, F.E.; Zheng, R.; Li, J.L.; Xu, D.Q. Quantitative trait loci for carcass traits on pig chromosomes 4, 6, 7, 8 and 13. J. Appl. Genet. 2007, 48, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Ponsuksili, S.; Zebunke, M.; Murani, E.; Trakooljul, N.; Krieter, J.; Puppe, B.; Schwerin, M.; Wimmers, K. Integrated Genome-wide association and hypothalamus eQTL studies indicate a link between the circadian rhythm-related gene PER1 and coping behavior. Sci. Rep. 2015, 5, 16264. [Google Scholar] [CrossRef] [PubMed]
- Desautes, C.; Bidanelt, J.P.; Milant, D.; Iannuccelli, N.; Amigues, Y.; Bourgeois, F.; Caritez, J.C.; Renard, C.; Chevalet, C.; Mormede, P. Genetic linkage mapping of quantitative trait loci for behavioral and neuroendocrine stress response traits in pigs. J. Anim. Sci. 2002, 80, 2276–2285. [Google Scholar] [CrossRef] [PubMed]
- Harmegnies, N.; Davin, F.; De Smet, S.; Buys, N.; Georges, M.; Coppieters, W. Results of a whole-genome quantitative trait locus scan for growth, carcass composition and meat quality in a porcine four-way cross. Anim. Genet. 2006, 37, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Hou, L.; Zhang, X.; Huang, M.; Mao, H.; Chen, H.; Ma, J.; Chen, C.; Ai, H.; Ren, J.; et al. A meta analysis of genome-wide association studies for limb bone lengths in four pig populations. BMC Genet. 2015, 16, 95. [Google Scholar] [CrossRef][Green Version]
- Bailey, N.W.; Marie-Orleach, L.; Moore, A.J. Indirect genetic effects in behavioral ecology: Does behavior play a special role in evolution? Behav. Ecol. 2018, 29, 1–11. [Google Scholar] [CrossRef]
- Camerlink, I.; Ursinus, W.W.; Bartels, A.C.; Bijma, P.; Bolhuis, J.E. Indirect Genetic Effects for Growth in Pigs Affect Behaviour and Weight Around Weaning. Behav. Genet. 2018, 48, 413–420. [Google Scholar] [CrossRef]
- Pierani, A.; Moran-Rivard, L.; Sunshine, M.J.; Littman, D.R.; Goulding, M.; Jessell, T.M. Control of interneuron fate in the developing spinal cord by the progenitor homeodomain protein Dbx1. Neuron 2001, 29, 367–384. [Google Scholar] [CrossRef]
- Sokolowski, K.; Esumi, S.; Hirata, T.; Kamal, Y.; Tran, T.; Lam, A.; Oboti, L.; Brighthaupt, S.C.; Zaghlula, M.; Martinez, J.; et al. Specification of select hypothalamic circuits and innate behaviors by the embryonic patterning gene dbx1. Neuron 2015, 86, 403–416. [Google Scholar] [CrossRef]
- Proskorovski-Ohayon, R.; Kadir, R.; Michalowski, A.; Flusser, H.; Perez, Y.; Hershkovitz, E.; Sivan, S.; Birk, O.S. PAX7 mutation in a syndrome of failure to thrive, hypotonia, and global neurodevelopmental delay. Hum. Mutat. 2017, 38, 1671–1683. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Qi, Y.; Sun, Z. The Role of Sonic Hedgehog Pathway in the Development of the Central Nervous System and Aging-Related Neurodegenerative Diseases. Front. Mol. Biosci. 2021, 8, 711710. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 2009, 27, 519–550. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Aleyasin, H.; Stavarache, M.A.; Li, L.; Cathomas, F.; Parise, L.F.; Lin, H.Y.; Burnett, C.J.; Aubry, A.; Flanigan, M.E.; et al. Neuromodulatory effect of interleukin 1beta in the dorsal raphe nucleus on individual differences in aggression. Mol. Psychiatry 2021, 27, 2563–2579. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef]
- Vallabhapurapu, S.; Karin, M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009, 27, 693–733. [Google Scholar] [CrossRef]
