Insights into the Evolution of Aphid Mitogenome Features from New Data and Comparative Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Isolation
2.2. Mitogenome Sequencing, Assembly and Annotation
2.3. Phylogenetic Analysis and Evolutionary Pattern of Tandem Repeats
2.4. Gene Arrangement Analysis
3. Results
3.1. General Organization of Three Aphid Mitogenomes
| Subfamily | Tribe | Species | Length (bp) | Accession Number | Reference |
|---|---|---|---|---|---|
| Outgroup | Adelges tsugae | 16,056 | MT263947 | [43] | |
| Lachninae | Stomaphidini | Stomaphis sinisalicis | 17,109 | NC_053790 | [22] |
| Tuberolachnini | Nippolachnus piri | 17,033 | OL069343 | This study | |
| Hormaphidinae | Cerataphidini | Ceratovacuna keduensis | 16,138 | OL069341 | This study |
| Cerataphidini | Pseudoregma bambucicola | 16,632 | NC_044640 | [25] | |
| Cerataphidini | Pseudoregma panicola | 16,059 | OL069342 | This study | |
| Hormaphidini | Hamamelistes spinosus | 15,089 | MT010853 | [44] | |
| Hormaphidini | Hormaphis betulae | 15,088 | NC_029495 | [45] | |
| Nipponaphidini | Schizoneuraphis gallarum | 14,990 | NC_053624 | [46] | |
| Eriosomatinae | Eriosomatini | Eriosoma lanigerum | 15,640 | NC_033352 | [47] |
| Eriosomatini | Paracolopha morrisoni | 16,330 | NC_045103 | [48] | |
| Fordini | Baizongia pistaciae | 15,602 | NC_035314 | [23] | |
| Fordini | Floraphis choui | 15,308 | NC_035310 | [49] | |
| Fordini | Floraphis meitanensis | 15,301 | NC_035316 | [49] | |
| Fordini | Kaburagia rhusicola ensigallis | 16,164 | MF043984 | [23] | |
| Fordini | Kaburagia rhusicola ovatirhusicola | 16,184 | MF043985 | [23] | |
| Fordini | Kaburagia rhusicola ovogallis | 16,164 | MF043986 | [23] | |
| Fordini | Kaburagia rhusicola rhusicola | 16,159 | MF043987 | [23] | |
| Fordini | Meitanaphis elongallis | 16,191 | NC_035315 | [49] | |
| Fordini | Meitanaphis flavogallis | 16,150 | NC_035312 | [49] | |
| Fordini | Meitanaphis microgallis | 16,191 | NC_047419 | [50] | |
| Fordini | Melaphis rhois | 15,436 | NC_036065 | [51] | |
| Fordini | Nurudea ibofushi | 16,054 | NC_035311 | [23] | |
| Fordini | Nurudea shiraii | 15,389 | NC_035301 | [23] | |
| Fordini | Nurudea yanoniella | 15,858 | NC_035313 | [49] | |
| Fordini | Schlechtendalia chinensis | 16,047 | NC_032386 | [52] | |
| Fordini | Schlechtendalia peitan | 15,609 | NC_035302 | [23] | |
| Greenideinae | Cervaphidini | Cervaphis quercus | 15,272 | NC_024926 | [53] |
| Greenideini | Eutrichosiphum pasaniae | 16,500 | NC_054157 | [54] | |
| Greenideini | Greenidea ficicola | 17,361 | NC_048525 | [55] | |
| Greenideini | Greenidea psidii | 16,202 | NC_041198 | [56] | |
| Greenideini | Mollitrichosiphum tenuicorpus | 15,727 | NC_054348 | [57] | |
| Schoutedeniini | Schoutedenia ralumensis | 16,051 | MT381994 | [58] | |
| Chaitophorinae | Chaitophorini | Periphyllus diacerivorus | 16,418 | MZ665537 | Direct submission |
| Calaphidinae | Panaphidini | Appendiseta robiniae | 15,049 | NC_042165 | [59] |
