Toxocara canis Infection Alters mRNA Expression Profiles of Peripheral Blood Mononuclear Cells in Beagle Dogs at the Lung Infection Period
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Animal and Sample Collection
2.3. RNA Extraction and RNA Sequencing Analysis
2.4. GO Annotation and KEGG Pathway Enrichment Analysis
2.5. Quantitative Real Time RT-PCR (qRT-PCR)
3. Results
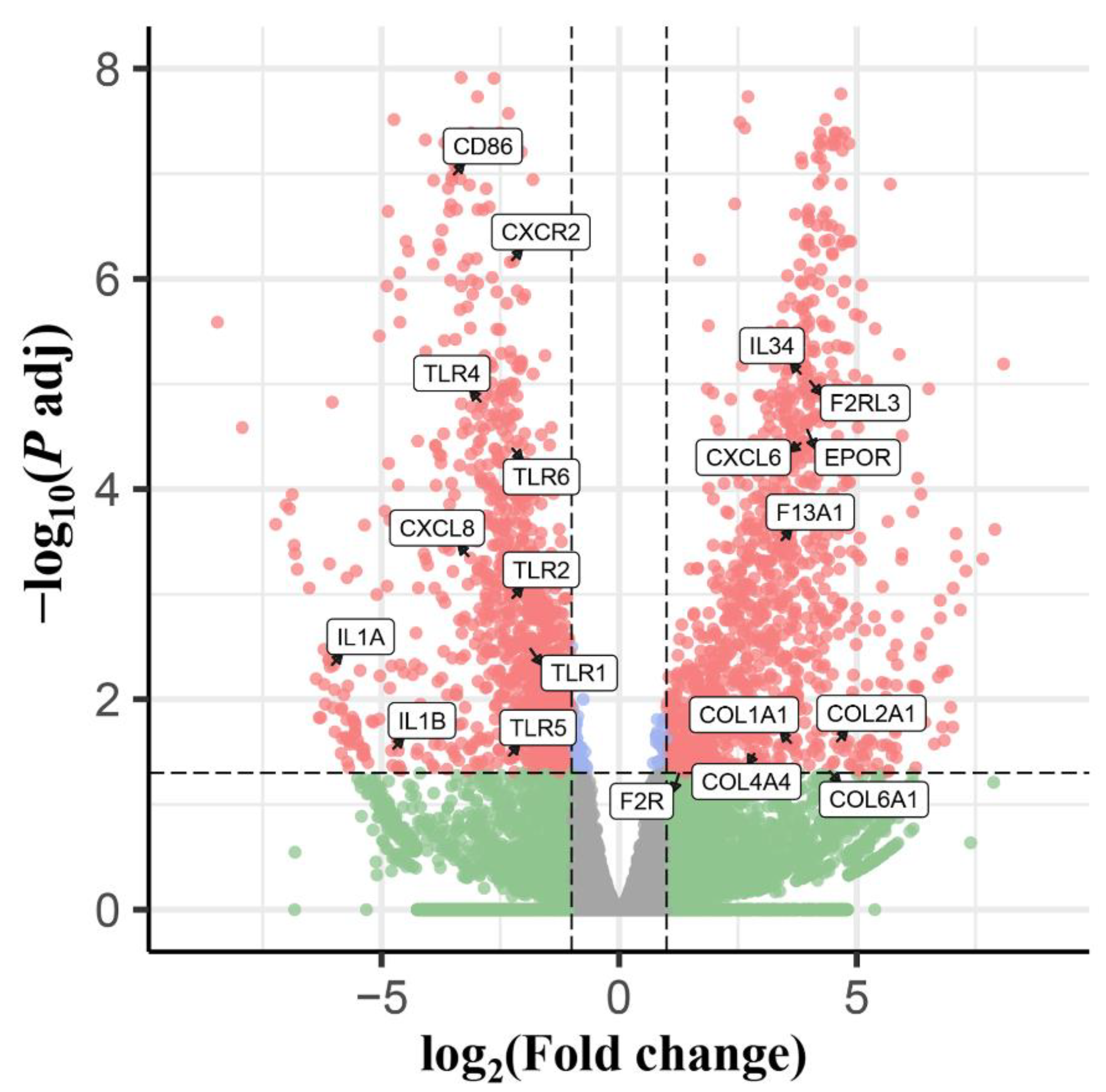
3.1. Confirmation of T. canis Infection and Overview of RNA Sequencing
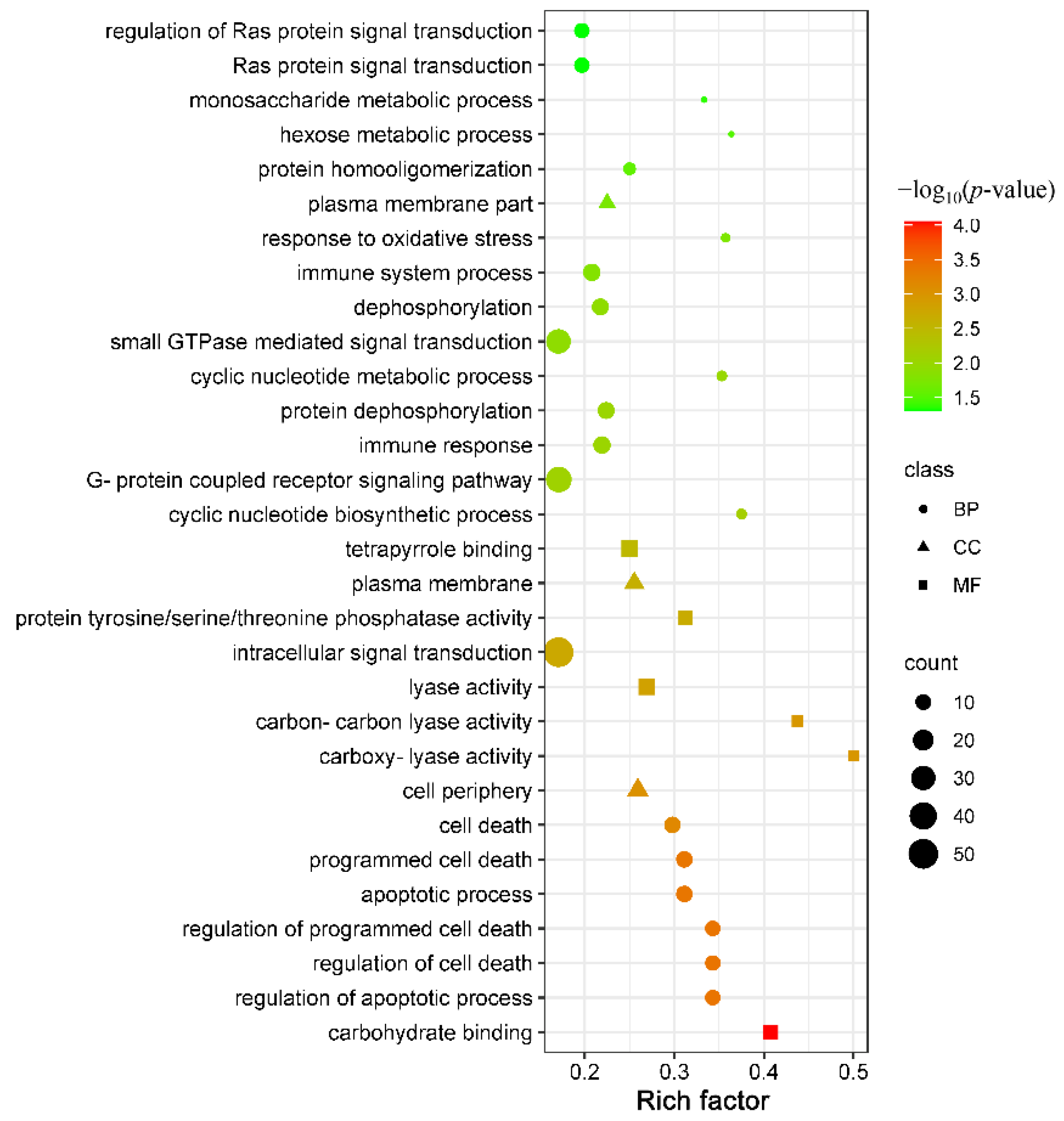
3.2. GO Annotation and KEGG Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, G.; Holland, C.V.; Wang, T.; Hofmann, A.; Fan, C.K.; Maizels, R.M.; Hotez, P.J.; Gasser, R.B. Human toxocariasis. Lancet Infect. Dis. 2018, 18, e14–e24. [Google Scholar] [CrossRef]
- Rostami, A.; Ma, G.; Wang, T.; Koehler, A.V.; Hofmann, A.; Chang, B.C.H.; Macpherson, C.N.; Gasser, R.B. Human toxocariasis—A look at a neglected disease through an epidemiological ‘prism’. Infect. Genet. Evol. 2019, 74, 104002. [Google Scholar] [CrossRef]
- Overgaauw, P.A. Aspects of Toxocara epidemiology: Toxocarosis in dogs and cats. Crit. Rev. Microbiol. 1997, 23, 233–251. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, Q.; Liu, G.H.; Zheng, W.B.; Hong, S.J.; Sugiyama, H.; Zhu, X.Q.; Elsheikha, H.M. Toxocariasis: A silent threat with a progressive public health impact. Infect. Dis. Poverty 2018, 7, 59. [Google Scholar] [CrossRef] [Green Version]
- Fakhri, Y.; Gasser, R.B.; Rostami, A.; Fan, C.K.; Ghasemi, S.M.; Javanian, M.; Bayani, M.; Armoon, B.; Moradi, B. Toxocara eggs in public places worldwide—A systematic review and meta-analysis. Environ. Pollut. 2018, 242, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Rostami, A.; Riahi, S.M.; Holland, C.V.; Taghipour, A.; Khalili-Fomeshi, M.; Fakhri, Y.; Omrani, V.F.; Hotez, P.J.; Gasser, R.B. Seroprevalence estimates for toxocariasis in people worldwide: A systematic review and meta-analysis. PLoS Negl. Trop. Dis. 2019, 13, e0007809. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Peng, H.J. Current epidemic situation of human toxocariasis in China. Adv. Parasitol. 2020, 109, 433–448. [Google Scholar] [PubMed]