- Gupta, S.C.; Sundaram, C.; Reuter, S.; Aggarwal, B.B. Inhibiting NF-κB activation by small molecules as a therapeutic strategy. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2010, 1799, 775–787. [Google Scholar] [CrossRef]
- Singh, S.; Singh, T.G. Role of Nuclear Factor Kappa B (NF-kappaB) Signalling in Neurodegenerative Diseases: An Mechanistic Approach. Curr. Neuropharmacol. 2020, 18, 918–935. [Google Scholar] [CrossRef]
- Levy, R.; Dubois, B. Apathy and the functional anatomy of the prefrontal cortex-basal ganglia circuits. Cereb. Cortex 2006, 16, 916–928. [Google Scholar] [CrossRef]
- Le Heron, C.; Apps, M.A.J.; Husain, M. The anatomy of apathy: A neurocognitive framework for amotivated behaviour. Neuropsychologia 2018, 118, 54–67. [Google Scholar] [CrossRef]
- Hines, L.M.; Hoffman, P.L.; Bhave, S.; Saba, L.; Kaiser, A.; Snell, L.; Goncharov, I.; LeGault, L.; Dongier, M.; Grant, B.; et al. A sex-specific role of type VII adenylyl cyclase in depression. J. Neurosci. 2006, 26, 12609–12619. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Ota, N.; Peng, I.; Refino, C.J.; Danilenko, D.M.; Caplazi, P.; Ouyang, W. IL-17RC is required for IL-17A–and IL-17F–dependent signaling and the pathogenesis of experimental autoimmune encephalomyelitis. J. Immunol. 2010, 184, 4307–4316. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, X.; Zhong, Y. Interleukin-17A: The Key Cytokine in Neurodegenerative Diseases. Front. Aging. Neurosci. 2020, 12, 566922. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Hsu, L.-C.; Liu, H.; Bankston, L.A.; Iimura, M.; Kagnoff, M.F.; Eckmann, L.; Karin, M. Nod2 mutation in Crohn’s disease potentiates NF-κB activity and IL-1ß processing. Science 2005, 307, 734–738. [Google Scholar] [CrossRef]
- Abmayr, S.M.; Pavlath, G.K. Myoblast fusion: Lessons from flies and mice. Development 2012, 139, 641–656. [Google Scholar] [CrossRef]
- Bryson-Richardson, R.J.; Currie, P.D. The genetics of vertebrate myogenesis. Nat. Rev. Genet. 2008, 9, 632–646. [Google Scholar] [CrossRef]
- Rehfeldt, C.; Fiedler, I.; Dietl, G.; Ender, K. Myogenesis and postnatal skeletal muscle cell growth as influenced by selection. Livest. Prod. Sci. 2000, 66, 177–188. [Google Scholar] [CrossRef]
- Bult, C.J.; Blake, J.A.; Smith, C.L.; Kadin, J.A.; Richardson, J.E.; Mouse Genome Database, G. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019, 47, D801–D806. [Google Scholar] [CrossRef]
- Galbiati, F.; Engelman, J.A.; Volonte, D.; Zhang, X.L.; Minetti, C.; Li, M.; Hou, H.; Kneitz, B.; Edelmann, W.; Lisanti, M.P. Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophin-glycoprotein complex, and t-tubule abnormalities. J. Biol. Chem. 2001, 276, 21425–21433. [Google Scholar] [CrossRef]
- Liu, M.; Li, B.; Shi, T.; Huang, Y.; Liu, G.E.; Lan, X.; Lei, C.; Chen, H. Copy number variation of bovine SHH gene is associated with body conformation traits in Chinese beef cattle. J. Appl. Genet. 2019, 60, 199–207. [Google Scholar] [CrossRef]
- Stearns, F.W. One hundred years of pleiotropy: A retrospective. Genetics 2010, 186, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, R.T.; Niehrs, C. Fibroblast growth factor signaling during early vertebrate development. Endocr. Rev. 2005, 26, 63–77. [Google Scholar] [CrossRef]
- McCarthy, N.; Sidik, A.; Bertrand, J.Y.; Eberhart, J.K. An Fgf-Shh signaling hierarchy regulates early specification of the zebrafish skull. Dev. Biol. 2016, 415, 261–277. [Google Scholar] [CrossRef]