| Panaphidini | Therioaphis trifolii | 16,068 | MK766411 | [60] | |
| Aphidinae | Aphidini | Aphis aurantii | 15,469 | MN397939 | [61] |
| Aphidini | Aphis citricidus | 16,763 | NC_043903 | [24] | |
| Aphidini | Aphis craccivora | 15,308 | NC_031387 | [62] | |
| Aphidini | Aphis fabae mordvilkoi | 15,346 | NC_039988 | [59] | |
| Aphidini | Aphis glycines | 17,954 | NC_045236 | [63] | |
| Aphidini | Aphis gossypii | 15,869 | NC_024581 | [64] | |
| Aphidini | Aphis spiraecola | 16,500 | NC_053819 | [65] | |
| Aphidini | Hyalopterus pruni | 15,410 | NC_050904 | [66] | |
| Aphidini | Melanaphis sacchari | 15,111 | MW811104 | Direct submission | |
| Aphidini | Rhopalosiphum nymphaeae | 15,594 | NC_046740 | [67] | |
| Aphidini | Schizaphis graminum | 15,721 | NC_006158 | [11] | |
| Macrosiphini | Acyrthosiphon pisum | 16,971 | NC_011594 | Direct submission | |
| Macrosiphini | Brevicoryne brassicae | 15,927 | NC_056270 | [68] | |
| Macrosiphini | Cavariella salicicola | 16,317 | NC_022682 | [42] | |
| Macrosiphini | Chaetosiphon fragaefolii | 16,108 | LC590896 | [69] | |
| Macrosiphini | Diuraphis noxia | 15,784 | NC_022727 | [70] | |
| Macrosiphini | Indomegoura indica | 15,220 | NC_045897 | [71] | |
| Macrosiphini | Myzus persicae | 17,382 | NC_029727 | [59] | |
| Macrosiphini | Neotoxoptera formosana | 15,642 | MW534268 | [72] | |
| Macrosiphini | Sitobion avenae | 15,180 | NC_024683 | [73] | |
| Macrosiphini | Uroleucon erigeronense | 15,691 | MZ695840 | [74] |
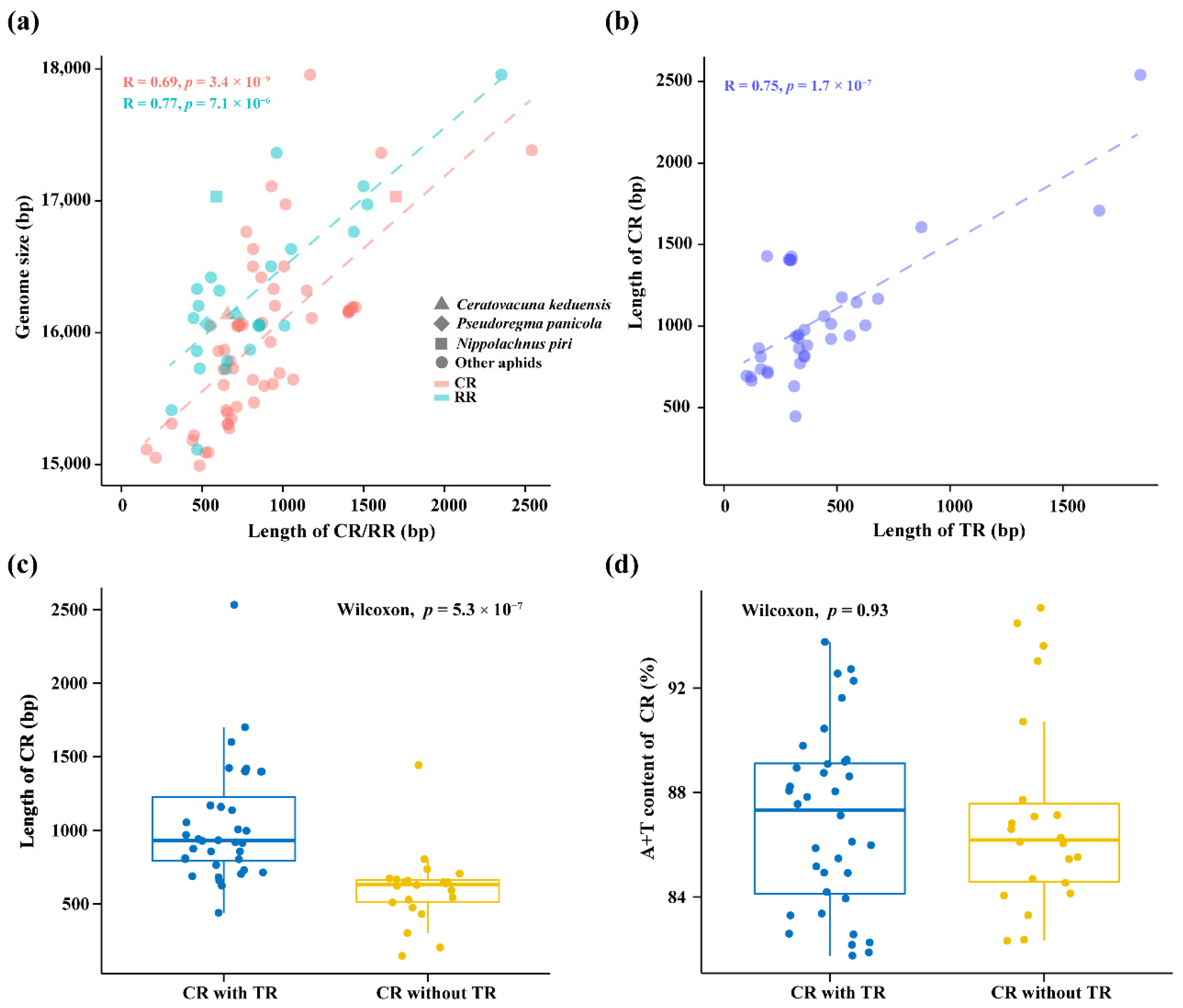
3.2. Comparative Analysis of Non-Coding Regions
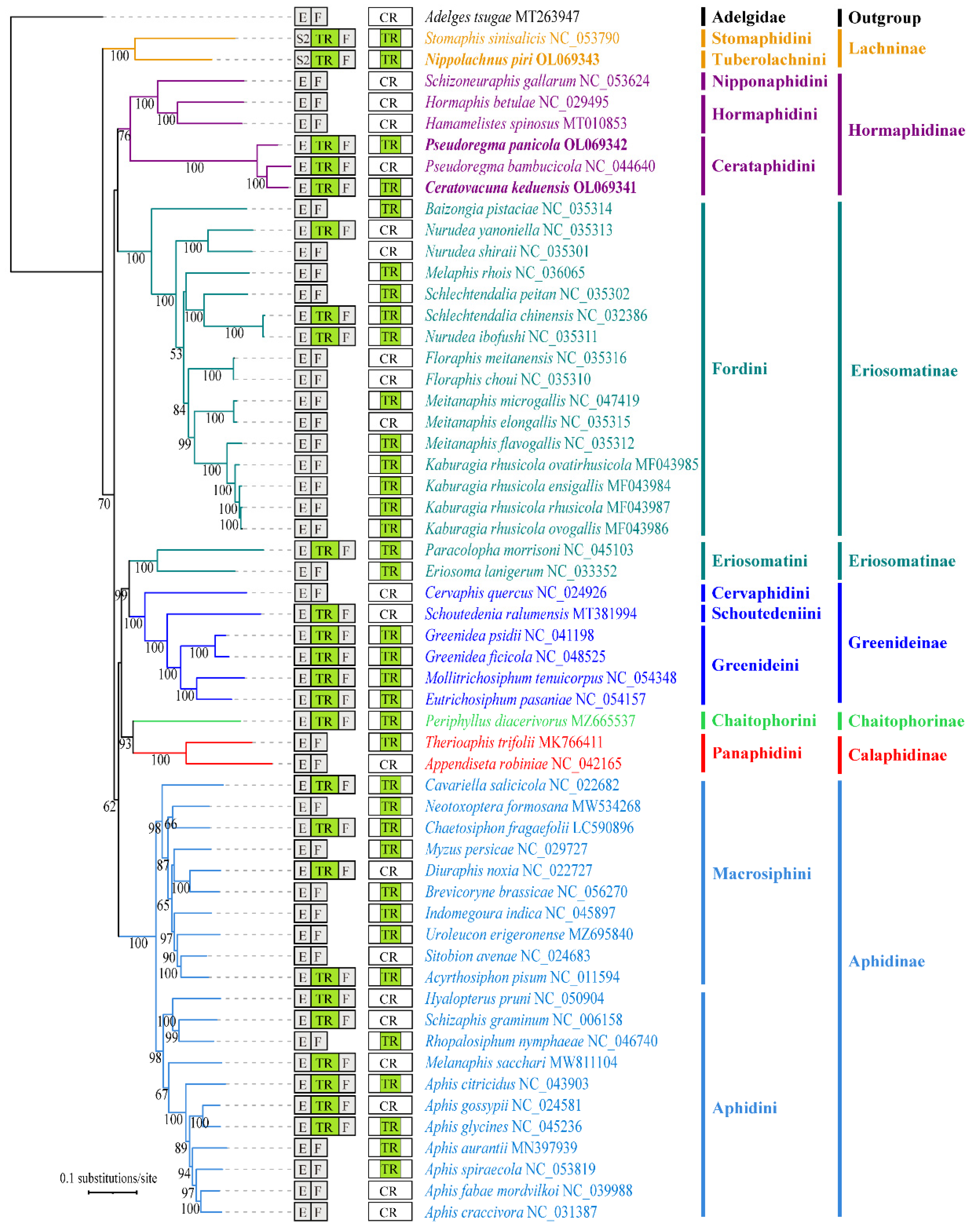
3.3. Gene Rearrangement of Aphid Mitogenomes
4. Discussion
4.1. Effect of Non-Coding Regions on Mitogenome Size Variation in Aphids
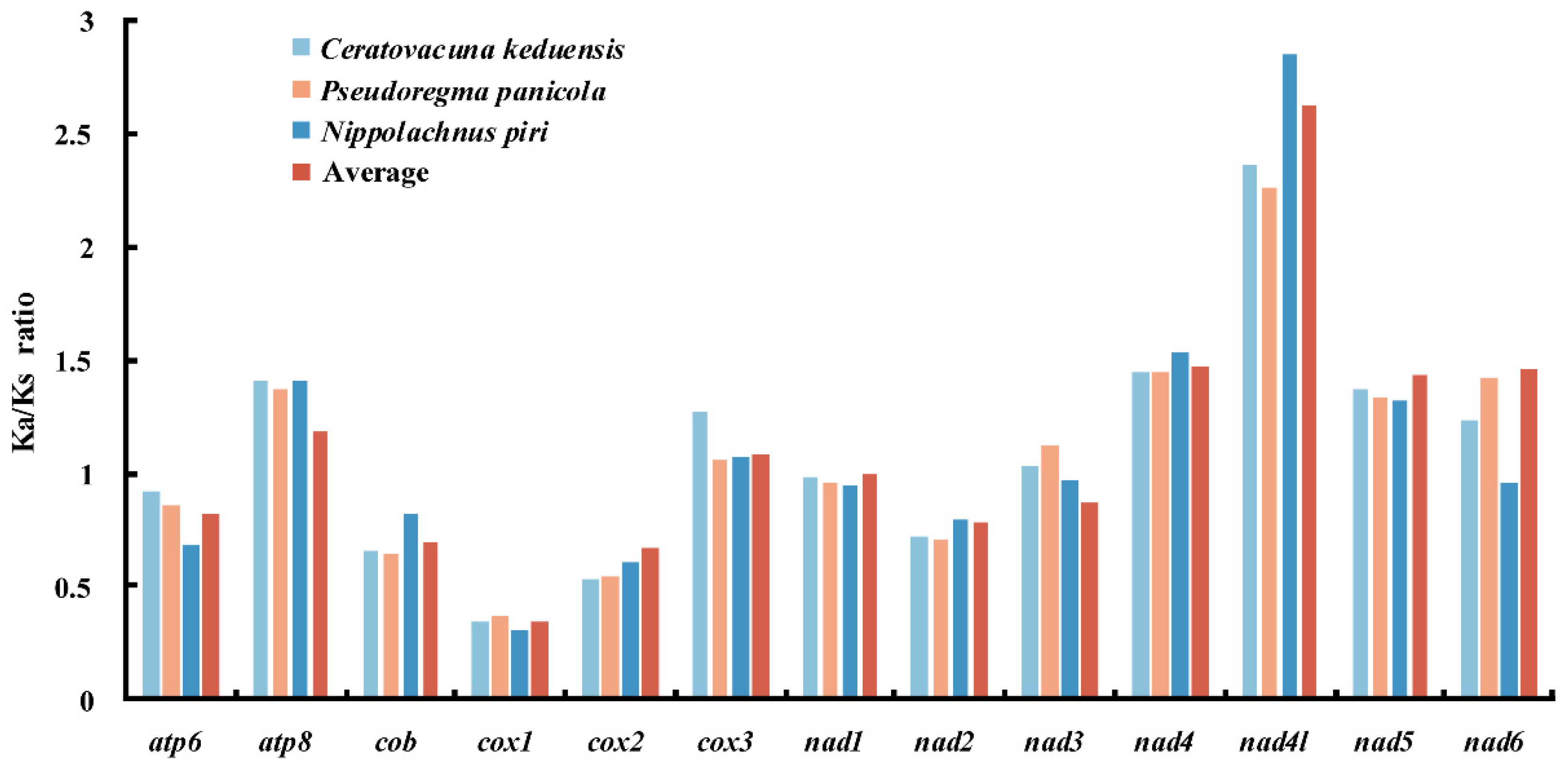
4.2. Phylogenetic Patterns of Tandem Repeats in Aphid Mitogenomes
4.3. Evolution of Gene Rearrangement in Aphid Mitogenomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Newmeyer, D.D.; Ferguson-Miller, S. Mitochondria: Releasing power for life and unleashing the machineries of death. Cell 2003, 112, 481–490. [Google Scholar] [CrossRef]
- Burger, G.; Gray, M.W.; Lang, B.F. Mitochondrial genomes: Anything goes. Trends Genet. 2003, 19, 709–716. [Google Scholar] [CrossRef]
- Boore, J.L.; Lavrov, D.V.; Brown, W.M. Gene translocation links insects and crustaceans. Nature 1998, 392, 667–668. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. Phylogenetic position of the Pentastomida and (pan)crustacean relationships. Proc. R. Soc. Lond. 2004, 271, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Aydemir, M.N.; Korkmaz, E.M. Comparative mitogenomics of Hymenoptera reveals evolutionary differences in structure and composition. Int. J. Biol. Macromol. 2020, 144, 460–472. [Google Scholar] [CrossRef] [PubMed]
- Mark, D.; Cameron, S.L.; Dowavic, J.I.; Austin, A.D.; Whiting, M.F. Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol. Biol. Evol. 2009, 26, 1607–1617. [Google Scholar] [CrossRef]
