Hereditary Basis of Coat Color and Excellent Feed Conversion Rate of Red Angus Cattle by Next-Generation Sequencing Data
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
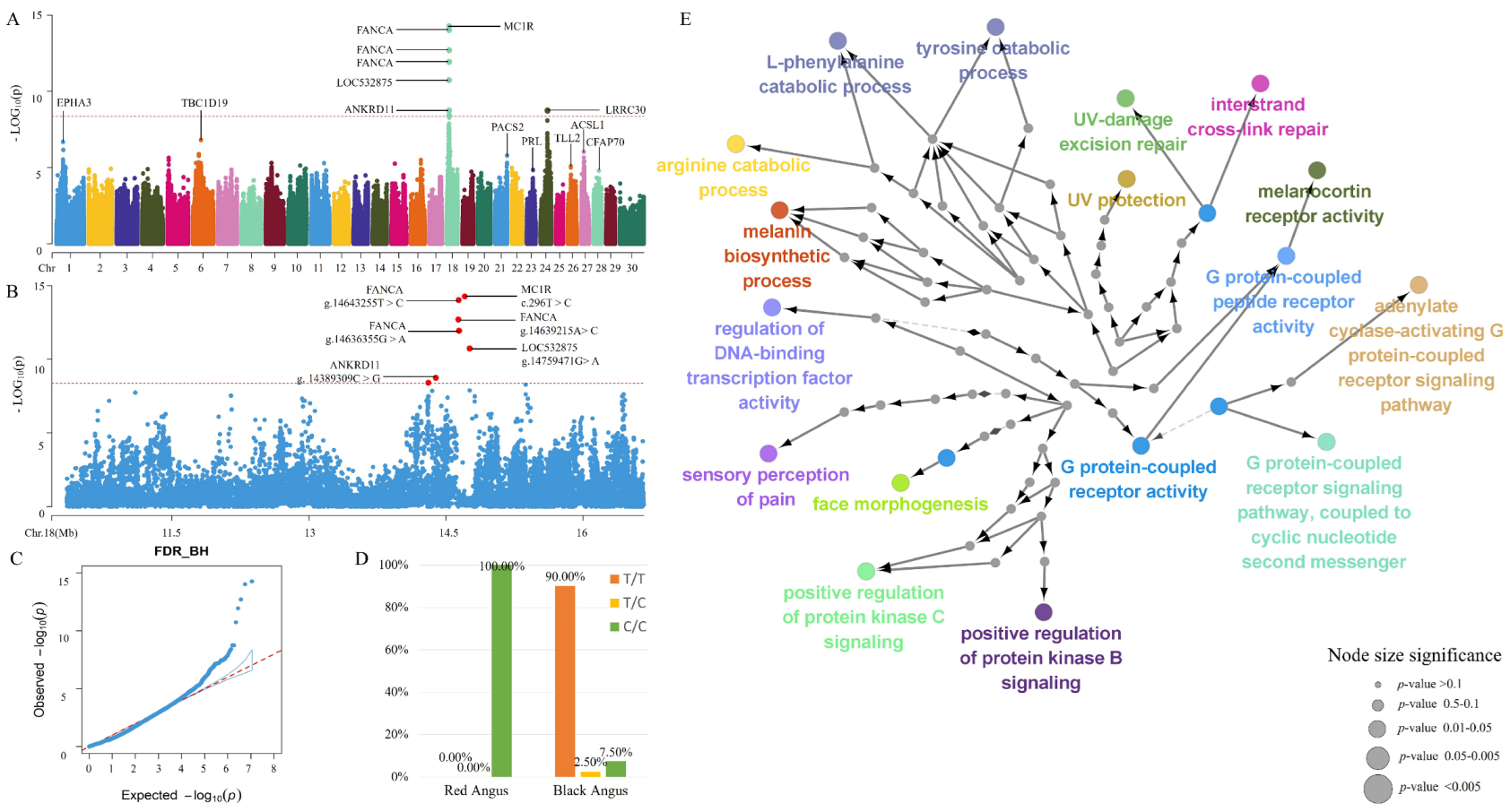
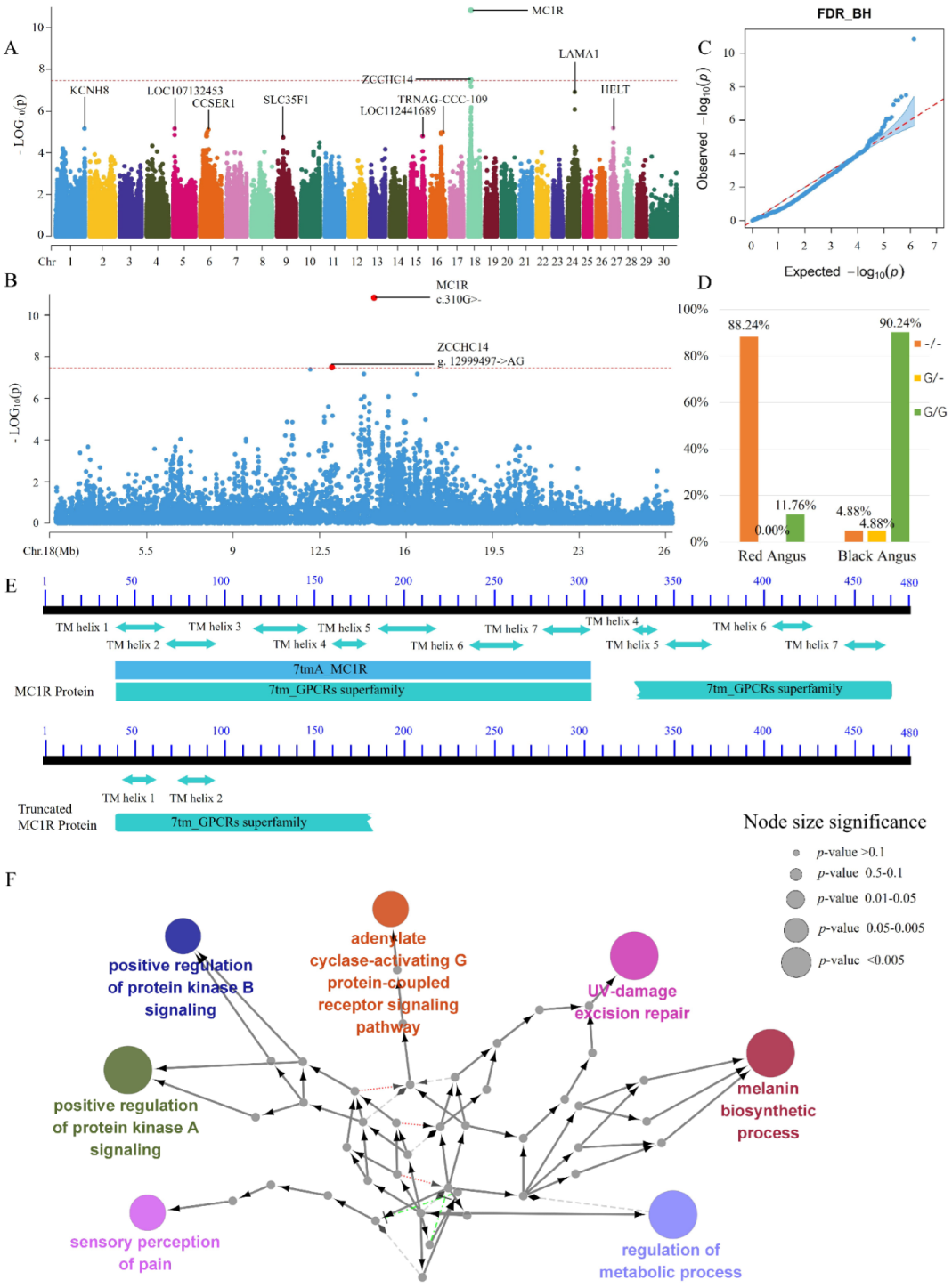
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meyerding, S.G.H.; Gentz, M.; Altmann, B.; Meier-Dinkel, L. Beef quality labels: A combination of sensory acceptance test, stated willingness to pay, and choice-based conjoint analysis. Appetite 2018, 127, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Aroeira, C.N.; de Almeida Torres Filho, R.; Fontes, P.R.; de Lemos Souza Ramos, A.; de Miranda Gomide, L.A.; Ladeira, M.M.; Ramos, E.M. Effect of freezing prior to aging on myoglobin redox forms and CIE color of beef from Nellore and Aberdeen Angus cattle. Meat Sci. 2017, 125, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Gagaoua, M.; Terlouw, E.M.C.; Picard, B. The study of protein biomarkers to understand the biochemical processes underlying beef color development in young bulls. Meat Sci. 2017, 134, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Tait, R.G., Jr.; Cushman, R.A.; McNeel, A.K.; Casas, E.; Smith, T.P.L.; Freetly, H.C.; Bennett, G.L. μ-Calpain (CAPN1), calpastatin (CAST), and growth hormone receptor (GHR) genetic effects on Angus beef heifer performance traits and reproduction. Theriogenology 2018, 113, 1–7. [Google Scholar] [CrossRef]
- Menegatti Zoca, S.; Shafii, B.; Price, W.; Utt, M.; Harstine, B.; McDonald, K.; Cruppe, L.; DeJarnette, M.; Peters, L.; Moraes Vasconcelos, J.L.; et al. Angus sire field fertility and in vitro sperm characteristics following use of different sperm insemination doses in Brazilian beef cattle. Theriogenology 2020, 147, 146–153. [Google Scholar] [CrossRef]
- Liu, T.; Wu, J.P.; Lei, Z.M.; Zhang, M.; Gong, X.Y.; Cheng, S.R.; Liang, Y.; Wang, J.F.; Senczuk, G.; Guerra, L.; et al. Fatty Acid Profile of Muscles from Crossbred Angus-Simmental, Wagyu-Simmental, and Chinese Simmental Cattles Fifteen Shades of Grey: Combined Analysis of Genome-Wide SNP Data in Steppe and Mediterranean Grey Cattle Sheds New Light on the Molecular Basis of Coat Color. Food Sci. Anim. Resour. 2020, 40, 563–577. [Google Scholar] [CrossRef]
