Molecular Cloning and Expression Analysis of Thyrotropin-Releasing Hormone, and Its Possible Role in Gonadal Differentiation in Rice Field eel Monopterus albus
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Fish and Collection of Samples
2.2. RNA Isolation and cDNA Synthesis
2.3. cDNA and Genomic Cloning of trh Gene
2.4. Bioinformatic Analysis of Sequences
2.5. Analysis of Quantitative Real-Time PCR (qRT-PCR)
2.6. Static Incubation Experiments
2.7. The Design of Intraperitoneal (IP) Injection Experiment
2.8. Serum Hormones Assays
2.9. Data Analysis
3. Results
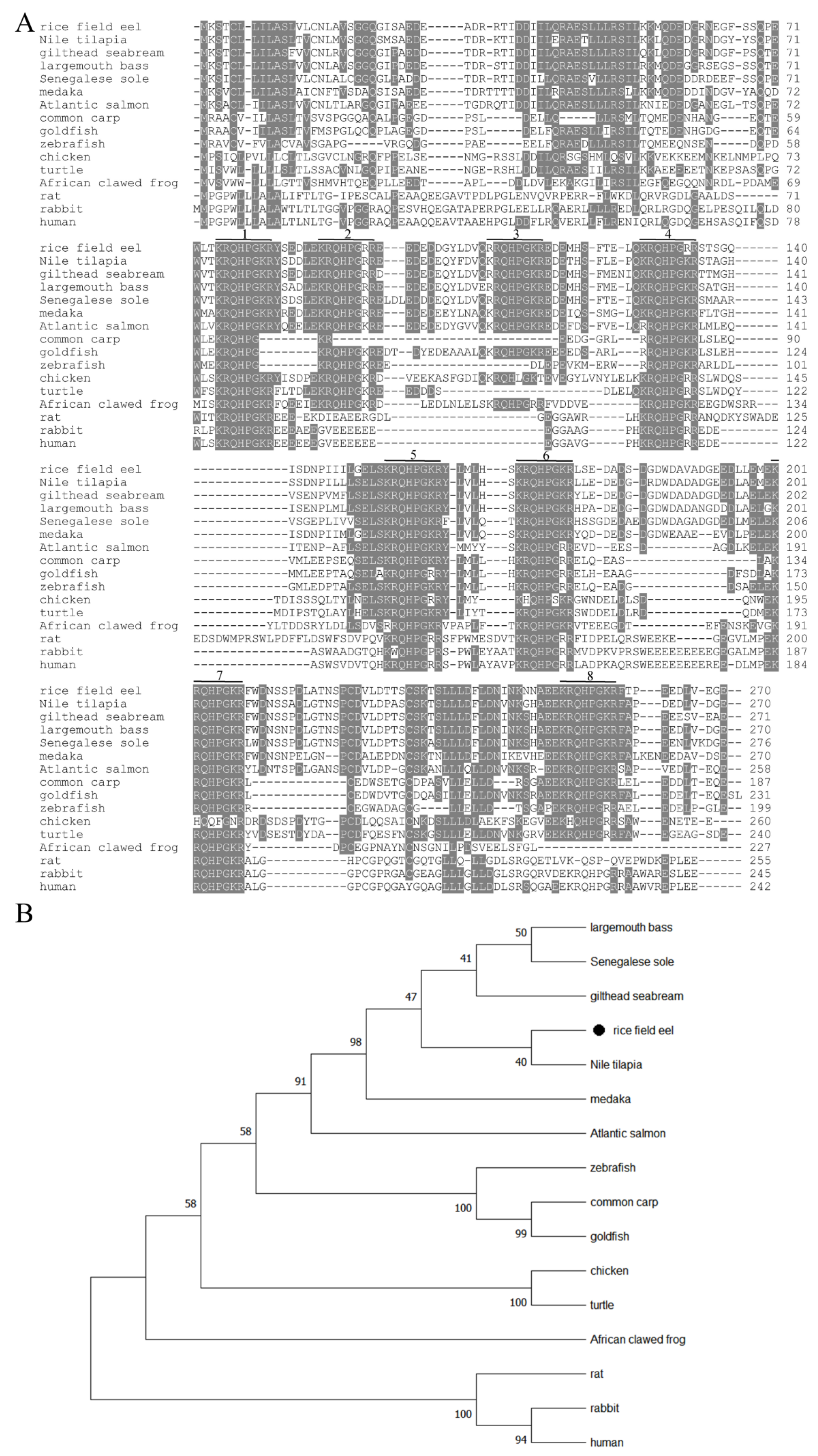
3.1. Molecular Characterization of trh
3.2. Sequence Alignment and Phylogenetic Tree Analysis
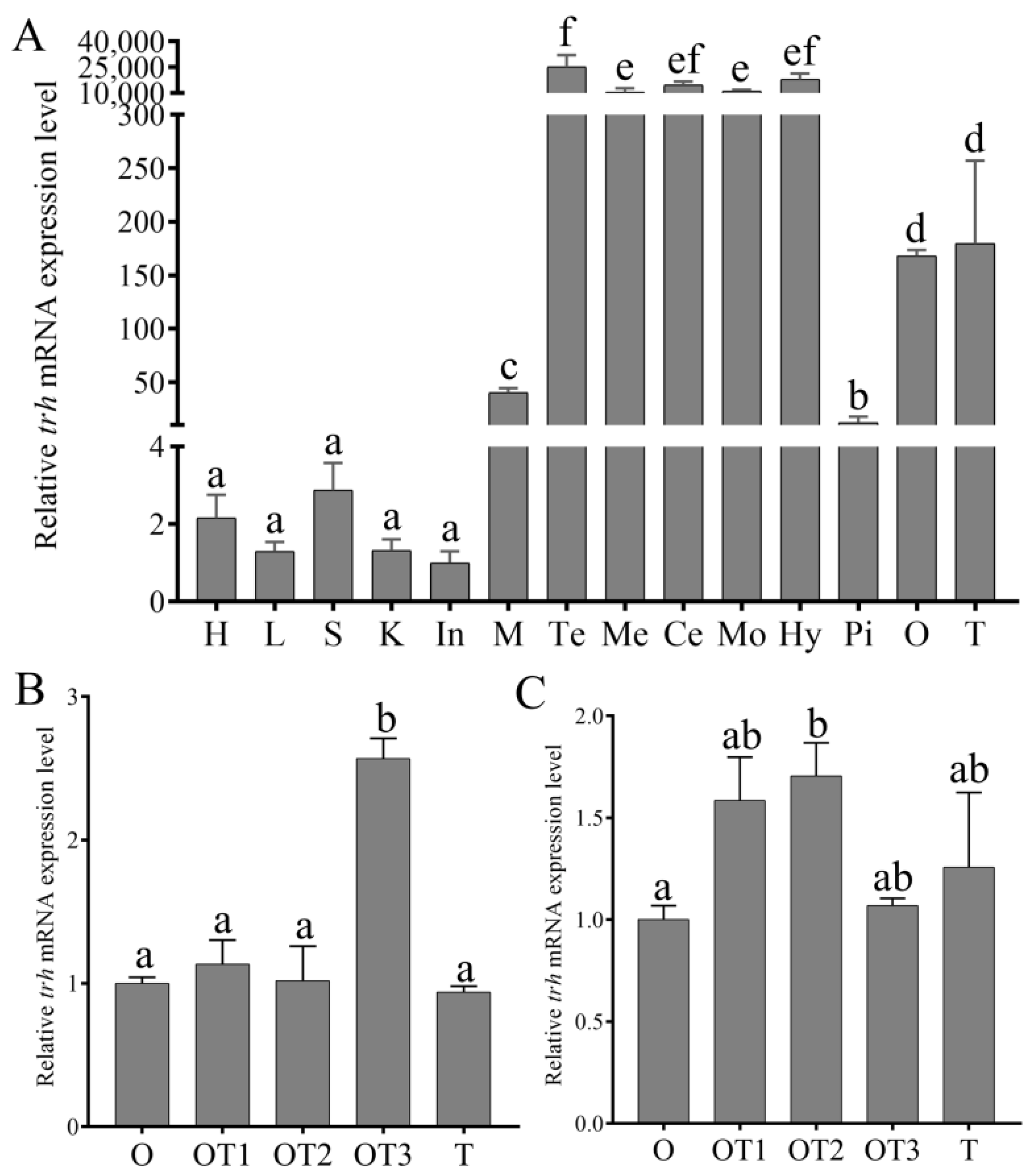
3.3. The Expression Pattern of trh mRNA in Various Tissues and during Sex Reversal
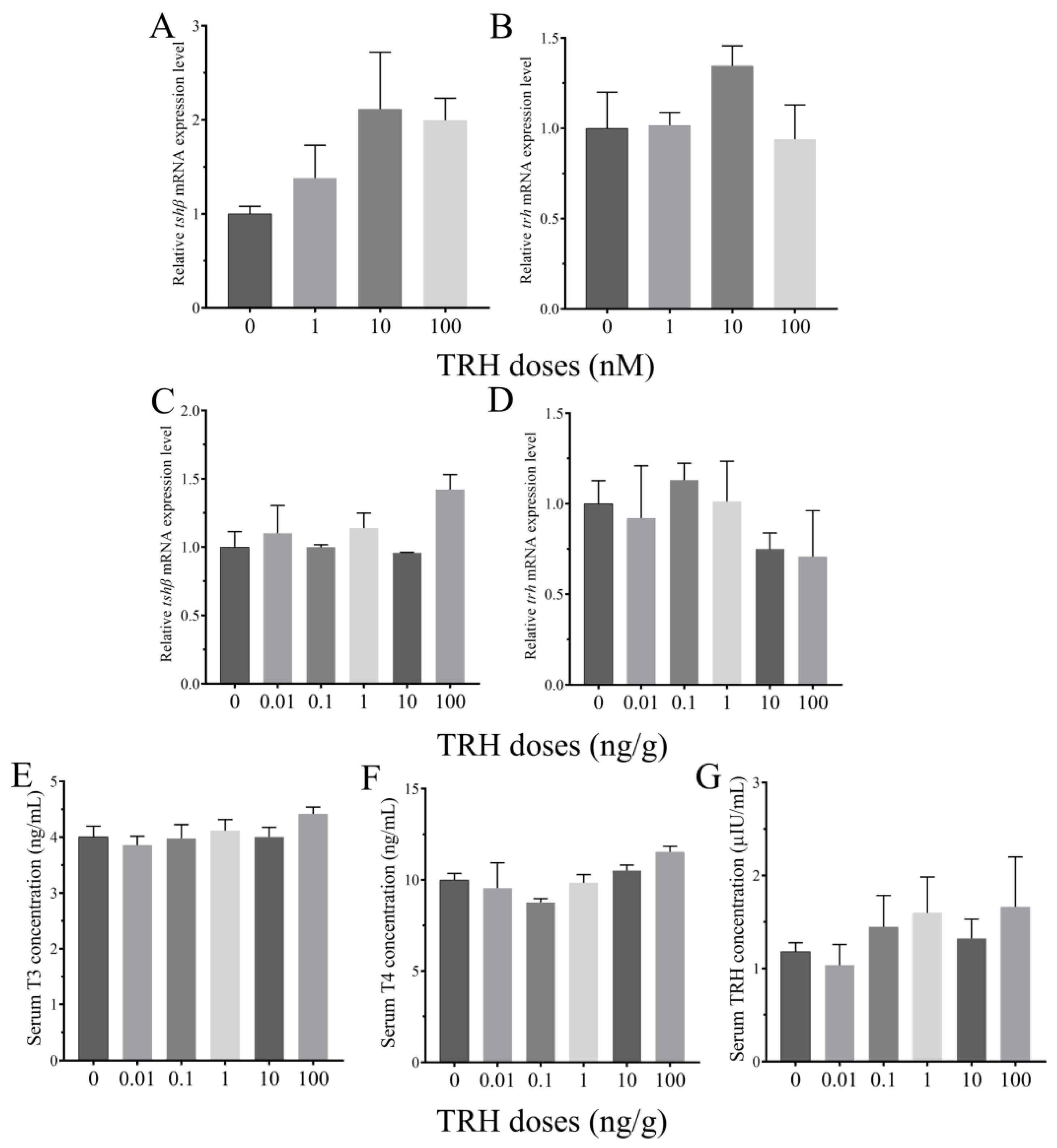
3.4. Roles of TRH on the Regulation of HPT Axis In Vitro and In Vivo
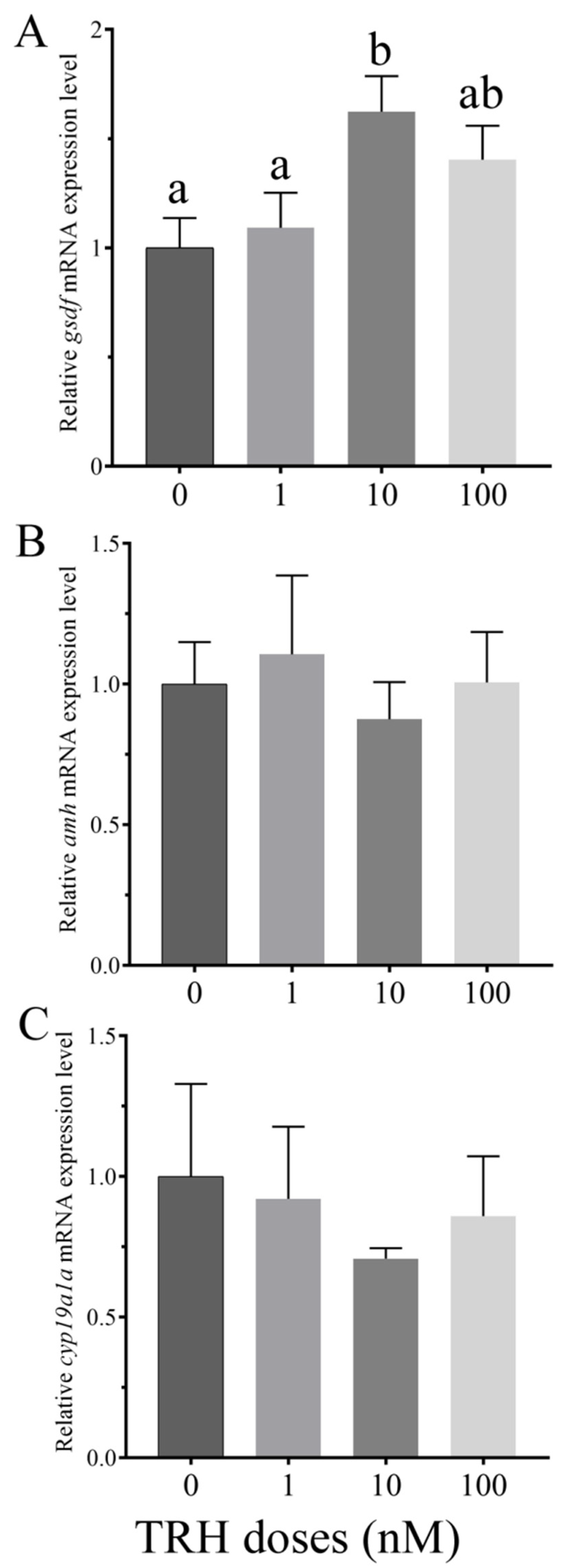
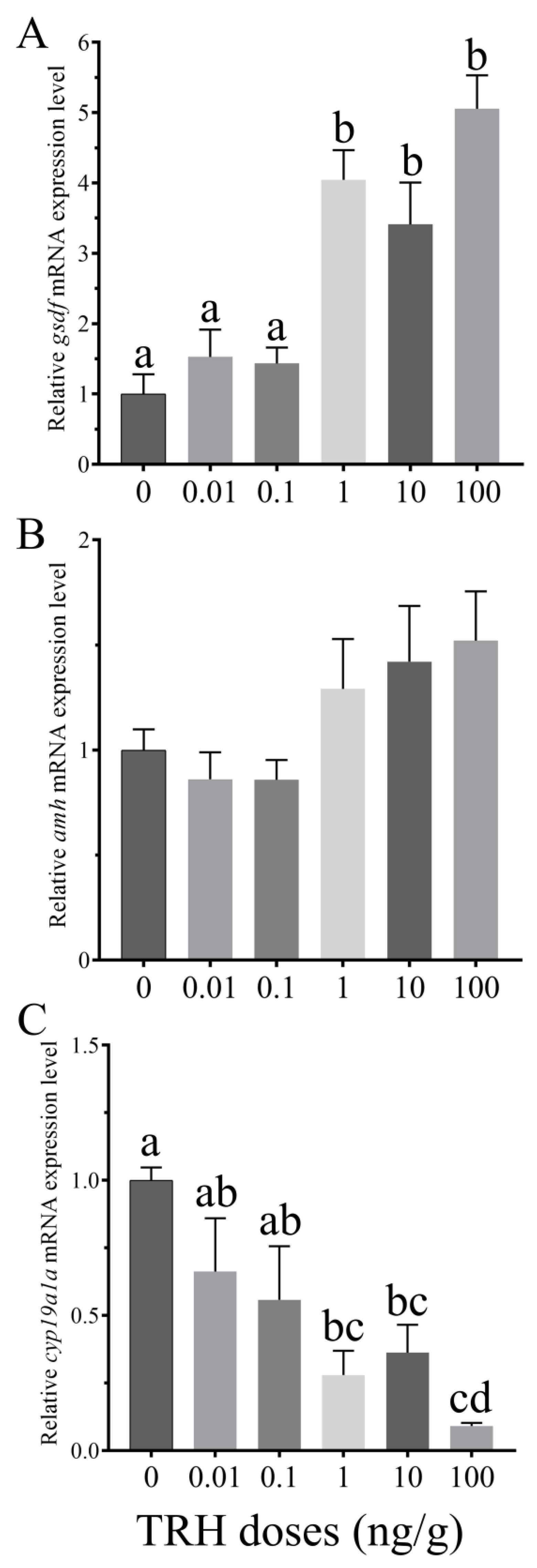
3.5. In Vitro Effects of TRH on the Expression of gsdf, amh and cyp19a1a
3.6. In Vivo Effects of TRH on the Expression of gsdf, amh and cyp19a1a
3.7. Effect of TRH Injection on Serum E2 and 11-KT Levles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galas, L.; Raoult, E.; Tonon, M.C.; Okada, R.; Jenks, B.G.; Castano, J.P.; Kikuyama, S.; Malagon, M.; Roubos, E.W.; Vaudry, H. TRH acts as a multifunctional hypophysiotropic factor in vertebrates. Gen. Comp. Endocrinol. 2009, 164, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Buckley, C.; MacDonald, E.E.; Tuziak, S.M.; Volkoff, H. Molecular cloning and characterization of two putative appetite regulators in winter flounder (Pleuronectes americanus): Preprothyrotropin-releasing hormone (TRH) and preproorexin (OX). Peptides 2010, 31, 1737–1747. [Google Scholar] [CrossRef] [PubMed]
- Deal, C.K.; Volkoff, H. Response of the thyroid axis and appetite-regulating peptides to fasting and overfeeding in goldfish (Carassius auratus). Mol. Cell. Endocrinol. 2021, 528, 111229. [Google Scholar] [CrossRef] [PubMed]
