Red Sea Bream Iridovirus (RSIV) Kinetics in Rock Bream (Oplegnathus fasciatus) at Various Fish-Rearing Seawater Temperatures
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Fish
2.2. Virus
2.3. Determination of RSIV Genome Copy Number
2.4. Iron Flocculation Method for RSIV Detection in Seawater
2.5. Experimental Infection at Different Rearing Water Temperatures and RSIV Concentrations
2.6. Rearing Water Temperature Shift after Experimental Viral Infection
2.7. Statistical Analysis
3. Results
3.1. RSIV Recovery Efficiency of the Iron Flocculation Method
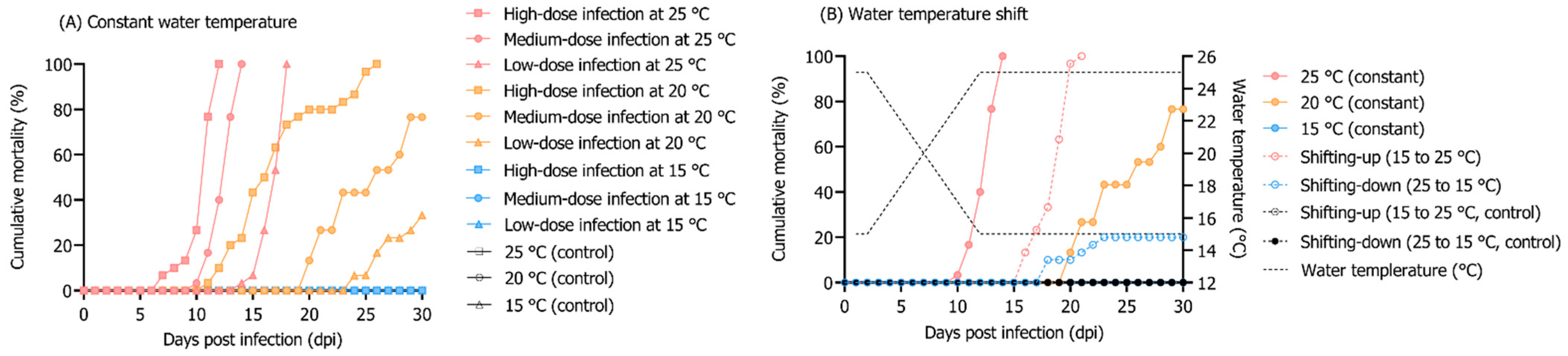
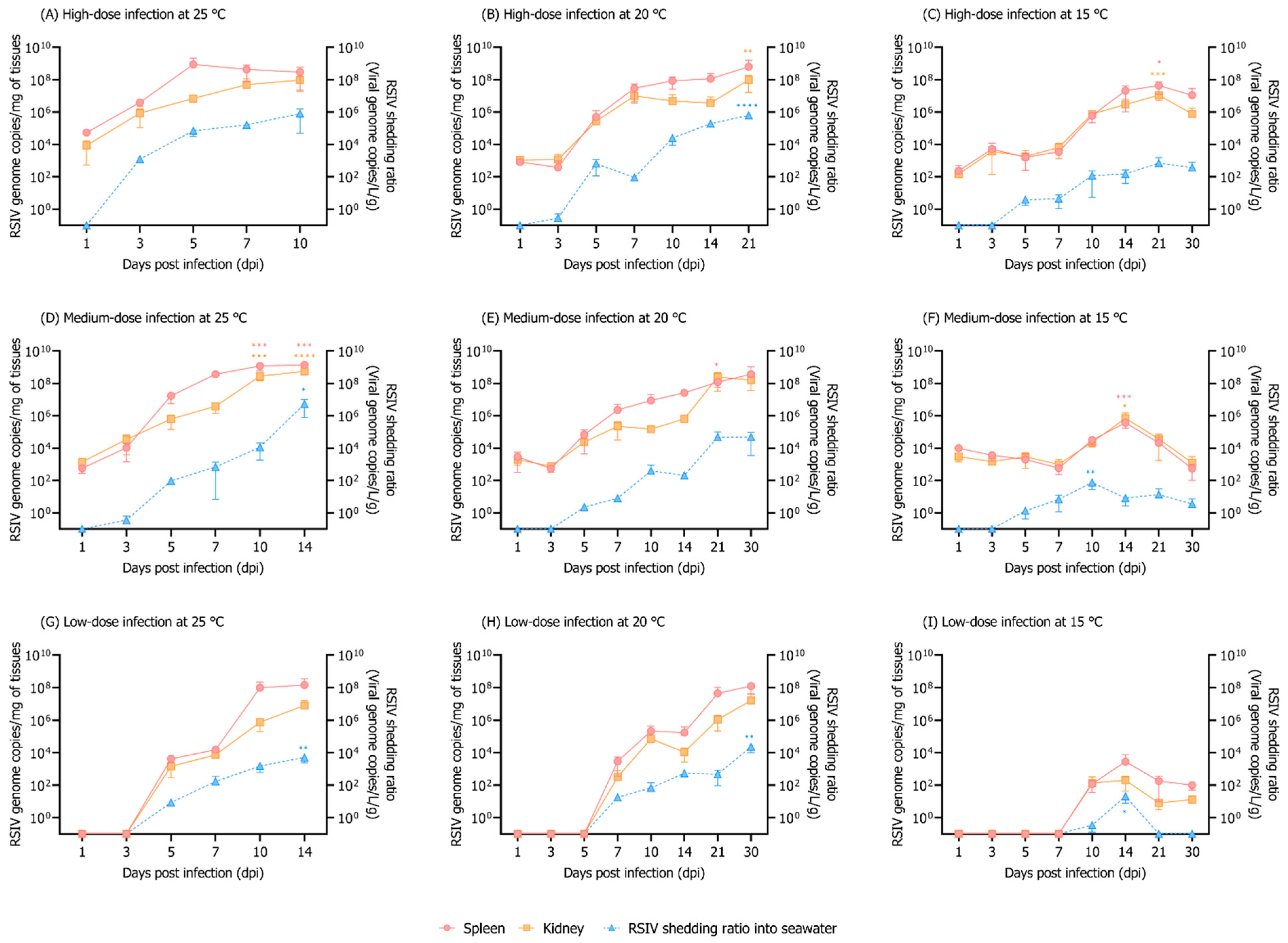
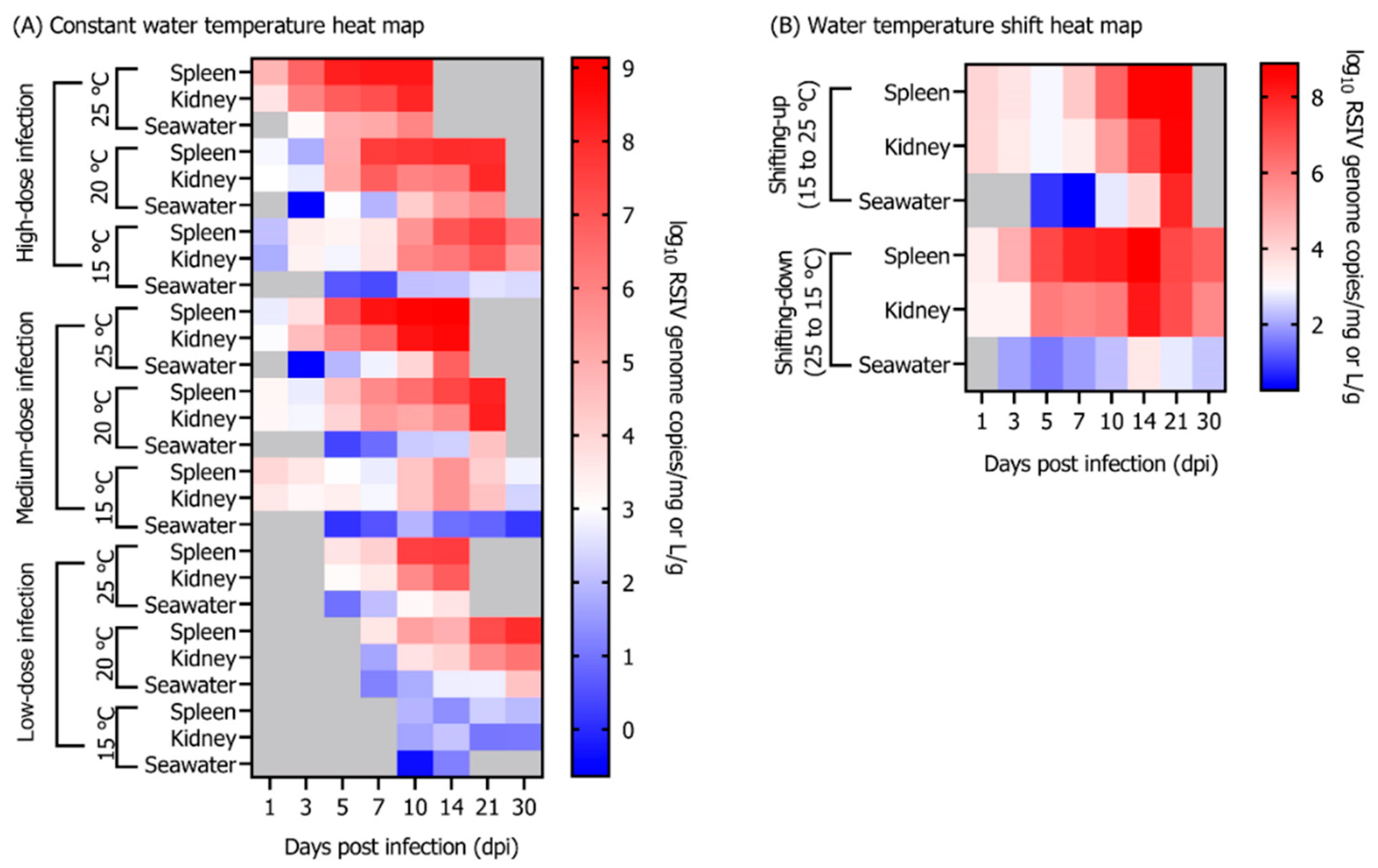
3.2. Influence of Different Rearing Water Temperatures and RSIV Concentration on RSIV Kinetics
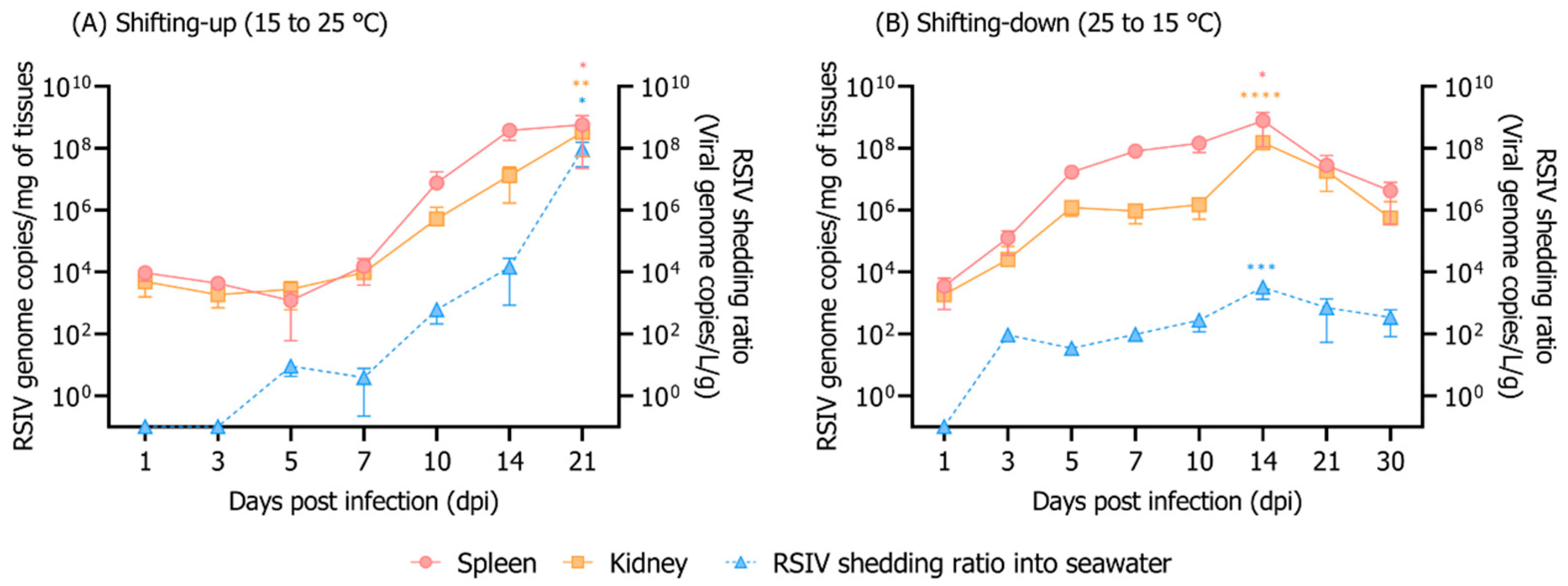
3.3. Influence of Rearing Water Temperature Shift on RSIV Kinetics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Inouye, K.; Yamano, K.; Maeno, Y.; Nakajima, K.; Matsuoka, M.; Wada, Y.; Sorimachi, M. Iridovirus infection of cultured red sea bream, Pagrus major. Fish Pathol. 1992, 27, 19–27. [Google Scholar] [CrossRef]
- Jancovich, J.K.; Chinchar, V.G.; Hyatt, A.; Miyazaki, T.; Williams, T.; Zhang, Q.Y. Iridoviridae. In Virus Taxonomy: 9th Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Academic Press: London, UK, 2012; pp. 207–210. [Google Scholar]
- He, J.G.; Wang, S.P.; Zeng, K.; Huang, Z.J.; Chan, S.M. Systemic disease caused by an iridovirus-like agent in cultured mandarinfish, Siniperca chuatsi (Basilewsky), in China. J. Fish Dis. 2000, 23, 219–222. [Google Scholar] [CrossRef]