- Tyagi, K.; Chakraborty, R.; Cameron, S.L.; Sweet, A.D.; Kumar, V. Rearrangement and evolution of mitochondrial genomes in Thysanoptera (Insecta). Sci. Rep. 2020, 10, 695. [Google Scholar] [CrossRef]
- Yoshizawa, K.; Johnson, K.P.; Sweet, A.D.; Yao, I.; Ferreira, R.L.; Cameron, S.L. Mitochondrial phylogenomics and genome rearrangements in the barklice (Insecta: Psocodea). Mol. Phylogenet. Evol. 2018, 119, 118–127. [Google Scholar] [CrossRef]
- Covacin, C.; Shao, R.; Cameron, S.L.; Barker, S.C. Extraordinary number of gene rearrangements in the mitochondrial genomes of lice (Phthiraptera: Insecta). Insect Mol. Biol. 2006, 15, 63–68. [Google Scholar] [CrossRef]
- Sweet, A.D.; Johnson, K.P.; Cao, Y.; de Moya, R.S.; Skinner, R.K.; Tan, M.; Herrera, S.V.; Cameron, S.L. Structure, gene order, and nucleotide composition of mitochondrial genomes in parasitic lice from Amblycera. Gene 2021, 768, 145312. [Google Scholar] [CrossRef]
- Thao, M.L.; Baumann, L.; Baumann, P. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha). BMC Evol. Biol. 2004, 4, 25. [Google Scholar] [CrossRef]
- Boore, J.L. The Duplication/Random Loss Model for Gene Rearrangement Exemplified by Mitochondrial Genomes of Deuterostome Animals; Springer: Dordrecht, The Netherlands, 2000; pp. 133–147. [Google Scholar]
- Lavrov, D.V.; Boore, J.L.; Brown, W.M. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: Duplication and nonrandom loss. Mol. Biol. Evol. 2002, 19, 163–169. [Google Scholar] [CrossRef]
- Lunt, D.H.; Hyman, B.C. Animal mitochondrial DNA recombination. Nature 1997, 387, 247. [Google Scholar] [CrossRef]
- Boore, J.L.; Collins, T.M.; Stanton, D.; Daehler, L.L.; Brown, W.M. Deducing the pattern of arthropod phylogeny from mitochondrial DNA rearrangements. Nature 1995, 376, 163–165. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Zhang, K.J.; Zhu, W.C.; Xia, R.; Zhang, Y.K.; Ding, X.L.; Liu, J.; Chen, D.S.; Du, Y.; Hong, X.Y. The complete mitochondrial genomes of two rice planthoppers, Nilaparvata lugens and Laodelphax striatellus: Conserved genome rearrangement in Delphacidae and discovery of new characteristics of atp8 and tRNA genes. BMC Genom. 2013, 14, 417. [Google Scholar] [CrossRef]
- Li, H.; Liu, H.; Shi, A.; Štys, P.; Zhou, X.; Cai, W. The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp. (Hemiptera: Enicocephalidae). PLoS ONE 2012, 7, e29419. [Google Scholar] [CrossRef]
- Hua, J.; Li, M.; Dong, P.; Cui, Y.; Xie, Q.; Bu, W. Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera). BMC Genom. 2008, 9, 610. [Google Scholar] [CrossRef]
- Liu, H.-L.; Chen, Q.-D.; Chen, S.; Pu, D.-Q.; Chen, Z.-T.; Liu, Y.-Y.; Liu, X. The highly rearranged mitochondrial genomes of three economically important scale insects and the mitochondrial phylogeny of Coccoidea (Hemiptera: Sternorrhyncha). PeerJ 2020, 8, e9932. [Google Scholar] [CrossRef]