- Burgus, R.; Dunn, T.F.; Desiderio, D.; Ward, D.N.; Vale, W.; Guillemin, R. Characterization of ovine hypothalamic hypophysiotropic TSH-releasing factor. Nature 1970, 226, 321–325. [Google Scholar] [CrossRef]
- Nair, R.M.; Barrett, J.F.; Bowers, C.Y.; Schally, A.V. Structure of porcine thyrotropin releasing hormone. Biochemistry 1970, 9, 1103–1106. [Google Scholar] [CrossRef]
- Wallis, M. Molecular evolution of the thyrotrophin-releasing hormone precursor in vertebrates: Insights from comparative genomics. J. Neuroendocrinol. 2010, 22, 608–619. [Google Scholar] [CrossRef]
- Chiamolera, M.I.; Wondisford, F.E. Thyrotropin-releasing hormone and the thyroid hormone feedback mechanism. Endocrinology 2009, 150, 1091–1096. [Google Scholar] [CrossRef]
- Nakajima, Y.; Yamada, M.; Taguchi, R.; Shibusawa, N.; Ozawa, A.; Tomaru, T.; Hashimoto, K.; Saito, T.; Tsuchiya, T.; Okada, S.; et al. NR4A1 (Nur77) mediates thyrotropin-releasing hormone-induced stimulation of transcription of the thyrotropin β gene: Analysis of TRH knockout mice. PLoS ONE 2012, 7, e40437. [Google Scholar] [CrossRef]
- Zeng, H.K.; Schimpf, B.A.; Rohde, A.D.; Pavlova, M.N.; Gragerov, A.; Bergmann, J.E. Thyrotropin-releasing hormone receptor 1-deficient mice display increased depression and anxiety-like behavior. Mol. Endocrinol. 2007, 21, 2795–2804. [Google Scholar] [CrossRef]
- Eales, J.G.; Himick, B.A. The effects of TRH on plasma thyroid hormone levels of rainbow trout (Salmo gairdneri) and arctic charr (Salvelinus alpinus). Gen. Comp. Endocrinol. 1988, 72, 333–339. [Google Scholar] [CrossRef]
- Chatterjee, A.; Hsieh, Y.L.; Yu, J.Y.L. Molecular cloning of cDNA encoding thyroid stimulating hormone β subunit of bighead carp Aristichthys nobilis and regulation of its gene expression. Mol. Cell. Endocrinol. 2001, 174, 1–9. [Google Scholar] [CrossRef]
