Differential mRNA Expression Profiling Reveals the Role of MiR-375 in Inflammation of Bovine Mammary Epithelial Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Transient Transfection of MiR-375 Inhibitor
2.3. Sample Collection and RNA Extraction
2.4. Constructing cDNA Libraries and High-Throughput Sequencing
2.5. Data Processing and Quality Control
2.6. Analysis of Differentially Expressed mRNAs
2.7. qPCR
2.8. Functional Annotation of DIE-mRNAs
3. Results
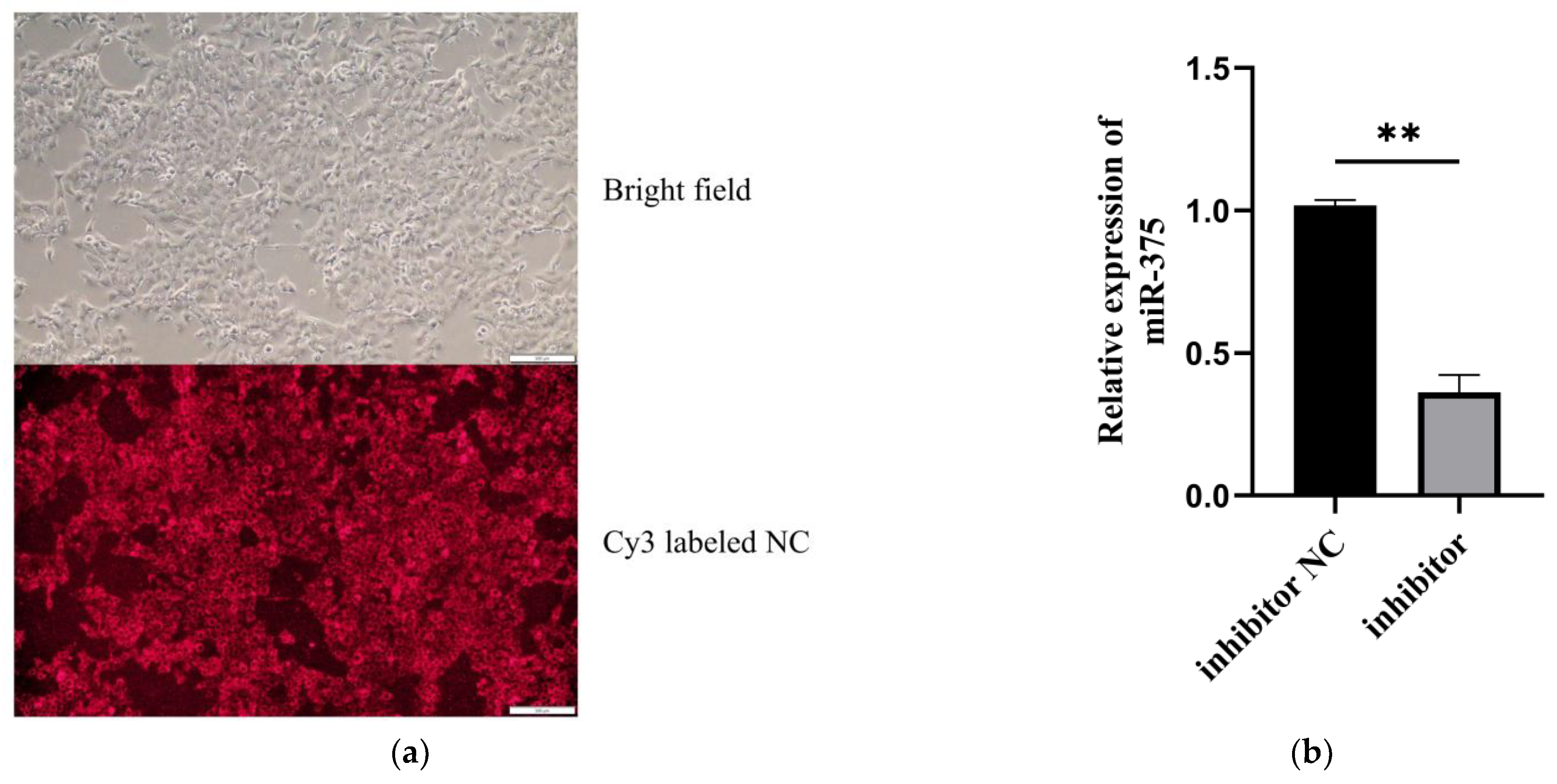
3.1. The Silencing Efficiency of MiR-375 in MAC-T
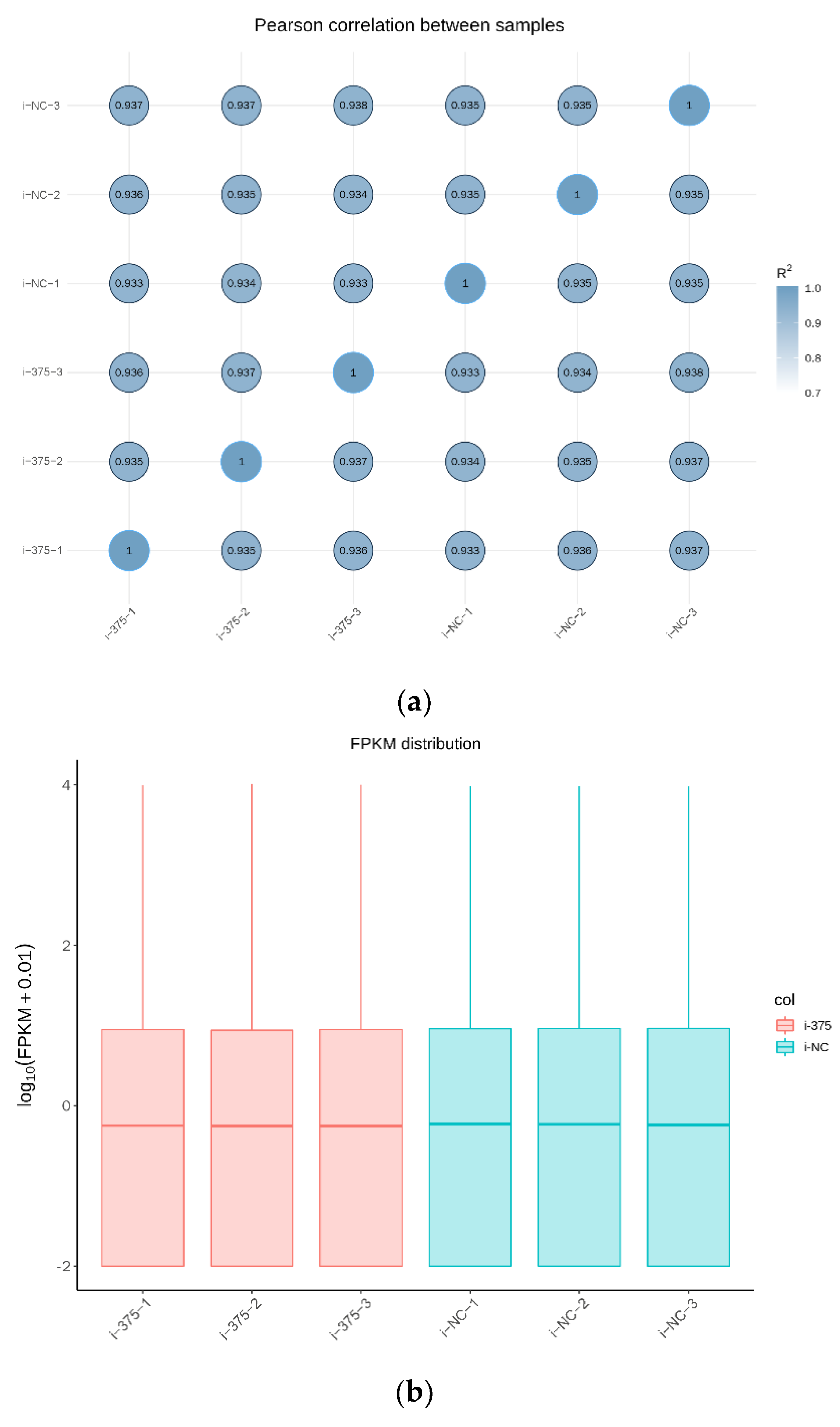
3.2. Sequence Data Analysis
3.3. Overall Distribution of mRNA Expression
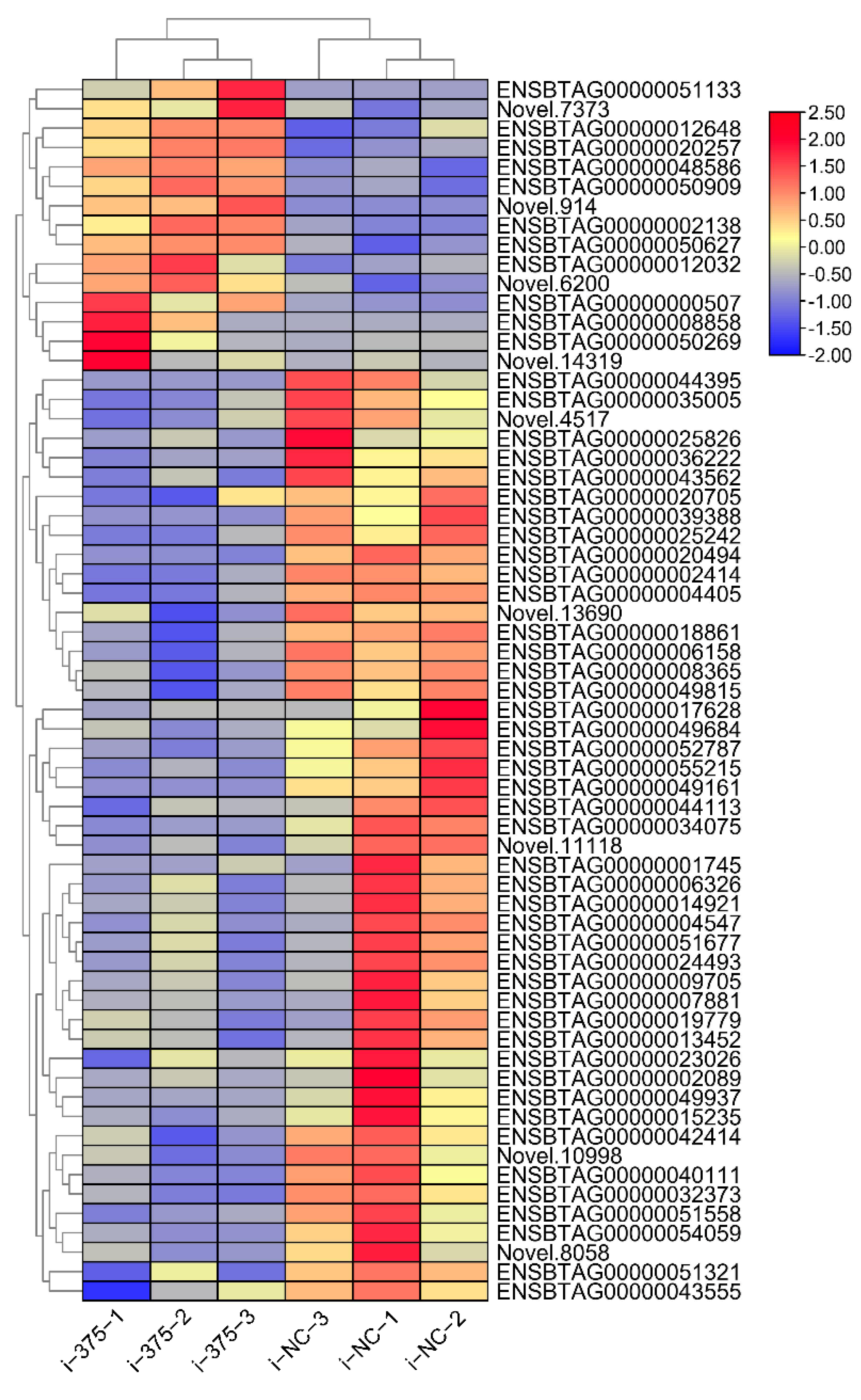
3.4. The Differently Expressed mRNAs in MAC-T after MiR-375 Inhibition
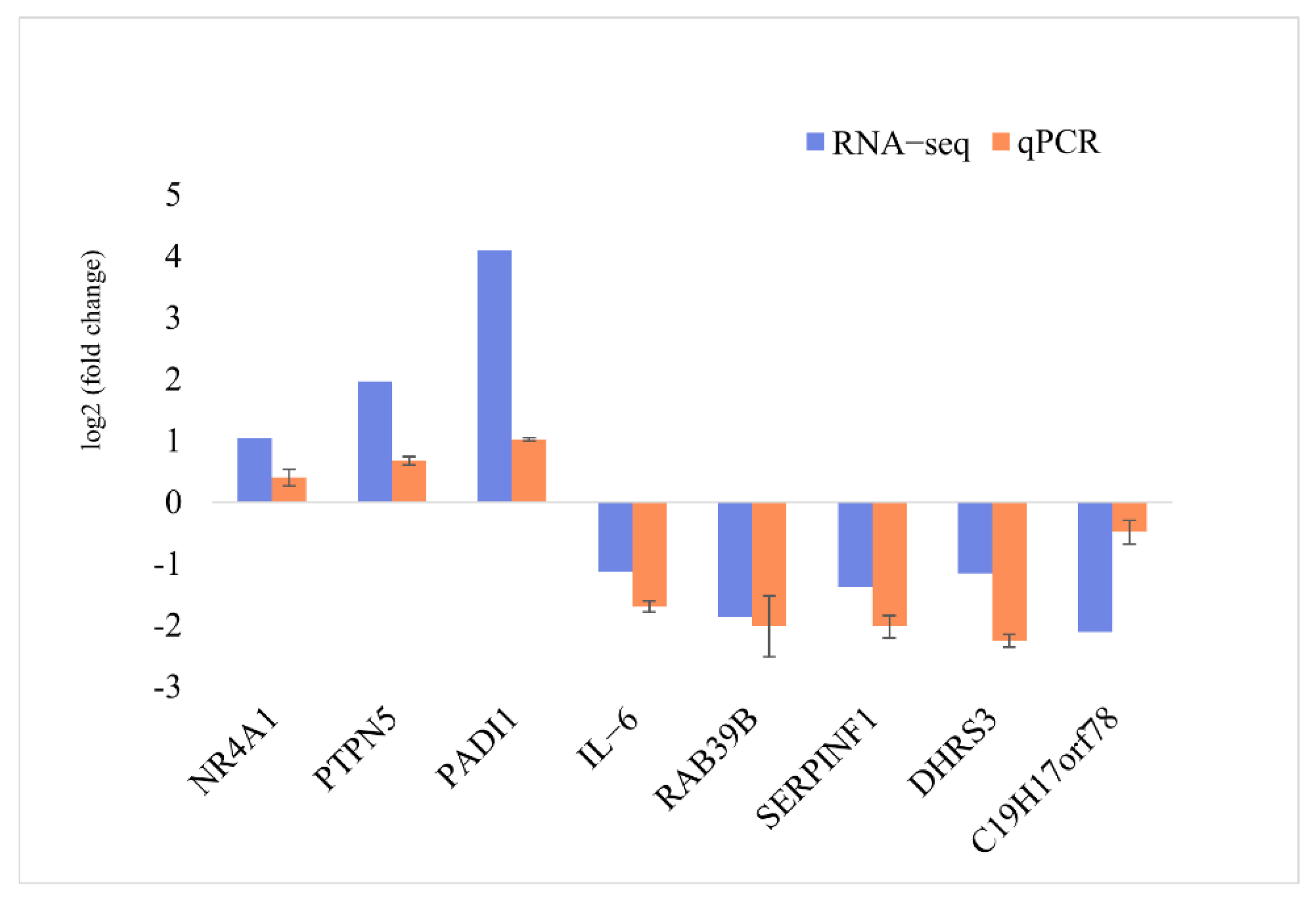
3.5. Validation of the DIE-mRNAs Using qPCR
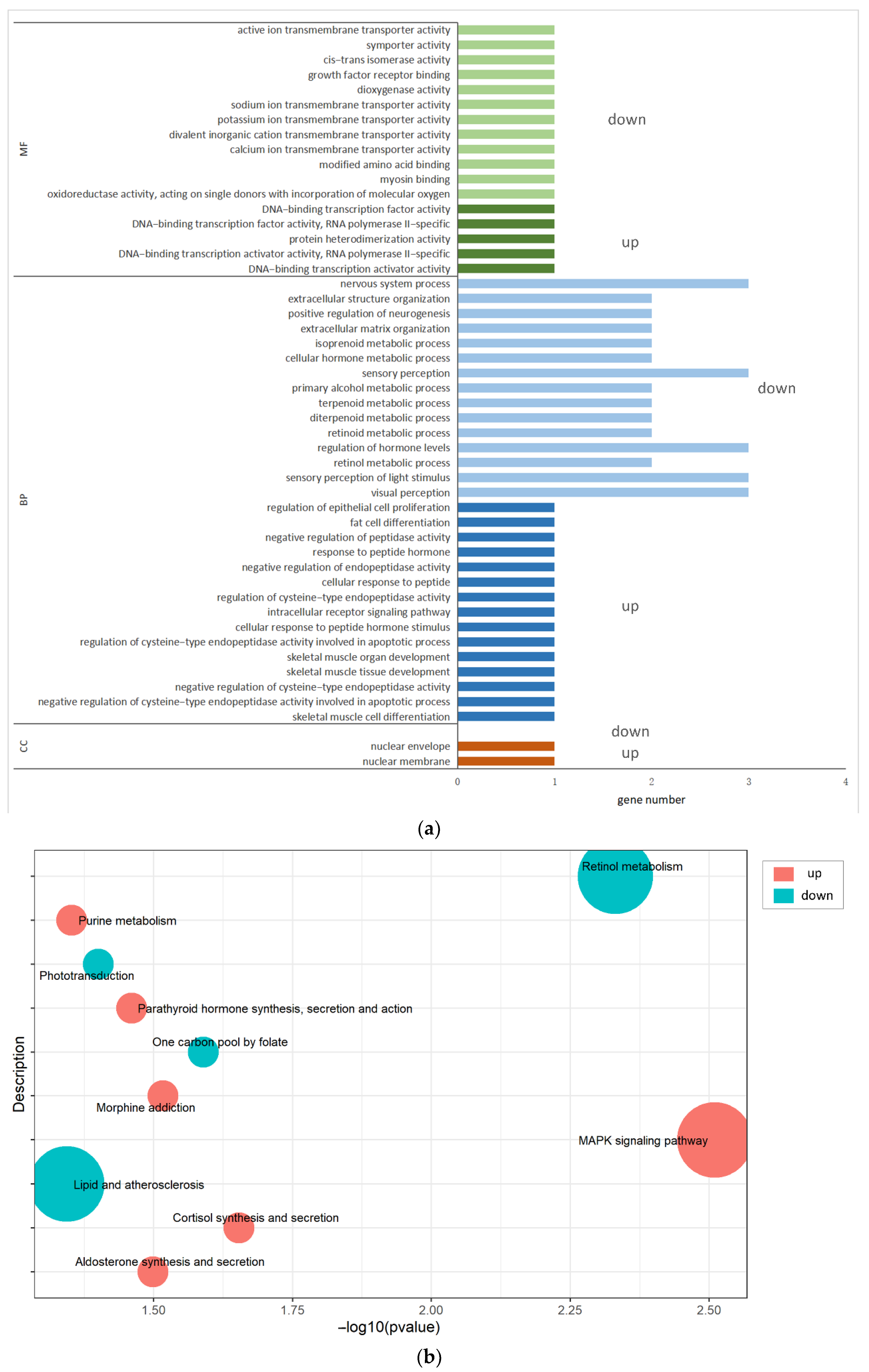
3.6. GO Enrichment and KEGG Analyses of the DIE-mRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharun, K.; Dhama, K.; Tiwari, R.; Gugjoo, M.B.; Iqbal Yatoo, M.; Patel, S.K.; Pathak, M.; Karthik, K.; Khurana, S.K.; Singh, R.; et al. Advances in therapeutic and managemental approaches of bovine mastitis: A comprehensive review. Vet. Q. 2021, 41, 107–136. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, A.; Imran, M. Causes, types, etiological agents, prevalence, diagnosis, treatment, prevention, effects on human health and future aspects of bovine mastitis. Anim. Health Res. Rev. 2020, 21, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, Y.; Kiku, Y.; Sugawara, K.; Tanabe, F.; Hayashi, T. Exfoliation rate of mammary epithelial cells in milk on bovine mastitis caused by Staphylococcus aureus is associated with bacterial load. Anim. Sci. J. 2018, 89, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Shen, L.; Jiang, J.; Xu, Q.; Luo, Z.; Luo, Q.; Yu, S.; Yao, X.; Ren, Z.; Hu, Y.; et al. Metabolomic profiles of bovine mammary epithelial cells stimulated by lipopolysaccharide. Sci. Rep. 2019, 9, 19131. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zhao, F.; Liu, J.; Liu, H. ASCT2 is involved in SARS-Mediated β-Casein synthesis of bovine mammary epithelial cells with methionine supply. J. Agr. Food Chem. 2020, 68, 13038–13045. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, P.; Iqbal, A.; Ali, S.; Gao, Z.; Pan, Z.; Xia, L.; Yin, F.; Zhao, Z. MiR-485 targets the DTX4 gene to regulate milk fat synthesis in bovine mammary epithelial cells. Sci. Rep. 2021, 11, 7623. [Google Scholar] [CrossRef]