- Boulet, A.M.; Capecchi, M.R. Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev. Biol. 2012, 371, 235–245. [Google Scholar]
- Kratochwil, K.; Galceran, J.; Tontsch, S.; Roth, W.; Grosschedl, R. FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1−/− mice. Genes Dev. 2002, 16, 3173–3185. [Google Scholar] [PubMed]
- Benoit, B.; Meugnier, E.; Castelli, M.; Chanon, S.; Vieille-Marchiset, A.; Durand, C.; Bendridi, N.; Pesenti, S.; Monternier, P.A.; Durieux, A.C.; et al. Fibroblast growth factor 19 regulates skeletal muscle mass and ameliorates muscle wasting in mice. Nat. Med. 2017, 23, 990–996. [Google Scholar] [CrossRef] [PubMed]
| Population | Variance Components 1 (PSD) | ||||||
|---|---|---|---|---|---|---|---|
| DUC | 1377 (352) | 75 (28) | 8792 (251) | 1709 (378) | 0.16 (0.04) | 0.19 (0.04) | 0.03 (0.20) |
| Crossbred | 3410 (621) | 72 (40) | 9376 (433) | 3625 (988) | 0.36 (0.05) | 0.39 (0.08) | −0.15 (0.27) |
| Breed | Genetic Effect | DUC | |
|---|---|---|---|
| DGE 2 | SGE 3 | ||
| Crossbred | DGE | 0.48 (0.19) | 0.77 (0.12) |
| SGE | −0.53 (0.23) | −0.27 (0.25) | |
| Term | Adjusted p-Value 1 | Candidate Gene |
|---|---|---|
| Inner ear morphogenesis (GO:0042472) | <0.05 | INSIG1, SLC9A3R1, SOX9, USH1G |
| Regulation of protein acetylation (GO:1901983) | <0.05 | CAMK1, PAXIP1, SETD5 |
| Regulation of systemic arterial blood pressure (GO:0003073) | <0.05 | NAV2, OXTR, SOD2 |
| Response to gamma radiation (GO:0010332) | <0.05 | FANCD2, GTF2H5, SOD2 |
| Cytoplasmic microtubule organization (GO:0031122) | <0.05 | CAV3, EZR, KIF19 |
| Cell differentiation in spinal cord (GO:0021515) | <0.05 | DBX1, PAX7, SHH |
| Neuron fate commitment (GO:0048663) | <0.05 | DBX1, PAX7, SHH |
| Regulation of mesenchymal cell proliferation (GO:0010464) | <0.05 | SHH, SOX9, TGFBR2 |
| Regulation of morphogenesis of a branching structure (GO:0060688) | <0.05 | CAV3, SHH, SOX9 |
| Positive regulation of mesenchymal cell proliferation (GO:0002053) | <0.01 | SHH, SOX9, TGFBR2 |
| Pancreas development (GO:0031016) | <0.05 | SHH, SOX9, VHL |
| Positive regulation of muscle cell differentiation (GO:0051149) | <0.05 | CAMK1, CAV3, SHH |
| Trachea development (GO:0060438) | <0.01 | SHH, SOX9, TGFBR2 |
| Gland morphogenesis (GO:0022612) | <0.01 | CAV3, SHH, SLC9A3R1, SOX9, TGFBR2 |
| Lung morphogenesis (GO:0060425) | <0.05 | SHH, SOX9, TGFBR2 |
| Vasculogenesis (GO:0001570) | <0.05 | PAXIP1, SHH, TGFBR2 |
| Neural crest cell development (GO:0014032) | <0.05 | ERBB4, SHH, SOX9 |
| Term | Adjusted p-Value 1 | Candidate Gene |
|---|---|---|
| Regulation of protein acetylation (GO:1901983) | <0.05 | BRD7, CAMK1, SETD5 |
| Response to gamma radiation (GO:0010332) | <0.05 | FANCD2, GTF2H5, SOD2 |
| Response to interleukin-1 (GO:0070555) | <0.01 | CCL19, CCL21, SOX9 |
| Granulocyte migration (GO:0097530) | <0.05 | CCL19, CCL21, IL17RC, JAGN1 |
| Cellular response to interleukin-1 (GO:0071347) | <0.05 | CCL19, CCL21, SOX9 |
| Production of molecular mediator involved in inflammatory response (GO:0002532) | <0.05 | ADCY7, IL17RC, NOD2 |
| Cytokine production involved in inflammatory response (GO:0002534) | <0.01 | ADCY7, IL17RC, NOD2 |
| Regulation of cytokine production involved in inflammatory response (GO:1900015) | <0.01 | ADCY7, IL17RC, NOD2 |
| NIK/NF-kappaB signaling (GO:0038061) | <0.05 | CCL19, CYLD, MAS1, NOD2 |
| Regulation of NIK/NF-kappaB signaling (GO:1901222) | <0.05 | CCL19, CYLD, MAS1, NOD2 |
| Positive regulation of NIK/NF-kappaB signaling (GO:1901224) | <0.05 | CCL19, MAS1, NOD2 |