- He, J.G.; Deng, M.; Weng, S.P.; Li, Z.; Zhou, S.Y.; Long, Q.X.; Wang, X.Z.; Chan, S.M. Complete genome analysis of the mandarin fish infectious spleen and kidney necrosis iridovirus. Virology 2001, 291, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.Y.; Wang, Y.G.; Yang, S.L.; Huang, J.; Wang, Q.Y. The first report of an iridovirus-like agent infection in farmed turbot, Scophthalmus maximus, in China. Aquaculture 2004, 236, 11–25. [Google Scholar] [CrossRef]
- Shi, C.Y.; Jia, K.T.; Yang, B.; Huang, J. Complete genome sequence of a Megalocytivirus (family Iridoviridae) associated with turbot mortality in China. Virol. J. 2010, 7, 159. [Google Scholar] [CrossRef] [PubMed]
- Gibson-Kueh, S.; Chee, D.; Chen, J.; Wang, Y.H.; Tay, S.; Leong, L.N.; Ng, M.L.; Jones, J.B.; Nicholls, P.K.; Ferguson, H.W. The pathology of ‘scale drop syndrome’ in Asian seabass, Lates calcarifer Bloch, a first description. J. Fish Dis. 2012, 35, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Kurita, J.; Nakajima, K. Megalocytiviruses. Viruses 2012, 4, 521–538. [Google Scholar] [CrossRef]
- OIE (World Organisation for Animal Health). Chapter 2.3.7. Red sea bream iridoviral disease. In Manual of Diagnostic Tests for Aquatic Animals; OIE: Paris, France, 2021. [Google Scholar]
- Sohn, S.G.; Choi, D.L.; Do, J.W.; Hwang, J.Y.; Park, J.W. Mass mortalities of cultured striped beakperch, Oplegnathus fasciatus by iridoviral infection. J. Fish Pathol. 2000, 13, 121–127. [Google Scholar]
- Kim, Y.J.; Jung, S.J.; Choi, T.J.; Kim, H.R.; Rajendran, K.V.; Oh, M.J. PCR amplification and sequence analysis of irido-like virus infecting fish in Korea. J. Fish Dis. 2002, 25, 121–124. [Google Scholar] [CrossRef]
- Kawato, Y.; Ito, T.; Kamaishi, T.; Fujiwara, A.; Ototake, M.; Nakai, T.; Nakajima, K. Development of red sea bream iridovirus concentration method in seawater by iron flocculation. Aquaculture 2016, 450, 308–312. [Google Scholar] [CrossRef]
- Watanabe, R.A.; Fryer, J.L.; Rohovec, J.S. Molecular filtration for recovery of waterborne viruses of fish. Appl. Environ. Microbiol. 1988, 54, 1606–1609. [Google Scholar] [CrossRef] [PubMed]