- Islam, M.A.; Takagi, M.; Fukuyama, K.; Komatsu, R.; Albarracin, L.; Nochi, T.; Suda, Y.; Ikeda-Ohtsubo, W.; Rutten, V.; Eden, W.V.; et al. Transcriptome analysis of the inflammatory responses of bovine mammary epithelial cells: Exploring immunomodulatory target genes for bovine mastitis. Pathogens 2020, 9, 200. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram, K.H.; Soleymani, F.S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Jin, W.; Ibeagha-Awemu, E.M.; Liang, G.; Beaudoin, F.; Zhao, X.; Guan, L.L. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC Genom. 2014, 15, 181. [Google Scholar] [CrossRef]
- Shen, B.; Zhang, L.; Lian, C.; Lu, C.; Zhang, Y.; Pan, Q.; Yang, R.; Zhao, Z. Deep sequencing and screening of differentially expressed MicroRNAs related to milk fat metabolism in bovine primary mammary epithelial cells. Int. J. Mol. Sci. 2016, 17, 200. [Google Scholar] [CrossRef]
- Chen, Z.; Xu, X.; Tan, T.; Chen, D.; Liang, H.; Sun, K.; Li, M.; Zhang, H.; Mao, Y.; Yang, Z. MicroRNA-145 regulates immune cytokines via targeting FSCN1 in Staphylococcus aureus-induced mastitis in dairy cows. Reprod. Domest. Anim. 2019, 54, 882–891. [Google Scholar] [CrossRef]
- Luoreng, Z.; Wei, D.; Wang, X. MiR-125b regulates inflammation in bovine mammary epithelial cells by targeting the NKIRAS2 gene. Vet. Res. 2021, 52, 122. [Google Scholar] [CrossRef]