- Han, Y.S.; Liao, I.C.; Tzeng, W.N.; Yu, J.Y.L. Cloning of the cDNA for thyroid stimulating hormone beta subunit and changes in activity of the pituitary-thyroid axis during silvering of the Japanese eel, Anguilla japonica. J. Mol. Endocrinol. 2004, 32, 179–194. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Melamed, P.; Eliahu, N.; Levavi-Sivan, B.; Ofir, M.; Farchi-Pisanty, O.; Rentier-Delrue, F.; Smal, J.; Yaron, Z.; Naor, Z. Hypothalamic and thyroidal regulation of growth hormone in tilapia. Gen. Comp. Endocrinol. 1995, 97, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Kagabu, Y.; Mishiba, T.; Okino, T.; Yanagisawa, T. Effects of thyrotropin-releasing hormone and its metabolites, cyclo(His-Pro) and TRH-OH, on growth hormone and prolactin synthesis in primary cultured pituitary cells of the common carp, Cyprinus carpio. Gen. Comp. Endocrinol. 1998, 111, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Larsen, D.A.; Swanson, P.; Dickey, J.T.; Rivier, J.; Dickhoff, W.W. In vitro thyrotropin-releasing activity of corticotropin-releasing hormone-family peptides in coho salmon, Oncorhynchus kisutch. Gen. Comp. Endocrinol. 1998, 109, 276–285. [Google Scholar] [CrossRef]
- Aoki, Y.; Takahashi, M.; Masuda, T.; Tsukamoto, T.; Iigo, M.; Yanagisawa, T. Molecular cloning of prepro-thyrotropin-releasing hormone cDNAs from the common carp Cyprinus carpio and goldfish Carassius auratus. Gen. Comp. Endocrinol. 2005, 141, 84–92. [Google Scholar] [CrossRef]
- Iziga, R.; Ponce, M.; Infante, C.; Rebordinos, L.; Canavate, J.P.; Manchado, M. Molecular characterization and gene expression of thyrotropin-releasing hormone in Senegalese sole (Solea senegalensis). Comp. Biochem. Physiol. B 2010, 157, 167–174. [Google Scholar] [CrossRef]
- Ruiz-Jarabo, I.; Martos-sitcha, J.A.; Barragan-Mendez, C.; Martinez-Rodriguez, G.; Mancera, J.M.; Arjona, F.J. Gene expression of thyrotropin- and corticotrophin-releasing hormones is regulated by environmental salinity in the euryhaline teleost Sparus aurata. Fish Physiol. Biochem. 2018, 44, 615–628. [Google Scholar] [CrossRef]
- Jiang, X.L.; Cai, Z.W.; Zhao, X.F.; Zhang, L.F.; Chen, Z.; Wang, Y.; Guo, X.L.; Xu, N.Y. Mapping, cDNA cloning and tissue expression of the porcine thyrotropin-releasing hormone receptor gene. Anim. Biotechnol. 2011, 22, 30–36. [Google Scholar] [CrossRef]