- Grant, A.A.; Jakob, E.; Richard, J.; Garver, K.A. Concentration of infectious aquatic rhabdoviruses from freshwater and seawater using ultrafiltration. J. Aquat. Anim. Health 2011, 23, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Nerland, A.H.; Skaar, C.; Eriksen, T.B.; Bleie, H. Detection of nodavirus in seawater from rearing facilities for Atlantic halibut Hippoglossus hippoglossus larvae. Dis. Aquat. Org. 2007, 73, 201–205. [Google Scholar] [CrossRef]
- Batts, W.N.; Winton, J.R. Concentration of hematopoietic necrosis virus from water sample by tangential flow filtration and polyethylene glycol precipitation. Can. J. Fish. Aquat. Sci. 2002, 46, 964–968. [Google Scholar] [CrossRef]
- Haramoto, E.; Kitajima, M.; Katayama, H.; Ito, T.; Ohgaki, S. Development of virus concentration methods for detection of koi herpesvirus in water. J. Fish Dis. 2009, 32, 297–300. [Google Scholar] [CrossRef]
- Honjo, M.N.; Minamoto, T.; Matsui, K.; Uchii, K.; Yamanaka, H.; Suzuki, A.A.; Kohmatsu, Y.; Iida, T.; Kawabata, Z. Quantification of cyprinid herpesvirus 3 in environmental water by using an external standard virus. Appl. Environ. Microbiol. 2010, 76, 161–168. [Google Scholar] [CrossRef]
- Ito, T.; Yoshiura, Y.; Kamaishi, T.; Nakajima, K. Dynamics of virus titer and quantity of red sea bream iridovirus DNA using natural and autoclaved artificial sea water under different water temperatures. Bull. Eur. Assoc. Fish Pathol. 2013, 33, 118–125. [Google Scholar]
- Bly, J.E.; Clem, L.W. Temperature and teleost immune functions. Fish Shellfish Immunol. 1992, 2, 159–171. [Google Scholar] [CrossRef]
- Alcorn, S.W.; Murray, A.L.; Pascho, R.J. Effects of rearing temperature on immune functions in sockeye salmon (Oncorhynchus nerka). Fish Shellfish Immunol. 2002, 12, 303–334. [Google Scholar] [CrossRef]