- Rostami, A.; Riahi, S.M.; Hofmann, A.; Ma, G.; Wang, T.; Behniafar, H.; Taghipour, A.; Fakhri, Y.; Spotin, A.; Chang, B.C.H.; et al. Global prevalence of Toxocara infection in dogs. Adv. Parasitol. 2020, 109, 561–583. [Google Scholar] [PubMed]
- Zheng, W.B.; Zou, Y.; Liu, G.H.; Zhu, X.Q. Epidemiology of Toxocara spp. in dogs and cats in mainland China, 2000–2019. Adv. Parasitol. 2020, 109, 843–860. [Google Scholar]
- Schnieder, T.; Laabs, E.M.; Welz, C. Larval development of Toxocara canis in dogs. Vet. Parasitol. 2011, 175, 193–206. [Google Scholar] [CrossRef]
- Miller, A.D. Pathology of larvae and adults in dogs and cats. Adv. Parasitol. 2020, 109, 537–544. [Google Scholar]
- Mazur-Melewska, K.; Jończyk, K.; Modlińska-Cwalińska, A.; Figlerowicz, M.; Służewski, W. Visceral larva migrans syndrome: Analysis of serum cytokine levels in children with hepatic lesions confirmed in radiological findings. Parasite Immunol. 2014, 36, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.X.; Zhou, D.H.; Liu, G.X.; Suo, X.; Zhu, X.Q. Transcriptomic analysis of porcine PBMCs infected with Toxoplasma gondii RH strain. Acta Trop. 2016, 154, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.B.; Zou, Y.; Zhu, X.Q.; Liu, G.H. Toxocara “omics” and the promises it holds for medicine and veterinary medicine. Adv. Parasitol. 2020, 109, 89–108. [Google Scholar] [PubMed]
- Zhu, X.Q.; Korhonen, P.K.; Cai, H.; Young, N.D.; Nejsum, P.; von Samson-Himmelstjerna, G.; Boag, P.R.; Tan, P.; Li, Q.; Min, J.; et al. Genetic blueprint of the zoonotic pathogen Toxocara canis. Nat. Commun. 2015, 6, 6145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.B.; Zou, Y.; He, J.J.; Elsheikha, H.M.; Liu, G.H.; Hu, M.H.; Wang, S.L.; Zhu, X.Q. Global profiling of lncRNAs-miRNAs-mRNAs reveals differential expression of coding genes and non-coding RNAs in the lung of Beagle dogs at different stages of Toxocara canis infection. Int. J. Parasitol. 2021, 51, 49–61. [Google Scholar] [CrossRef]
- Zheng, W.B.; Zou, Y.; Elsheikha, H.M.; Liu, G.H.; Hu, M.H.; Wang, S.L.; Zhu, X.Q. Serum metabolomic alterations in Beagle dogs experimentally infected with Toxocara canis. Parasit. Vectors 2019, 12, 447. [Google Scholar] [CrossRef] [Green Version]