- Wang, X.; Luoreng, Z.; Zan, L.; Li, F.; Li, N. Bovine miR-146a regulates inflammatory cytokines of bovine mammary epithelial cells via targeting the TRAF6 gene. J. Dairy Sci. 2017, 100, 7648–7658. [Google Scholar] [CrossRef]
- Cai, M.; Fan, W.; Li, X.; Sun, H.; Dai, L.; Lei, D.; Dai, Y.; Liao, Y. The regulation of Staphylococcus aureus-induced inflammatory responses in bovine mammary epithelial cells. Front. Vet. Sci. 2021, 8, 683886. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, M.; Kong, L.; Liu, J.; Wang, Y.; Song, C.; Chen, X.; Lai, M.; Fang, X.; Chen, H.; et al. MiR-204-5p promotes lipid synthesis in mammary epithelial cells by targeting SIRT1. Biochem. Biophys. Res. Commun. 2020, 533, 1490–1496. [Google Scholar] [CrossRef]
- Poy, M.N.; Eliasson, L.; Krutzfeldt, J.; Kuwajima, S.; Ma, X.; Macdonald, P.E.; Pfeffer, S.; Tuschl, T.; Rajewsky, N.; Rorsman, P.; et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432, 226–230. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, Q.; Li, M.; Jiang, S.; Wang, X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem. Biophys. Res. Commun. 2014, 444, 199–204. [Google Scholar] [CrossRef]
- Li, H.; Li, Z.; Pi, Y.; Chen, Y.; Mei, L.; Luo, Y.; Xie, J.; Mao, X. MicroRNA-375 exacerbates knee osteoarthritis through repressing chondrocyte autophagy by targeting ATG2B. Aging 2020, 12, 7248–7261. [Google Scholar] [CrossRef]