- Jung, M.H.; Jung, S.J.; Vinay, T.N.; Nikapitiya, C.; Kim, J.O.; Lee, J.H.; Lee, J.; Oh, M.J. Effects of water temperature on mortality in Megalocytivirus-infected rock bream Oplegnathus fasciatus (Temminck et Schlegel) and development of protective immunity. J. Fish. Dis. 2015, 38, 729–737. [Google Scholar] [CrossRef]
- Jun, L.J.; Jeong, J.B.; Kim, J.H.; Nam, J.H.; Shin, K.W.; Kim, J.K.; Kang, J.C.; Jeong, H.D. Influence of temperature shifts on the onset and development of red sea bream iridoviral disease in rock bream Oplegnathus fasciatus. Dis. Aquat. Org. 2009, 84, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.I.; Hwang, S.D.; Cho, M.Y.; Jung, S.H.; Kim, Y.C.; Jeong, H.D. A natural infection by the red sea bream iridovirus-type Megalocytivirus in the golden mandarin fish Siniperca scherzeri. J. Fish. Dis. 2018, 41, 1229–1233. [Google Scholar] [CrossRef] [PubMed]
- Snow, M.; Cunningham, C.O. Virulence and nucleotide sequence analysis of marine viral haemorrhagic septicaemia virus following in vivo passage in rainbow trout Onchorhynchus mykiss. Dis. Aquat. Organ. 2000, 42, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Mohr, P.G.; Moody, N.J.; Williams, L.M.; Hoad, J.; Cummins, D.M.; Davies, K.R.; StJ Crane, M. Molecular confirmation of infectious spleen and kidney necrosis virus (ISKNV) in farmed and imported ornamental fish in Australia. Dis. Aquat. Organ. 2015, 116, 103–110. [Google Scholar] [CrossRef]
- Godornes, C.; Leader, B.T.; Molini, B.J.; Centurion-Lara, A.; Lukehart, S.A. Quantitation of rabbit cytokine mRNA by real-time RT-PCR. Cytokine 2007, 38, 1–7. [Google Scholar] [CrossRef]
- He, J.G.; Zeng, K.; Weng, S.P.; Chan, S.M. Experimental transmission, pathogenicity and physical-chemical properties of infectious spleen and kidney necrosis virus (ISKNV). Aquaculture 2002, 204, 11–24. [Google Scholar] [CrossRef]