- Takamiya, S.; Mita, T. Large-scale purification of active liquid-cultured Caenorhabditis elegans using a modified baermann apparatus. Parasitol. Int. 2016, 65, 580–583. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, H.J.; Zhao, B.; Sun, Q.Q.; Cao, Y.Y.; Li, R.; Wu, X.X.; Weeda, S.; Li, L.; Ren, S.; et al. The RNA-seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation. J. Pineal Res. 2014, 56, 39–50. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. Clusterprofiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.B.; Zou, Y.; Liu, Q.; Hu, M.H.; Elsheikha, H.M.; Zhu, X.Q. Toxocara canis infection alters lncRNA and mRNA expression profiles of dog bone marrow. Front. Cell Dev. Biol. 2021, 9, 688128. [Google Scholar] [CrossRef] [PubMed]
- Maizels, R.M. Toxocara canis: Molecular basis of immune recognition and evasion. Vet. Parasitol. 2013, 193, 365–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuriakose, S.M.; Singh, R.; Uzonna, J.E. Host intracellular signaling events and pro-inflammatory cytokine production in African trypanosomiasis. Front. Immunol. 2016, 7, 181. [Google Scholar] [CrossRef] [Green Version]
- Goodridge, H.S.; Stepek, G.; Harnett, W.; Harnett, M.M. Signalling mechanisms underlying subversion of the immune response by the Filarial nematode secreted product ES-62. Immunology 2005, 115, 296–304. [Google Scholar] [CrossRef]
- Saradna, A.; Do, D.C.; Kumar, S.; Fu, Q.L.; Gao, P. Macrophage polarization and allergic asthma. Transl. Res. 2018, 191, 1–14. [Google Scholar] [CrossRef]
- Keewan, E.; Naser, S.A. Notch-1 signaling modulates macrophage polarization and immune defense against Mycobacterium avium paratuberculosis infection in inflammatory diseases. Microorganisms 2020, 8, 1006. [Google Scholar] [CrossRef]
- Chen, W.; Liu, Y.; Chen, J.; Ma, Y.; Song, Y.; Cen, Y.; You, M.; Yang, G. The Notch signaling pathway regulates macrophage polarization in liver diseases. Int. Immunopharmacol. 2021, 99, 107938. [Google Scholar] [CrossRef]
- Lopez-Lopez, S.; Romero de Avila, M.J.; Hernandez de Leon, N.C.; Ruiz-Marcos, F.; Baladron, V.; Nueda, M.L.; Laborda, J.; Garcia-Ramirez, J.J.; Monsalve, E.M.; Diaz-Guerra, M.J.M. Notch4 exhibits anti-inflammatory activity in activated macrophages by interfering with Interferon-gamma and TLR4 signaling. Front. Immunol. 2021, 12, 734966. [Google Scholar] [CrossRef]