- Mekuchi, M.; Saito, Y.; Aoki, Y.; Masuda, T.; Iigo, M.; Yanagisawa, T. Molecular cloning, gene structure, molecular evolution and expression analyses of thyrotropin-releasing hormone receptors from medaka (Oryzias latipes). Gen. Comp. Endocrinol. 2011, 170, 374–380. [Google Scholar] [CrossRef]
- Senthilkumaran, B.; Sudhakumari, C.; Mamta, S.; Raghuveer, K.; Swapna, I.; Murugananthkumar, R. “Brain sex differentiation” in teleosts: Emerging concepts with potential biomarkers. Gen. Comp. Endocrinol. 2015, 220, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Castaneda-Cortes, D.C.; Fernandino, J.I. Stress and sex determination in fish: From brain to gonads. Int. J. Dev. Biol. 2021, 65, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Fisheries Bureau of Agriculture Ministry of China. China Fishery Statistical Yearbook; Chinese Agricultural Press: Beijing, China, 2021; (In Chinese).
- Liu, C.K. Rudimentary hermaphroditism in the symbranchoid eel, Monopterus albus. Sinensia 1944, 15, 1–8. [Google Scholar]
- Jang, S.H.; Zhou, F.; Xia, L.X.; Zhao, W.; Cheng, H.H.; Zhou, R.J. Construction of a BAC library and identification of dmrt1 gene of the rice field eel, Monopterus albus. Biochem. Bioph. Res. Comumun. 2006, 348, 775–780. [Google Scholar] [CrossRef]
- Feng, K.; Luo, H.R.; Li, Y.M.; Chen, J.; Wang, Y.P.; Sun, Y.H.; Zhu, Z.Y.; Hu, W. High efficient gene targeting in rice field eel Monopterus albus by transcription activator-like effector nucleases. Sci. Bull. 2017, 62, 162–164. [Google Scholar] [CrossRef]
- Qu, X.C. Sex determination and control in eels. In Sex Control in Aquaculture, 1st ed.; Wang, H.P., Piferrer, F., Chen, S.L., Shen, Z.G., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2018; Volume 11, pp. 775–792. [Google Scholar] [CrossRef]
- Hu, Q.; Guo, W.; Gao, Y.; Tang, R.; Li, D.P. Reference gene selection for real-time RT-PCR normalization in rice field eel (Monopterus albus) during gonad development. Fish Physiol. Biochem. 2014, 40, 1721–1730. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2014; pp. 515–530. [Google Scholar]