- Taye, M.; Yoon, J.; Dessie, T.; Cho, S.; Oh, S.J.; Lee, H.K.; Kim, H. Deciphering signature of selection affecting beef quality traits in Angus cattle. Genes Genom. 2018, 40, 63–75. [Google Scholar] [CrossRef]
- Wolfger, B.; Quinn, C.J.; Torres, G.W.; Taylor, M.; Orsel, K. Comparison of feeding behavior between black and red Angus feeder heifers. Can. J. Anim. Sci. 2016, 96, 404–409. [Google Scholar] [CrossRef]
- Yum, S.Y.; Lee, S.J.; Kim, H.M.; Choi, W.J.; Park, J.H.; Lee, W.W.; Kim, H.S.; Kim, H.J.; Bae, S.H.; Lee, J.H.; et al. Efficient generation of transgenic cattle using the DNA transposon and their analysis by next-generation sequencing. Sci. Rep. 2016, 6, 27185. [Google Scholar] [CrossRef]
- Trigo, B.B.; Utsunomiya, A.T.H.; Fortunato, A.; Milanesi, M.; Torrecilha, R.B.P.; Lamb, H.; Nguyen, L.; Ross, E.M.; Hayes, B.; Padula, R.C.M.; et al. Variants at the ASIP locus contribute to coat color darkening in Nellore cattle. Genet. Sel. Evol. GSE 2021, 53, 40. [Google Scholar] [CrossRef]
- Pegolo, S.; Cecchinato, A.; Savoia, S.; Di Stasio, L.; Pauciullo, A.; Brugiapaglia, A.; Bittante, G.; Albera, A. Genome-wide association and pathway analysis of carcass and meat quality traits in Piemontese young bulls. Anim. Int. J. Anim. Biosci. 2020, 14, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P.; Liao, X.; Arantes, A.S.; De Pauw, M.; Coros, C.; Plastow, G.S.; Sargolzaei, M.; Crowley, J.J.; Basarab, J.A.; Schenkel, F.; et al. A large and diverse collection of bovine genome sequences from the Canadian Cattle Genome Project. GigaScience 2015, 4, 49. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Yan, H.; Neuhauser, C.; Slager, S.L. An analytical workflow for accurate variant discovery in highly divergent regions. BMC Genom. 2016, 17, 703. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608. [Google Scholar] [CrossRef]
- Henderson, C.R. Best linear unbiased estimation and prediction under a selection model. Biometrics 1975, 31, 423–447. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Wu, G.; Wang, L.; Li, D.; Tang, S.; Liang, J.; Mao, H.; Luo, H.; Zhang, Y. The role of MC1R gene in buffalo coat color. Sci. China. Life Sci. 2010, 53, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Goud, T.S.; Upadhyay, R.C.; Pichili, V.B.R.; Onteru, S.K.; Chadipiralla, K. Molecular characterization of coat color gene in Sahiwal versus Karan Fries bovine. J. Genet. Eng. Biotechnol. 