- Zheng, R.; Liu, H.; Zhou, Y.; Yan, D.; Chen, J.; Ma, D.; Feng, Y.; Qin, L.; Liu, F.; Huang, X.; et al. Notch4 negatively regulates the inflammatory response to Mycobacterium tuberculosis infection by inhibiting TAK1 activation. J. Infect. Dis. 2018, 218, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Ashour, D.S. Toll-like receptor signaling in parasitic infections. Expert Rev. Clin. Immunol. 2015, 11, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.C.; Lin, C.N.; Peng, S.Y.; Li, L.L.; Luo, T.Y.; Fan, C.K.; Lee, K.M. A study of immunomodulatory genes responses to macrophages of Schistosoma japonicum infection during different stages by microarray analysis. Acta Trop. 2013, 127, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Babu, S.; Blauvelt, C.P.; Kumaraswami, V.; Nutman, T.B. Diminished expression and function of TLR in lymphatic filariasis: A novel mechanism of immune dysregulation. J. Immunol. 2005, 175, 1170–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, J.D.; Langley, R.S.; Johnston, K.L.; Egerton, G.; Wanji, S.; Taylor, M.J. Wolbachia endosymbiotic bacteria of Brugia malayi mediate macrophage tolerance to TLR- and CD40-specific stimuli in a MyD88/TLR2-dependent manner. J. Immunol. 2006, 177, 1240–1249. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Chryssafidis, A.L.; Browne, J.A.; O’Sullivan, J.; McGettigan, P.A.; Mulcahy, G. Transcriptomic study on ovine immune responses to Fasciola hepatica infection. PLoS Negl. Trop. Dis. 2016, 10, e0005015. [Google Scholar] [CrossRef]
- Libby, P. Interleukin-1 beta as a target for atherosclerosis therapy: Biological basis of CANTOS and beyond. J. Am. Coll. Cardiol. 2017, 70, 2278–2289. [Google Scholar] [CrossRef]
- Dinarello, C.A. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev. 2018, 281, 8–27. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Q.; Li, S.; Chen, Z.; Tan, J.; Yao, J.; Duan, D. Low molecular weight fucoidan alleviates diabetic nephropathy by binding fibronectin and inhibiting ECM-receptor interaction in human renal mesangial cells. Int. J. Biol. Macromol. 2020, 150, 304–314. [Google Scholar] [CrossRef]