- Zhao, S.; Yu, C.; Xiang, K.; Yang, M.; Liu, Z.; Yang, B. MiR-375 inhibits autophagy and further promotes inflammation and apoptosis of acinar cells by targeting ATG7. Pancreas 2020, 49, 543–551. [Google Scholar] [CrossRef]
- Xu, J.; Song, W.; Lu, F.; Dai, Z.; Cao, L.; Lin, S. MiR-375 inhibits the proliferation and invasion of nasopharyngeal carcinoma cells by suppressing PDK1. Biomed. Res. Int. 2020, 2020, 9704245. [Google Scholar] [CrossRef]
- Zehentmayr, F.; Hauser-Kronberger, C.; Zellinger, B.; Hlubek, F.; Schuster, C.; Bodenhofer, U.; Fastner, G.; Deutschmann, H.; Steininger, P.; Reitsamer, R.; et al. Hsa-miR-375 is a predictor of local control in early stage breast cancer. Clin. Epigenetics 2016, 8, 28. [Google Scholar] [CrossRef]
- Luoreng, Z.; Wang, X.; Mei, C.; Zan, L. Comparison of microRNA Profiles between Bovine Mammary Glands Infected with Staphylococcus aureus and Escherichia coli. Int. J. Biol. Sci. 2018, 14, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Huynh, H.T.; Robitaille, G.; Turner, J.D. Establishment of bovine mammary epithelial cells (MAC-T): An in vitro model for bovine lactation. Exp. Cell Res. 1991, 197, 191–199. [Google Scholar] [CrossRef]