2021, 19, 22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Q.; Ye, S.; Faruque, M.O.; Yu, Y.; Sun, D.; Zhang, S.; Wang, Y. New variants in the melanocortin 1 receptor gene (MC1R) in Asian cattle. Anim. Genet. 2014, 45, 609–610. [Google Scholar] [CrossRef]
- Klungland, H.; Våge, D.I.; Gomez-Raya, L.; Adalsteinsson, S.; Lien, S. The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mamm. Genome 1995, 6, 636–639. [Google Scholar] [CrossRef] [PubMed]
- García-Borrón, J.C.; Sánchez-Laorden, B.L.; Jiménez-Cervantes, C. Melanocortin-1 receptor structure and functional regulation. Pigment. Cell Res. 2005, 18, 393–410. [Google Scholar] [CrossRef]
- Matsumoto, H.; Kojya, M.; Takamuku, H.; Kimura, S.; Kashimura, A.; Imai, S.; Yamauchi, K.; Ito, S. MC1R c.310G>- and c.871G > A determine the coat color of Kumamoto sub-breed of Japanese Brown cattle. Anim. Sci. J. 2020, 91, e13367. [Google Scholar] [CrossRef]
- Chen, S.; Han, C.; Miao, X.; Li, X.; Yin, C.; Zou, J.; Liu, M.; Li, S.; Stawski, L.; Zhu, B.; et al. Targeting MC1R depalmitoylation to prevent melanomagenesis in redheads. Nat. Commun. 2019, 10, 877. [Google Scholar] [CrossRef]
- Zorina-Lichtenwalter, K.; Lichtenwalter, R.N.; Zaykin, D.V.; Parisien, M.; Gravel, S.; Bortsov, A.; Diatchenko, L. A study in scarlet: MC1R as the main predictor of red hair and exemplar of the flip-flop effect. Hum. Mol. Genet. 2019, 28, 2093–2106. [Google Scholar] [CrossRef]
- Kimble, D.C.; Lach, F.P.; Gregg, S.Q.; Donovan, F.X.; Flynn, E.K.; Kamat, A.; Young, A.; Vemulapalli, M.; Thomas, J.W.; Mullikin, J.C.; et al. A comprehensive approach to identification of pathogenic FANCA variants in Fanconi anemia patients and their families. Hum. Mutat. 2018, 39, 237–254. [Google Scholar] [CrossRef]
- Nie, D.; Cao, P.; Wang, F.; Zhang, J.; Liu, M.; Zhang, W.; Liu, L.; Zhao, H.; Teng, W.; Tian, W.; et al. Analysis of overlapping heterozygous novel submicroscopic CNVs and FANCA-VPS9D1 fusion transcripts in a Fanconi anemia patient. J. Hum. Genet. 2019, 64, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, E.; Ravera, S.; Vaccaro, D.; Cuccarolo, P.; Bartolucci, M.; Panfoli, I.; Dufour, C.; Degan, P. Mitochondrial respiratory complex I defects in Fanconi anemia. Trends Mol. Med. 2013, 19, 513–514. [Google Scholar] [CrossRef] [PubMed]