- Lu, C.; Huang, X.; Deng, J. The challenge of Coccidae (Hemiptera: Coccoidea) mitochondrial genomes: The case of Saissetia coffeae with novel truncated tRNAs and gene rearrangements. Int. J. Biol. Macromol. 2020, 158, 854–864. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Q.; Lu, C.; Deng, J.; Huang, X. The first complete mitochondrial genome of Lachninae species and comparative genomics provide new insights into the evolution of gene rearrangement and the repeat region. Insects 2021, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Harris, A.J.; Dikow, R.B.; Ma, E.; Zhong, Y.; Wen, J. Another look at the phylogenetic relationships and intercontinental biogeography of eastern Asian-North American Rhus gall aphids (Hemiptera: Aphididae: Eriosomatinae): Evidence from mitogenome sequences via genome skimming. Mol. Phylogenet. Evol. 2017, 117, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.D.; Lang, N.; Tao, Y.; He, W.; Wang, J.J. The mitochondrial genome of the brown citrus aphid Aphis (Toxoptera) citricidus: Insights into the repeat regions in aphids and phylogenetic implications. Int. J. Biol. Macromol. 2019, 136, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Deng, J.; Liu, Q.; Huang, X. The mitochondrial genome of a social aphid, Pseudoregma bambucicola (Hemiptera: Aphididae: Hormaphidinae). Mitochondr. DNA B 2019, 4, 2100–2101. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse1, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; zu Siederdissen, C.H.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 37, 573–580. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Bernt, M.; Merkle, D.; Ramsch, K.; Fritzsch, G.; Perseke, M.; Bernhard, D.; Schlegel, M.; Stadler, P.F.; Middendorf, M. CREx: Inferring genomic rearrangements based on common intervals. Bioinformatics 2007, 23, 2957–2958. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecule of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, X.-L.; Qiao, G.-X. Comparative analysis of mitochondrial genomes of five aphid species (Hemiptera: Aphididae) and phylogenetic implications. PLoS ONE 2013, 8, e77511. [Google Scholar] [CrossRef]
- Yeh, H.-T.; Ko, C.-C.; Wu, L.-W. The first complete mitochondrial genome of Adelges tsugae Annand (Hemiptera: Adelgidae). Mitochondrial DNA B 2020, 5, 2288–2290. [Google Scholar] [CrossRef]
- Lu, Z.-H.; Liang, Y.-K.; Wen, J.; Ren, Z.-M. Complete mitochondrial genome of the witch-hazel leaf gall aphid Hamamelistes spinosus (Hemiptera: Aphididae: Hormaphidinae). Mitochondr. DNA B 2020, 5, 1388–1389. [Google Scholar] [CrossRef]
- Nong, X.; Liu, Y.; Wang, L.; Zhong, S.; Yu, X.; Xie, Y. Mitochondrial genome of Hormaphis betulae and its comparative analysis with Pseudoregma bambucicola (Hemiptera: Hormaphidinae). Mitochondr. DNA B 2020, 5, 906–907. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Li, C.; Qiao, G.X.; Chen, J. The complete mitochondrial genome of Schizoneuraphis gallarum van der Goot, 1917 (Hemiptera: Aphididae: Hormaphidinae). Mitochondr. DNA B 2021, 6, 2982–2983. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, L.; Liu, Y.; Chen, J.; Qiao, G. General methods to obtain and analyze the complete mitochondrial genome of aphid species: Eriosoma lanigerum (Hemiptera: Aphididae) as an example. Zool. Syst. 2016, 41, 123–132. [Google Scholar] [CrossRef]