- Min, J.G.; Jeong, Y.J.; Jeong, M.A.; Kim, J.O.; Hwang, J.Y.; Kwon, M.G.; Kim, K.I. Experimental transmission of red sea bream iridovirus (RSIV) between rock bream (Oplegnathus fasciatus) and rockfish (Sebastes schlegelii). J. Fish Pathol. 2021, 34, 1–7. [Google Scholar]
- Graham, D.A.; Frost, P.; McLaughlin, K.; Rowley, H.M.; Gabestad, I.; Gordon, A.; McLoughlin, M.F. A comparative study of marine salmonid alphavirus subtypes 1–6 using an experimental cohabitation challenge model. J. Fish Dis. 2011, 34, 273–286. [Google Scholar] [CrossRef]
- Jarungsriapisit, J.; Moore, L.J.; Taranger, G.L.; Nilsen, T.O.; Morton, H.C.; Fiksdal, I.U.; Stefansson, S.; Fjelldal, P.G.; Evensen, Ø.; Patel, S. Atlantic salmon (Salmo salar L.) post-smolts challenged two or nine weeks after seawater-transfer show differences in their susceptibility to salmonid alphavirus subtype 3 (SAV3). Virol. J. 2016, 13, 66. [Google Scholar] [CrossRef][Green Version]
- Urquhart, K.; Murray, A.G.; Gregory, A.; O’Dea, M.; Munro, L.A.; Smail, D.A.; Shanks, A.M.; Raynard, R.S. Estimation of infectious dose and viral shedding rates for infectious pancreatic necrosis virus in Atlantic salmon, Salmo salar L., post-smolts. J. Fish Dis. 2008, 31, 879–887. [Google Scholar] [CrossRef]
- Oh, S.Y.; Nishizawa, T. Establishment of rock bream Oplegnathus fasciatus embryo (RoBE-4) cells with cytolytic infection of red seabream iridovirus (RSIV). J. Virol. Methods 2016, 238, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.H.; Nikapitiya, C.; Vinay, T.N.; Lee, J.H.; Jung, S.J. Rock bream iridovirus (RBIV) replication in rock bream (Oplegnathus fasciatus) exposed for different time periods to susceptible water temperatures. Fish Shellfish Immunol. 