- Sumpter, R., Jr.; Sirasanagandla, S.; Fernández, Á.F.; Wei, Y.; Dong, X.; Franco, L.; Zou, Z.; Marchal, C.; Lee, M.Y.; Clapp, D.W.; et al. Fanconi Anemia Proteins Function in Mitophagy and Immunity. Cell 2016, 165, 867–881. [Google Scholar] [CrossRef] [PubMed]
- Moreno, O.M.; Paredes, A.C.; Suarez-Obando, F.; Rojas, A.; Andersson, L.; Haley, C.S.; Ellegren, H.; Knott, S.A.; Johansson, M.; Andersson, K.; et al. An update on Fanconi anemia: Clinical, cytogenetic and molecular approaches (Review) Genetic mapping of quantitative trait loci for growth and fatness in pigs. Biomed. Rep. 2021, 15, 74. [Google Scholar] [CrossRef]
- Sreedhar, A.; Aguilera-Aguirre, L.; Singh, K.K. Mitochondria in skin health, aging, and disease. Cell Death Dis. 2020, 11, 444. [Google Scholar] [CrossRef]
- Niraj, J.; Färkkilä, A.; D’Andrea, A.D.; Bhat, B.; Singh, A.; Iqbal, Z.; Kaushik, J.K.; Rao, A.R.; Ahmad, S.M.; Bhat, H.; et al. The Fanconi Anemia Pathway in Cancer Comparative transcriptome analysis reveals the genetic basis of coat color variation in Pashmina goat. Annu. Rev. Cancer Biol. 2019, 3, 457–478. [Google Scholar] [CrossRef]
- Jarrett, S.G.; Wolf Horrell, E.M.; Boulanger, M.C.; D’Orazio, J.A. Defining the Contribution of MC1R Physiological Ligands to ATR Phosphorylation at Ser435, a Predictor of DNA Repair in Melanocytes. J. Investig. Dermatol. 2015, 135, 3086–3095. [Google Scholar] [CrossRef]
- Lee, A.Y. Skin Pigmentation Abnormalities and Their Possible Relationship with Skin Aging. Int. J. Mol. Sci. 2021, 22, 40. [Google Scholar] [CrossRef]
- Casari, G.; De Fusco, M.; Ciarmatori, S.; Zeviani, M.; Mora, M.; Fernandez, P.; De Michele, G.; Filla, A.; Cocozza, S.; Marconi, R.; et al. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 1998, 93, 973–983. [Google Scholar] [CrossRef]
- Muid, K.A.; Kimyon, Ö.; Reza, S.H.; Karakaya, H.C.; Koc, A. Characterization of long living yeast deletion mutants that lack mitochondrial metabolism genes DSS1, PPA2 and AFG3. Gene 2019, 706, 172–180. [Google Scholar] [CrossRef]