| Term | Adjusted p-Value 1 | Candidate Gene |
|---|---|---|
| Response to gamma radiation (GO:0010332) | <0.01 | FANCD2, GTF2H5, SOD2 |
| Positive regulation of muscle cell differentiation (GO:0051149) | <0.05 | CAMK1, CAV3, FOXP1 |
| Negative regulation of stress-activated protein kinase signaling cascade (GO:0070303) | <0.01 | AIDA, DUSP10, EZR |
| p38MAPK cascade (GO:0038066) | <0.01 | CAV3, DUSP10, EZR |
| Negative regulation of stress-activated MAPK cascade (GO:0032873) | <0.01 | AIDA, DUSP10, EZR |
| Negative regulation of MAP kinase activity (GO:0043407) | <0.05 | AIDA, CAV3, DUSP10 |
| Regulation of p38MAPK cascade (GO:1900744) | <0.01 | CAV3, DUSP10, EZR |
| Lymphocyte migration (GO:0072676) | <0.01 | CCL19, CCL21, MIA3 |
| Response to interleukin-1 (GO:0070555) | <0.05 | CCL19, CCL21, IRAK2 |
| Granulocyte migration (GO:0097530) | <0.01 | CCL19, CCL21, IL17RC, JAGN1 |
| Cellular response to interleukin-1 (GO:0071347) | <0.01 | CCL19, CCL21, IRAK2 |
| Regulation of leukocyte apoptotic process (GO:2000106) | <0.05 | CCL19, CCL21, TCP1, VHL |
| Negative regulation of leukocyte apoptotic process (GO:2000107) | <0.01 | CCL19, CCL21, VHL |
| Term | Adjusted p-Value 1 | Candidate Gene |
|---|---|---|
| Vasculogenesis (GO:0001570) | <0.01 | PAXIP1, SHH, TGFBR2 |
| Cranial skeletal system development (GO:1904888) | <0.01 | FGF4, INSIG1, TGFBR2 |
| Response to gamma radiation (GO:0010332) | <0.01 | FANCD2, GTF2H5, SOD2 |
| Positive regulation of muscle cell differentiation (GO:0051149) | <0.01 | CAMK1, CAV3, SHH |
| Mammary gland epithelium development (GO:0061180) | <0.01 | CAV3, CCND1, ERBB4 |
| Regulation of protein acetylation (GO:1901983) | <0.01 | CAMK1, PAXIP1, SETD5 |
| Neural crest cell development (GO:0014032) | <0.01 | ERBB4, FGF19, SHH |
| Neural crest cell migration (GO:0001755) | <0.01 | ERBB4, FGF19, SHH |
| Melanoma (KEGG:05218) | <0.01 | CCND1, FGF19, FGF3, FGF4 |
| Gastric cancer (KEGG:05226) | <0.01 | CCND1, FGF19, FGF3, FGF4, SHH, TGFBR2 |
| Response to fibroblast growth factor (GO:0071774) | <0.01 | FGF19, FGF3, FGF4 |
| Cellular response to fibroblast growth factor stimulus (GO:0044344) | <0.01 | FGF19, FGF3, FGF4 |
| Fibroblast growth factor receptor signaling pathway (GO:0008543) | <0.01 | FGF19, FGF3, FGF4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seong, H.-S.; Kim, Y.-S.; Sa, S.-J.; Jeong, Y.; Hong, J.-K.; Cho, E.-S. Genetic Parameter Estimation and Genome-Wide Association Analysis of Social Genetic Effects on Average Daily Gain in Purebreds and Crossbreds. Animals 2022, 12, 2300. https://doi.org/10.3390/ani12172300
Seong H-S, Kim Y-S, Sa S-J, Jeong Y, Hong J-K, Cho E-S. Genetic Parameter Estimation and Genome-Wide Association Analysis of Social Genetic Effects on Average Daily Gain in Purebreds and Crossbreds. Animals. 2022; 12(17):2300. https://doi.org/10.3390/ani12172300
Chicago/Turabian StyleSeong, Ha-Seung, Young-Sin Kim, Soo-Jin Sa, Yongdae Jeong, Joon-Ki Hong, and Eun-Seok Cho. 2022. "Genetic Parameter Estimation and Genome-Wide Association Analysis of Social Genetic Effects on Average Daily Gain in Purebreds and Crossbreds" Animals 12, no. 17: 2300. https://doi.org/10.3390/ani12172300
APA StyleSeong, H.-S., Kim, Y.-S., Sa, S.-J., Jeong, Y., Hong, J.-K., & Cho, E.-S. (2022). Genetic Parameter Estimation and Genome-Wide Association Analysis of Social Genetic Effects on Average Daily Gain in Purebreds and Crossbreds. Animals, 12(17), 2300. https://doi.org/10.3390/ani12172300