- Lee, J.; Park, J.; Lee, H.; Park, J.; Lee, W. The complete mitochondrial genome of Paracolopha morrisoni (Baker, 1919) (Hemiptera: Aphididae). Mitochondr. DNA B 2019, 4, 3037–3039. [Google Scholar] [CrossRef]
- Ren, Z.; von Dohlen, C.D.; Harris, A.J.; Dikow, R.B.; Su, X.; Wen, J. Congruent phylogenetic relationships of Melaphidina aphids (Aphididae: Eriosomatinae: Fordini) according to nuclear and mitochondrial DNA data with taxonomic implications on generic limits. PLoS ONE 2019, 14, e0213181. [Google Scholar] [CrossRef]
- Liang, Y.-K.; Wen, J.; Ren, Z.-M. Complete mitochondrial genome of Rhus gall aphid Meitanaphis microgallis (Hemiptera: Aphididae: Eriosomatinae). Mitochondr. DNA B 2019, 4, 2363–2364. [Google Scholar] [CrossRef]
- Ren, Z.-M.; Wen, J. Complete mitochondrial genome of the North American Rhus gall aphid Melaphis rhois (Hemiptera: Aphididae: Eriosomatinae). Mitochondr. DNA B 2017, 2, 169–170. [Google Scholar] [CrossRef]
- Ren, Z.-M.; Bai, X.; Harris, A.J.; Wen, J. Complete mitochondrial genome of the Rhus gall aphid Schlechtendalia chinensis (Hemiptera: Aphididae: Eriosomatinae). Mitochondr. DNA B 2016, 1, 849–850. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, X.-L.; Qiao, G.-X. The complete mitochondrial genome of Cervaphis quercus (Insecta: Hemiptera: Aphididae: Greenideinae). Insect Sci. 2014, 21, 278–290. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jiang, L.Y.; Zhang, X.; Jing, C.; Qiao, G.X. The complete mitochondrial genome of Eutrichosiphum pasaniae (Okajima, 1908) (Hemiptera: Aphididae: Greenideinae). Mitochondr. DNA B 2020, 5, 3650–3651. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, H.; Deng, J.; Lin, X.; Huang, X. The complete mitochondrial genome of Greenidea ficicola (Hemiptera: Aphididae: Greenideinae), a pest of Ficus. Mitochondr. DNA B 2020, 5, 254–256. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.; Qin, M.; Jiang, L.Y.; Qiao, G.X. The mitochondrial genome of Greenidea psidii van der Goot (Hemiptera: Aphididae: Greenideinae) and comparisons with other Aphididae aphids. Int. J. Biol. Macromol. 2019, 122, 824–832. [Google Scholar] [CrossRef]
- Li, C.; Jiang, L.Y.; Qiao, G.X.; Chen, J. Complete mitochondrial genome of Mollitrichosiphum tenuicorpus (Okajima, 1908) (Hemiptera: Aphididae: Greenideinae). Mitochondr. DNA B 2021, 6, 361–362. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, L.; Zhang, X.; Qiao, G. The complete mitochondrial genome of Schoutedenia ralumensis Rübsaamen, 1905 (Hemiptera: Aphididae: Greenideinae). Mitochondr. DNA B 2020, 5, 2217–2218. [Google Scholar] [CrossRef]
- Voronova, N.V.; Levykina, S.; Warner, D.; Shulinski, R.; Bandarenka, Y.; Zhorov, D. Characteristic and variability of five complete aphid mitochondrial genomes: Aphis fabae mordvilkoi, Aphis craccivora, Myzus persicae, Therioaphis tenera and Appendiseta robiniae (Hemiptera; Sternorrhyncha; Aphididae). Int. J. Biol. Macromol. 2020, 149, 187–206. [Google Scholar] [CrossRef]