- Davies, K.L.; Camm, E.J.; Atkinson, E.V.; Lopez, T.; Forhead, A.J.; Murray, A.J.; Fowden, A.L. Development and thyroid hormone dependence of skeletal muscle mitochondrial function towards birth. J. Physiol. 2020, 598, 2453–2468. [Google Scholar] [CrossRef] [PubMed]
- Dhanabalan, K.; Mzezewa, S.; Huisamen, B.; Lochner, A. Mitochondrial Oxidative Phosphorylation Function and Mitophagy in Ischaemic/Reperfused Hearts from Control and High-Fat Diet Rats: Effects of Long-Term Melatonin Treatment. Cardiovasc. Drugs Ther. 2020, 34, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Toyomizu, M.; Kikusato, M.; Kawabata, Y.; Azad, M.A.; Inui, E.; Amo, T. Meat-type chickens have a higher efficiency of mitochondrial oxidative phosphorylation than laying-type chickens. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2011, 159, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.; Kress, W.; Schoser, B.; Hehr, U.; Müller, C.R.; Rost, S. Identification of variants in MBNL1 in patients with a myotonic dystrophy-like phenotype. Eur. J. Hum. Genet. 2016, 24, 1467–1472. [Google Scholar] [CrossRef] [PubMed][Green Version]
- André, L.M.; Ausems, C.R.M.; Wansink, D.G.; Wieringa, B. Abnormalities in Skeletal Muscle Myogenesis, Growth, and Regeneration in Myotonic Dystrophy. Front. Neurol. 2018, 9, 368. [Google Scholar] [CrossRef]
- Braz, S.O.; Acquaire, J.; Gourdon, G.; Gomes-Pereira, M. Of Mice and Men: Advances in the Understanding of Neuromuscular Aspects of Myotonic Dystrophy. Front. Neurol. 2018, 9, 519. [Google Scholar] [CrossRef]
- Renna, L.V.; Bosè, F.; Brigonzi, E.; Fossati, B.; Meola, G.; Cardani, R. Aberrant insulin receptor expression is associated with insulin resistance and skeletal muscle atrophy in myotonic dystrophies. PLoS ONE 2019, 14, e0214254. [Google Scholar] [CrossRef]
- Luck, K.; Kim, D.K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef]
- Ganassi, M.; Badodi, S.; Wanders, K.; Zammit, P.S.; Hughes, S.M. Myogenin is an essential regulator of adult myofibre growth and muscle stem cell homeostasis. eLife 2020, 9, e60445. [Google Scholar] [CrossRef]