- Kapustin, Y.; Souvorov, A.; Tatusova, T.; Lipman, D. Splign: Algorithms for computing spliced alignments with identification of paralogs. Bio. Direct 2008, 3, 20. [Google Scholar] [CrossRef]
- Armenteros, J.J.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2-△△Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Tse, M.C.L.; Wong, G.K.P.; Xiao, P.; Cheng, C.H.K.; Chan, M. Down-regulation of goldfish (Carassius auratus) prolactin gene expression by dopamine and thyrotropin releasing hormone. Gen. Comp. Endocrinol. 2008, 155, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Van Goor, F.; Goldberg, J.I.; Wong, A.O.L.; Jobin, R.M.; Chang, J.P. Morphological identification of live gonadotropin, growth-hormone, and prolactin cells in goldfish (Carassius auratus) pituitary-cell cultures. Cell Tissue Res. 1994, 276, 253–261. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.; Liu, Z.X.; Zhang, L.H.; Zhang, W.M. Epigenetic modifications during sex change repress gonadotropin stimulation of cyp19a1a in a teleost rice field eel (Monopterus albus). Endocrinology 2013, 154, 2881–2890. [Google Scholar] [CrossRef]
- Feng, K.; Luo, H.R.; Hou, M.X.; Li, Y.M.; Chen, J.; Zhu, Z.Y.; Hu, W. Alternative splicing of GnRH2 and GnRH2-associated peptide plays roles in gonadal differentiation of the rice field eel, Monopterus albus. Gen. Comp. Endocrinol. 2018, 267, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Abbott, M.; Volkoff, H. Thyrotropin releasing hormone (TRH) in goldfish (Carassius auratus): Role in the regulation of feeding and locomotor behaviors and interactions with the orexin system and cocaine- and amphetamine regulated transcript (CART). Horm. Behav. 2011, 59, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Vandenborne, K.; Roelens, S.A.; Darras, V.M.; Kuhn, E.R.; Van der Geyten, S. Cloning and hypothalamic distribution of the chicken thyrotropin-releasing hormone precursor cDNA. J. Endocrinol. 2005, 186, 387–396. [Google Scholar] [CrossRef]
- Lechan, R.M.; Wu, P.; Jackson, I.M.D.; Wolf, H.; Cooperman, S.; Mandel, G.; Goodman, R.D. Thyrotropin-releasing hormone precursor: Characterization in rat brain. Science 1986, 231, 159–161. [Google Scholar] [CrossRef]