- Wang, J.; Hu, Q.; Yang, J.; Luoreng, Z.; Wang, X.; Ma, Y.; Wei, D. Differential expression profiles of lncRNA following LPS-induced inflammation in bovine mammary epithelial cells. Front. Vet. Sci. 2021, 8, 758488. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hu, Q.; Wang, J.; Ren, Q.; Wang, X.; Luoreng, Z.; Wei, D.; Ma, Y. RNA-Seq reveals the role of miR-29c in regulating inflammation and oxidative stress of bovine mammary epithelial cells. Front. Vet. Sci. 2022, 9, 865415. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative Real-Time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Bougarn, S.; Cunha, P.; Gilbert, F.B.; Meurens, F.; Rainard, P. Technical note: Validation of candidate reference genes for normalization of quantitative PCR in bovine mammary epithelial cells responding to inflammatory stimuli. J. Dairy Sci. 2011, 94, 2425–2430. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Arthur, J.S.C.; Ley, S.C. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 2013, 13, 679–692. [Google Scholar] [CrossRef]
- Kabiri, Z.; Greicius, G.; Zaribafzadeh, H.; Hemmerich, A.; Counter, C.M.; Virshup, D.M. Wnt signaling suppresses MAPK-driven proliferation of intestinal stem cells. J. Clin. Investig. 2018, 128, 3806–3812. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, W.; Liu, T.; Feng, X.; Yang, N.; Zhou, H. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. 2015, 35, 600–604. [Google Scholar] [CrossRef]