- Liu, X.; Wei, S.; He, J.; Song, F.; Cai, W. Complete mitochondrial genome of the spotted alfalfa aphid, Therioaphis trifolii (Hemipera: Aphididae). Mitochondr. DNA B 2019, 4, 3260–3261. [Google Scholar] [CrossRef]
- Pu, D.; Liu, C.; Liu, H.; Chen, Z.T.; Huang, Q. Complete mitochondrial genome of Sichuan’s population of Aphis aurantii (Hemiptera: Aphididae). Mitochondr. DNA B 2020, 5, 2119–2120. [Google Scholar] [CrossRef]
- Song, N.; Zhang, H.; Li, H.; Cai, W. All 37 mitochondrial genes of aphid Aphis craccivora obtained from transcriptome sequencing: Implications for the evolution of aphids. PLoS ONE 2016, 11, e0157857. [Google Scholar] [CrossRef]
- Song, H.; Donthu, R.K.; Hall, R.; Hon, L.; Weber, E.; Badger, J.H.; Giordano, R. Description of soybean aphid (Aphis glycines Matsumura) mitochondrial genome and comparative mitogenomics of Aphididae (Hemiptera: Sternorrhyncha). Int. J. Mol. Sci. 2019, 113, 103208. [Google Scholar] [CrossRef]
- Zhang, S.; Luo, J.; Wang, C.; Lv, L.; Li, C.; Jiang, W.; Cui, J.; Rajput, L.B. Complete mitochondrial genome of Aphis gossypii Glover (Hemiptera: Aphididae). Mitochondr. DNA A 2016, 27, 854–855. [Google Scholar] [CrossRef]
- Du, Y.; Liu, X.; Wang, Y.; Lu, Z. Characterization and phylogenetic analysis of the complete mitochondrial genome of Aphis spiraecola (Hemiptera: Aphididae). Mitochondr. DNA B 2019, 4, 3303–3304. [Google Scholar] [CrossRef]
- Liang, Y.; Du, Z.; Song, F.; He, J. The complete mitochondrial genome of the mealy plum aphid, Hyalopterus pruni (Hemiptera: Aphididae). Mitochondr. DNA B 2020, 5, 3685–3687. [Google Scholar] [CrossRef]
- Park, J.; Kim, Y.; Xi, H.; Park, J.; Lee, W. The complete mitochondrial genome of Rhopalosiphum nymphaeae (Linnaeus, 1761) (Hemiptera: Aphididae). Mitochondr. DNA B 2020, 5, 1613–1615. [Google Scholar] [CrossRef]
- Li, A.; Fang, Z.; Weng, Q. The complete mitochondrial genome of Brevicoryne brassicae (Hemiptera: Aphididae). Mitochondrial DNA B 2021, 6, 974–975. [Google Scholar] [CrossRef]
- Acosta, M.; Alcantar, D.; Alier-Reyes, I.; Alvarez, C.; Arroyo, C.B.; Calderon, D.; Cardenas, D.; Castro, A.R.; Companion, J.K.; Constante, C.; et al. The complete mitochondrial genome of the strawberry aphid Chaetosiphon fragaefolii Cockerell, 1901 (Hemiptera: Aphididae) from California, USA. Mitochondr. DNA B 2021, 6, 2373–2375. [Google Scholar] [CrossRef]
- Zhang, B.; Ma, C.; Edwards, O.; Fuller, S.; Kang, L. The mitochondrial genome of the Russian wheat aphid Diuraphis noxia: Large repetitive sequences between trnE and trnF in aphids. Gene 2014, 533, 253–260. [Google Scholar] [CrossRef]
- Hong, B.; Zhang, F.; Hu, Z.; Zhao, H. The complete mitochondrial genome of Indomegoura indica (Hemiptera: Aphididae). Mitochondr. DNA B 2019, 4, 882–883. [Google Scholar] [CrossRef]