- Yamada, M.; Radovick, S.; Wondisford, F.E.; Nakayama, Y.; Weintraub, B.D.; Wilber, J.F. Cloning and structure of human genomic DNA and hypothalamic cDNA encoding human preprothyrotropin-releasing hormone. Mol. Endocrinol. 1990, 4, 551–556. [Google Scholar] [CrossRef]
- Bulant, M.; Richter, K.; Kuchler, K.; Kreil, G. A cDNA from brain of Xenopus laevis coding for a new precursor of thyrotropin-releasing hormone. FEBS Lett. 1992, 296, 292–296. [Google Scholar] [CrossRef]
- Aoki, Y.; Masuda, T.; Iigo, M.; Yanagisawa, T. Molecular cloning of prepro-thyrotropin-releasing hormone cDNA from medaka (Oryzias latipes). Gen. Comp. Endocrinol. 2007, 150, 364–370. [Google Scholar] [CrossRef]
- Kuchler, K.; Richter, K.; Trnovsky, J.; Egger, R.; Kreil, G. Two precursors of thyrotropin-releasing hormone from skin of Xenopus laevis. J. Biol. Chem. 1990, 265, 11731–11733. [Google Scholar] [CrossRef]
- Singh, O.; Pradhan, D.R.; Nagalakashmi, B.; Kumar, S.; Mitra, S.; Sagarkar, S.; Sakharkar, A.J.; Lechan, R.M.; Singru, P.S. Thyrotropin-releasing hormone (TRH) in the brain and pituitary of the teleost, Clarias batrachus and its role in regulation of hypophysiotropic dopamine neurons. J. Comp. Neurol. 2019, 527, 1070–1101. [Google Scholar] [CrossRef] [PubMed]
- Ojima, D.; Iwata, M. Central administration of growth hormone-releasing hormone and corticotropin-releasing hormone stimulate downstream movement and thyroxine secretion in fall-smolting coho salmon (Oncorhynchus kisutch). Gen. Comp. Endocrinol. 2010, 168, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Bromage, N.R. The effects of mammalian thyrotropin-releasing hormone on the pituitary-thyroid axis of teleost fish. Gen. Comp. Endocrinol. 1975, 25, 292–297. [Google Scholar] [CrossRef]
- Matz, S.P.; Hofeldt, G.T. Immunohistochemical localization of corticotropin-releasing factor in the brain and corticotropin-releasing factor and thyrotropin-stimulating hormone in the pituitary of chinook salmon (Oncorhynchus tshawytscha). Gen. Comp. Endocrinol. 1999, 114, 151–160. [Google Scholar] [CrossRef]