- Gnazzo, M.; Lepri, F.R.; Dentici, M.L.; Capolino, R.; Pisaneschi, E.; Agolini, E.; Rinelli, M.; Alesi, V.; Versacci, P.; Genovese, S.; et al. KBG syndrome: Common and uncommon clinical features based on 31 new patients. Am. J. Med. Genet. Part A 2020, 182, 1073–1083. [Google Scholar] [CrossRef]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Go, C.D.; Knight, J.D.R.; Rajasekharan, A.; Rathod, B.; Hesketh, G.G.; Abe, K.T.; Youn, J.Y.; Samavarchi-Tehrani, P.; Zhang, H.; Zhu, L.Y.; et al. A proximity-dependent biotinylation map of a human cell. Nature 2021, 595, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shen, X.; Cao, X.; He, H.; Han, S.; Chen, Y.; Cui, C.; Wei, Y.; Wang, Y.; Li, D.; et al. HDAC4 Regulates the Proliferation, Differentiation and Apoptosis of Chicken Skeletal Muscle Satellite Cells. Animals 2020, 10, 84. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Won, K.J.; Lee, K.P.; Jung, S.H.; Baek, S.; Chung, H.W.; Choi, W.S.; Lee, H.M.; Lee, B.H.; Jeon, B.H.; et al. Angiotensin II facilitates neointimal formation by increasing vascular smooth muscle cell migration: Involvement of APE/Ref-1-mediated overexpression of sphingosine-1-phosphate receptor 1. Toxicol. Appl. Pharmacol. 2018, 347, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, L.; Song, Y.; Zhang, M.; Shan, D.; Liu, Y.; Fang, M.; Lv, F.; Xiao, R.P.; Zhang, Y. β-arrestin 2 mediates cardiac ischemia-reperfusion injury via inhibiting GPCR-independent cell survival signalling. Cardiovasc. Res. 2017, 113, 1615–1626. [Google Scholar] [CrossRef]
- Rao, S.; Mondragón, L.; Pranjic, B.; Hanada, T.; Stoll, G.; Köcher, T.; Zhang, P.; Jais, A.; Lercher, A.; Bergthaler, A.; et al. AIF-regulated oxidative phosphorylation supports lung cancer development. Cell Res. 2019, 29, 579–591. [Google Scholar] [CrossRef]
- Song, C.; Mitter, S.K.; Qi, X.; Beli, E.; Rao, H.V.; Ding, J.; Ip, C.S.; Gu, H.; Akin, D.; Dunn, W.A., Jr.; et al. Oxidative stress-mediated NFκB phosphorylation upregulates p62/SQSTM1 and promotes retinal pigmented epithelial cell survival through increased autophagy. PLoS ONE 2017, 12, e0171940. [Google Scholar] [CrossRef]
- Noda, S.; Suzuki, Y.; Yamamura, H.; Giles, W.R.; Imaizumi, Y. Roles of LRRC26 as an auxiliary γ1-subunit of large-conductance Ca(2+)-activated K(+) channels in bronchial smooth muscle cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 318, L366–L375. [Google Scholar] [CrossRef]