- Song, Y.F.; Zhang, H.; Zeng, C.; Ye, S.; Yang, M.; Liu, J.F. Complete mitochondrial genome of Neotoxoptera formosana (Takahashi, 1921) (Hemiptera: Aphididae), with the phylogenetic analysis. Mitochondr. DNA B 2021, 6, 1706–1707. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zheng, J.; Liang, L.; Fuller, S.; Ma, C.-S. The complete mitochondrial genome of Sitobion avenae (Hemiptera: Aphididae). Mitochondr. DNA A 2016, 27, 945–946. [Google Scholar] [CrossRef]
- Park, J.; Lee, W. The complete mitochondrial genome of Uroleucon erigeronense (Thomas, 1878) (Hemiptera: Aphididae). Mitochondr. DNA B 2022, 7, 84–86. [Google Scholar] [CrossRef]
- Saito, S.; Tamura, K.; Aotsuka, T. Replication origin of mitochondrial DNA in insects. Genetics 2005, 171, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Crozier, R.H.; Crozier, Y.C. The mitochondrial genome of the honeybee Apis mellifera: Complete sequence and genome organization. Genetics 1993, 133, 97–117. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, J.; Jiang, L.-Y.; Qiao, G.-X. Hemipteran mitochondrial genomes: Features, structures and implications for phylogeny. Int. J. Mol. Sci. 2015, 16, 12382–12404. [Google Scholar] [CrossRef]
- Dowton, M.; Castro, L.R.; Austin, A.D. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: The examination of genome ‘morphology’. Invertebr. Syst. 2002, 16, 345–356. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, L.Y.; Qiao, G.X. A total-evidence phylogenetic analysis of Hormaphidinae (Hemiptera: Aphididae), with comments on the evolution of galls. Cladistics 2014, 30, 26–66. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.L.; Xiang-Yu, J.G.; Ren, S.S.; Zhang, R.L.; Zhang, Y.P.; Qiao, G.X. Molecular phylogeny and divergence times of Hormaphidinae (Hemiptera: Aphididae) indicate Late Cretaceous tribal diversification. Zool. J. Linn. Soc. 2012, 165, 73–87. [Google Scholar] [CrossRef][Green Version]
- Castro, L.R.; Austin, A.D.; Dowton, M. Contrasting rates of mitochondrial molecular evolution in parasitic Diptera and Hymenoptera. Mol. Biol. Evol. 2002, 19, 1100–1113. [Google Scholar] [CrossRef]
- Rui, C.; Favret, C.; Jiang, L.; Zhe, W.; Qiao, G. An aphid lineage maintains a bark-feeding niche while switching to and diversifying on conifers. Cladistics 2016, 32, 555–572. [Google Scholar] [CrossRef]
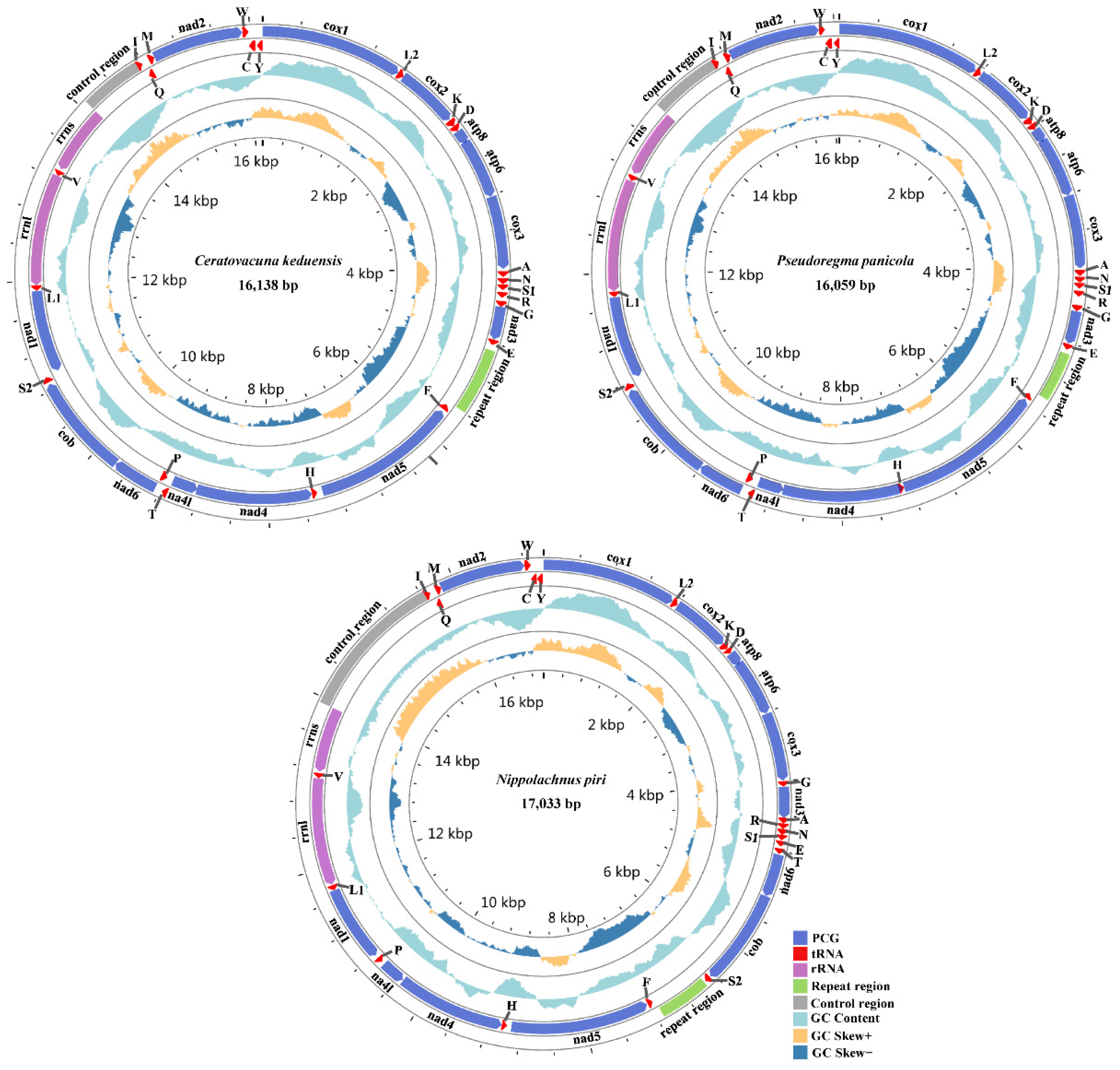

| Regions | Size (bp) | Nucleotides Composition (%) | AT-Skew | GC-Skew | |||||
|---|---|---|---|---|---|---|---|---|---|
| T | C | A | G | A + T | G + C | ||||
| Full genome | |||||||||
| C. keduensis | 16,138 | 39.84 | 9.64 | 45.15 | 5.36 | 84.99 | 15.00 | 0.062 | −0.285 |
| P. panicola | 16,059 | 39.82 | 9.66 | 45.15 | 5.37 | 84.97 | 15.03 | 0.063 | −0.285 |
| N. piri | 17,033 | 37.67 | 10.53 | 46.22 | 5.58 | 83.89 | 16.11 | 0.102 | −0.307 |
| PCGs 1 | |||||||||
| C. keduensis | 10,935 | 48.42 | 8.65 | 35.38 | 7.54 | 83.80 | 16.20 | −0.156 | −0.068 |
| P. panicola | 11,028 | 48.82 | 8.37 | 35.07 | 7.74 | 83.89 | 16.11 | −0.164 | −0.039 |