- Lee, H.J.; Jang, M.; Kim, H.; Kwak, W.; Park, W.; Hwang, J.Y.; Lee, C.K.; Jang, G.W.; Park, M.N.; Kim, H.C.; et al. Comparative transcriptome analysis of adipose tissues reveals that ECM-receptor interaction is involved in the depot-specific adipogenesis in cattle. PLoS ONE 2013, 8, e66267. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.J.; Tao, J.; Sheng, L.; Hu, X.; Rong, R.M.; Xu, M.; Zhu, T.Y. Twist2 promotes kidney cancer cell proliferation and invasion by regulating ITGA6 and CD44 expression in the ECM-receptor interaction pathway. OncoTargets Ther. 2016, 9, 1801–1812. [Google Scholar] [CrossRef] [PubMed]
- Mathew-Steiner, S.S.; Roy, S.; Sen, C.K. Collagen in wound healing. Bioengineering 2021, 8, 63. [Google Scholar] [CrossRef] [PubMed]


| Sample | Raw Reads | Clean Reads | Clean Bases | Q20 | Q30 | GC Content |
|---|---|---|---|---|---|---|
| AS96hC1 | 46,080,628 | 44,799,936 | 6.72G | 97.29% | 92.89% | 49.98% |
| AS96hC2 | 46,016,190 | 44,600,408 | 6.69G | 97.58% | 93.58% | 52.14% |
| AS96hC3 | 43,874,812 | 42,545,422 | 6.38G | 97.51% | 93.32% | 48.84% |
| AS96hT1 | 44,745,214 | 43,474,512 | 6.52G | 97.47% | 93.31% | 48.98% |
| AS96hT2 | 45,812,738 | 44,663,176 | 6.70G | 97.83% | 94.07% | 52.66% |
| AS96hT3 | 46,227,832 | 44,959,256 | 6.74G | 97.49% | 93.26% | 49.18% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, L.; Zou, Y.; Xu, Y.; Li, H.-Y.; Xie, S.-C.; Zhu, X.-Q.; Zheng, W.-B. Toxocara canis Infection Alters mRNA Expression Profiles of Peripheral Blood Mononuclear Cells in Beagle Dogs at the Lung Infection Period. Animals 2022, 12, 1517. https://doi.org/10.3390/ani12121517
Cai L, Zou Y, Xu Y, Li H-Y, Xie S-C, Zhu X-Q, Zheng W-B. Toxocara canis Infection Alters mRNA Expression Profiles of Peripheral Blood Mononuclear Cells in Beagle Dogs at the Lung Infection Period. Animals. 2022; 12(12):1517. https://doi.org/10.3390/ani12121517
Chicago/Turabian StyleCai, Lang, Yang Zou, Yue Xu, Hao-Yu Li, Shi-Chen Xie, Xing-Quan Zhu, and Wen-Bin Zheng. 2022. "Toxocara canis Infection Alters mRNA Expression Profiles of Peripheral Blood Mononuclear Cells in Beagle Dogs at the Lung Infection Period" Animals 12, no. 12: 1517. https://doi.org/10.3390/ani12121517
APA StyleCai, L., Zou, Y., Xu, Y., Li, H.-Y., Xie, S.-C., Zhu, X.-Q., & Zheng, W.-B. (2022). Toxocara canis Infection Alters mRNA Expression Profiles of Peripheral Blood Mononuclear Cells in Beagle Dogs at the Lung Infection Period. Animals, 12(12), 1517. https://doi.org/10.3390/ani12121517