- Wang, J.; Guo, C.; Wei, Z.; He, X.; Kou, J.; Zhou, E.; Yang, Z.; Fu, Y. Morin suppresses inflammatory cytokine expression by downregulation of nuclear factor-κB and mitogen-activated protein kinase (MAPK) signaling pathways in lipopolysaccharide-stimulated primary bovine mammary epithelial cells. J. Dairy Sci. 2016, 99, 3016–3022. [Google Scholar] [CrossRef]
- Maxwell, M.A.; Muscat, G.E.O. The NR4A subgroup: Immediate early response genes with pleiotropic physiological roles. Nucl. Recept. Signal. 2006, 4, e002. [Google Scholar] [CrossRef]
- Rodriguez-Calvo, R.; Tajes, M.; Vazquez-Carrera, M. The NR4A subfamily of nuclear receptors: Potential new therapeutic targets for the treatment of inflammatory diseases. Expert Opin. Ther. Targets 2017, 21, 291–304. [Google Scholar] [CrossRef]
- Pei, L.; Castrillo, A.; Chen, M.; Hoffmann, A.; Tontonoz, P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J. Biol. Chem. 2005, 280, 29256–29262. [Google Scholar] [CrossRef]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-kappaB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Mitchell, J.P.; Carmody, R.J. NF-κB and the transcriptional control of inflammation. Int. Rev. Cell Mol. Biol. 2018, 335, 41–84. [Google Scholar] [CrossRef]