| N. piri | 10,917 | 47.16 | 9.21 | 35.36 | 8.27 | 82.51 | 17.49 | −0.143 | −0.054 |
| 1st codon | |||||||||
| C. keduensis | 3645 | 40.47 | 8.70 | 40.36 | 10.48 | 80.82 | 19.18 | −0.001 | 0.093 |
| P. panicola | 3676 | 40.72 | 8.79 | 39.96 | 10.53 | 80.69 | 19.31 | −0.009 | 0.090 |
| N. piri | 3639 | 39.19 | 8.71 | 41.08 | 11.02 | 80.27 | 19.73 | 0.024 | 0.117 |
| 2nd codon | |||||||||
| C. keduensis | 3645 | 53.58 | 13.31 | 22.66 | 10.45 | 76.24 | 23.76 | −0.406 | −0.120 |
| P. panicola | 3676 | 53.67 | 13.00 | 22.96 | 10.36 | 76.63 | 23.37 | −0.401 | −0.113 |
| N. piri | 3639 | 52.98 | 13.49 | 22.95 | 10.58 | 75.93 | 24.07 | −0.396 | −0.121 |
| 3rd codon | |||||||||
| C. keduensis | 3645 | 51.22 | 3.95 | 43.13 | 1.70 | 94.35 | 5.65 | −0.086 | −0.398 |
| P. panicola | 3676 | 52.07 | 3.32 | 42.27 | 2.34 | 94.34 | 5.66 | −0.104 | −0.173 |
| N. piri | 3639 | 49.30 | 5.44 | 42.04 | 3.22 | 91.34 | 8.66 | −0.079 | −0.257 |
| tRNAs | |||||||||
| C. keduensis | 1466 | 42.02 | 5.59 | 44.41 | 7.98 | 86.43 | 13.57 | 0.028 | 0.176 |
| P. panicola | 1481 | 41.93 | 5.74 | 44.23 | 8.10 | 86.16 | 13.84 | 0.027 | 0.171 |
| N. piri | 1486 | 41.05 | 5.52 | 45.63 | 7.81 | 86.68 | 13.32 | 0.053 | 0.172 |
| rRNAs | |||||||||
| C. keduensis | 2035 | 44.62 | 4.77 | 40.93 | 9.68 | 85.55 | 14.45 | 0.014 | 0.340 |
| P. panicola | 2033 | 44.61 | 4.82 | 41.12 | 9.44 | 85.74 | 14.26 | −0.041 | 0.246 |
| N. piri | 2075 | 45.59 | 4.72 | 39.08 | 10.60 | 84.67 | 15.33 | −0.077 | 0.384 |
| Repeat region | |||||||||
| C. keduensis | 715 | 43.22 | 10.91 | 42.10 | 3.78 | 85.31 | 14.69 | −0.013 | −0.486 |
| P. panicola | 528 | 43.37 | 11.93 | 40.91 | 3.79 | 84.28 | 15.72 | −0.029 | −0.518 |
| N. piri | 589 | 32.26 | 10.53 | 55.52 | 1.70 | 87.78 | 12.22 | 0.265 | −0.722 |
| Control region | |||||||||
| C. keduensis | 657 | 47.95 | 3.50 | 45.81 | 2.74 | 93.76 | 6.24 | −0.023 | −0.122 |
| P. panicola | 728 | 47.66 | 3.98 | 45.05 | 3.30 | 92.72 | 7.28 | −0.028 | −0.094 |
| N. piri | 1699 | 40.14 | 7.53 | 45.97 | 6.36 | 86.11 | 13.89 | 0.068 | −0.085 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Lu, C.; Liu, Q.; Zou, T.; Qiao, G.; Huang, X. Insights into the Evolution of Aphid Mitogenome Features from New Data and Comparative Analysis. Animals 2022, 12, 1970. https://doi.org/10.3390/ani12151970
Zhang H, Lu C, Liu Q, Zou T, Qiao G, Huang X. Insights into the Evolution of Aphid Mitogenome Features from New Data and Comparative Analysis. Animals. 2022; 12(15):1970. https://doi.org/10.3390/ani12151970
Chicago/Turabian StyleZhang, Hui, Congcong Lu, Qian Liu, Tianmin Zou, Gexia Qiao, and Xiaolei Huang. 2022. "Insights into the Evolution of Aphid Mitogenome Features from New Data and Comparative Analysis" Animals 12, no. 15: 1970. https://doi.org/10.3390/ani12151970
APA StyleZhang, H., Lu, C., Liu, Q., Zou, T., Qiao, G., & Huang, X. (2022). Insights into the Evolution of Aphid Mitogenome Features from New Data and Comparative Analysis. Animals, 12(15), 1970. https://doi.org/10.3390/ani12151970