- Tovo-Neto, A.; Rodrigues, M.S.; Habibi, H.R.; Nobrega, R.H. Thyroid hormone actions on male reproductive system of teleost fish. Gen. Comp. Endocrinol. 2018, 265, 230–236. [Google Scholar] [CrossRef]
- Kaneko, H.; Ijiri, S.; Kobayashi, T.; Izumi, H.; Kuramochi, Y.; Wang, D.S.; Mizuno, S.; Nagahama, Y. Gonadal soma-derived factor (gsdf), a TGF-beta superfamily gene, induces testis differentiation in the teleost fish Oreochromis niloticus. Mol. Cell. Endocrinol. 2015, 415, 87–99. [Google Scholar] [CrossRef]
- Pfennig, F.; Standke, A.; Gutzeit, H.O. The role of Amh signaling in teleost fish—Multiple functions not restricted to the gonads. Gen. Comp. Endocrinol. 2015, 223, 87–107. [Google Scholar] [CrossRef]
- Guiguen, Y.; Fostier, A.; Piferrer, F.; Chang, C.F. Ovarian aromatase and estrogens: A pivotal role for gonadal sex differentiation and sex change in fish. Gen. Comp. Endocrinol. 2010, 165, 352–366. [Google Scholar] [CrossRef]
- Hu, Q.; Guo, W.; Gao, Y.; Tang, R.; Li, D.P. Molecular cloning and characterization of amh and dax1 genes and their expression during sex inversion in rice field eel Monopterus albus. Sci. Rep. 2015, 5, 16667. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.F.; Wang, C.L.; Chen, X.W.; Guan, G.J. Identification of gonadal soma-derived factor involvement in Monopterus albus (protogynous rice field eel) sex change. Mol. Biol. Rep. 2016, 43, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, W.M.; Yang, H.Y.; Zhou, W.L.; Hu, C.Q.; Zhang, L.H. Two cytochrome P450 aromatase genes in the hermaphrodite rice field eel Monopterus albus: mRNA expression during ovarian development and sex change. J. Endocrinol. 2008, 199, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, Q.H.; Xiao, Y.H.; Xu, S.H.; Wang, X.Y.; Yang, J.K.; Song, Z.C.; You, F.; Li, J. Effects of environmental stress (sex steroids and heat) during sex differentiation in Japanese flounder (Paralichthys olivaceus): Insight from germ cell proliferation and gsdf-amh-cyp19a1a expression. Aquaculture 2020, 515, 734536. [Google Scholar] [CrossRef]


| Name | Primers (5′→3′) | Use |
|---|---|---|