- Evanson, K.W.; Bannister, J.P.; Leo, M.D.; Jaggar, J.H. LRRC26 is a functional BK channel auxiliary γ subunit in arterial smooth muscle cells. Circ. Res. 2014, 115, 423–431. [Google Scholar] [CrossRef]
- More, D.A.; Kumar, A. SRSF3: Newly discovered functions and roles in human health and diseases. Eur. J. Cell Biol. 2020, 99, 151099. [Google Scholar] [CrossRef]
- Cowper, A.E.; Cáceres, J.F.; Mayeda, A.; Screaton, G.R. Serine-arginine (SR) protein-like factors that antagonize authentic SR proteins and regulate alternative splicing. J. Biol. Chem. 2001, 276, 48908–48914. [Google Scholar] [CrossRef] [PubMed]
- Buskin, A.; Zhu, L.; Chichagova, V.; Basu, B.; Mozaffari-Jovin, S.; Dolan, D.; Droop, A.; Collin, J.; Bronstein, R.; Mehrotra, S.; et al. Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nat. Commun. 2018, 9, 4234. [Google Scholar] [CrossRef] [PubMed]
- Nikonova, E.; Kao, S.Y.; Spletter, M.L. Contributions of alternative splicing to muscle type development and function. Semin. Cell Dev. Biol. 2020, 104, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Nakka, K.; Ghigna, C.; Gabellini, D.; Dilworth, F.J. Diversification of the muscle proteome through alternative splicing. Skelet. Muscle 2018, 8, 8. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Huang, Y.; Wang, S.; Zhang, L.; Gao, H.; Zhao, Y.; E, G. Hereditary Basis of Coat Color and Excellent Feed Conversion Rate of Red Angus Cattle by Next-Generation Sequencing Data. Animals 2022, 12, 1509. https://doi.org/10.3390/ani12121509
He Y, Huang Y, Wang S, Zhang L, Gao H, Zhao Y, E G. Hereditary Basis of Coat Color and Excellent Feed Conversion Rate of Red Angus Cattle by Next-Generation Sequencing Data. Animals. 2022; 12(12):1509. https://doi.org/10.3390/ani12121509
Chicago/Turabian StyleHe, Yongmeng, Yongfu Huang, Shizhi Wang, Lupei Zhang, Huijiang Gao, Yongju Zhao, and Guangxin E. 2022. "Hereditary Basis of Coat Color and Excellent Feed Conversion Rate of Red Angus Cattle by Next-Generation Sequencing Data" Animals 12, no. 12: 1509. https://doi.org/10.3390/ani12121509
APA StyleHe, Y., Huang, Y., Wang, S., Zhang, L., Gao, H., Zhao, Y., & E, G. (2022). Hereditary Basis of Coat Color and Excellent Feed Conversion Rate of Red Angus Cattle by Next-Generation Sequencing Data. Animals, 12(12), 1509. https://doi.org/10.3390/ani12121509