2017, 70, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Andersen, L.; Hodneland, K.; Nylund, A. No influence of oxygen levels on pathogenesis and virus shedding in Salmonid alphavirus (SAV)-challenged Atlantic salmon (Salmo salar L.). Virol. J. 2010, 7, 198. [Google Scholar] [CrossRef] [PubMed]
- Kawato, Y.; Mekata, T.; Inada, M.; Ito, T. Application of Environmental DNA for Monitoring Red Sea Bream Iridovirus at a Fish Farm. Microbiol. Spectr. 2021, 9, e00796-21. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.-H.; Choi, K.-M.; Joo, M.-S.; Kang, G.; Woo, W.-S.; Sohn, M.-Y.; Son, H.-J.; Kwon, M.-G.; Kim, J.-O.; Kim, D.-H.; et al. Red Sea Bream Iridovirus (RSIV) Kinetics in Rock Bream (Oplegnathus fasciatus) at Various Fish-Rearing Seawater Temperatures. Animals 2022, 12, 1978. https://doi.org/10.3390/ani12151978
Kim K-H, Choi K-M, Joo M-S, Kang G, Woo W-S, Sohn M-Y, Son H-J, Kwon M-G, Kim J-O, Kim D-H, et al. Red Sea Bream Iridovirus (RSIV) Kinetics in Rock Bream (Oplegnathus fasciatus) at Various Fish-Rearing Seawater Temperatures. Animals. 2022; 12(15):1978. https://doi.org/10.3390/ani12151978
Chicago/Turabian StyleKim, Kyung-Ho, Kwang-Min Choi, Min-Soo Joo, Gyoungsik Kang, Won-Sik Woo, Min-Young Sohn, Ha-Jeong Son, Mun-Gyeong Kwon, Jae-Ok Kim, Do-Hyung Kim, and et al. 2022. "Red Sea Bream Iridovirus (RSIV) Kinetics in Rock Bream (Oplegnathus fasciatus) at Various Fish-Rearing Seawater Temperatures" Animals 12, no. 15: 1978. https://doi.org/10.3390/ani12151978
APA StyleKim, K.-H., Choi, K.-M., Joo, M.-S., Kang, G., Woo, W.-S., Sohn, M.-Y., Son, H.-J., Kwon, M.-G., Kim, J.-O., Kim, D.-H., & Park, C.-I. (2022). Red Sea Bream Iridovirus (RSIV) Kinetics in Rock Bream (Oplegnathus fasciatus) at Various Fish-Rearing Seawater Temperatures. Animals, 12(15), 1978. https://doi.org/10.3390/ani12151978