| trh-F1 | TTCACAGCCAGTGGGAACC | cDNA fragment cloning |
| trh-R1 | ATAGAGGCAACATCGCAAAAG | |
| trh-F2 | TCCGTGCGTAATTGTCCTCC | |
| trh-R2 | CTGGCCACAATGTCCTTGAA | |
| oligo-dT adaptor | GACTCGAGTCGACATCGA(T)17 | adaptor primers for RACE |
| adaptor | GACTCGAGTCGACATCGA | |
| 3′RACE outer | GCAACTGTCCACAAGCCTGAT | 3′RACE cloning |
| 3′RACE inner | TCCCAAAGGACAAGCATCAA | |
| 5′RACE outer | CGTCTATGGTCCTTCGGTCC | 5′RACE cloning |
| 5′RACE inner | GCACGAGAGAAGCCAAGATG | |
| trh-Q-F1 | CGGACCGAAGGACCATAGAC | real-time PCR |
| trh-Q-R1 | GATGCTGTCGCTTTGTCAGC | |
| tshβ-Q-F1 | GCTGTTCCCACGTGTTTACC | |
| tshβ-Q-R1 | GGAAGCGTGGGCCAAGAATA | |
| gsdf-Q-F1 | ATACCCACATCCTGCACAACG | |
| gsdf-Q-R1 | TGAAGATCGCATTTGGAGAG | |
| amh-Q-F1 | TGCTCCCGACGTTTGAAG | |
| amh-Q-R1 | GAATGTTGCCCTCTGATGC | |
| cyp19a1a-Q-F1 | TACTCAGCAGGTCATCAGCG | |
| cyp19a1a-Q-R1 | TCTCATTGACAGGTACACCA | |
| ef1a-Q-F1 | CGCTGCTGTTTCCTTCGTCC | |
| ef1a-Q-R1 | TTGCGTTCAATCTTCCATCCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, K.; Su, J.; Wu, Z.; Su, S.; Yao, W. Molecular Cloning and Expression Analysis of Thyrotropin-Releasing Hormone, and Its Possible Role in Gonadal Differentiation in Rice Field eel Monopterus albus. Animals 2022, 12, 1691. https://doi.org/10.3390/ani12131691
Feng K, Su J, Wu Z, Su S, Yao W. Molecular Cloning and Expression Analysis of Thyrotropin-Releasing Hormone, and Its Possible Role in Gonadal Differentiation in Rice Field eel Monopterus albus. Animals. 2022; 12(13):1691. https://doi.org/10.3390/ani12131691
Chicago/Turabian StyleFeng, Ke, Jialin Su, Zhengli Wu, Shengqi Su, and Weizhi Yao. 2022. "Molecular Cloning and Expression Analysis of Thyrotropin-Releasing Hormone, and Its Possible Role in Gonadal Differentiation in Rice Field eel Monopterus albus" Animals 12, no. 13: 1691. https://doi.org/10.3390/ani12131691
APA StyleFeng, K., Su, J., Wu, Z., Su, S., & Yao, W. (2022). Molecular Cloning and Expression Analysis of Thyrotropin-Releasing Hormone, and Its Possible Role in Gonadal Differentiation in Rice Field eel Monopterus albus. Animals, 12(13), 1691. https://doi.org/10.3390/ani12131691