- Ismail, H.; Mofarrahi, M.; Echavarria, R.; Harel, S.; Verdin, E.; Lim, H.W.; Jin, Z.; Sun, J.; Zeng, H.; Hussain, S.N.A. Angiopoietin-1 and vascular endothelial growth factor regulation of leukocyte adhesion to endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1707–1716. [Google Scholar] [CrossRef]
- Li, L.; Liu, Y.; Chen, H.; Li, F.; Wu, J.; Zhang, H.; He, J.; Xing, Y.; Chen, Y.; Wang, W.; et al. Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation. Nat. Chem. Biol. 2015, 11, 339–346. [Google Scholar] [CrossRef]
- Hanna, R.N.; Shaked, I.; Hubbeling, H.G.; Punt, J.A.; Wu, R.; Herrley, E.; Zaugg, C.; Pei, H.; Geissmann, F.; Ley, K.; et al. NR4A1 (Nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circ. Res. 2012, 110, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Song, J.; Zhao, M. NR4A1 regulates cerebral ischemia-induced brain injury by regulating neuroinflammation through interaction with NF-κB/p65. Biochem. Biophys. Res. Commun. 2019, 518, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, S.; He, X.; Guo, P.; Lu, X.; Wang, J.; Li, J.; Wu, H. Nur77-mediated TRAF6 signalling protects against LPS-induced sepsis in mice. J. Inflamm. 2016, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wilmanns, M.; Thornton, J.; Köhn, M. Elucidating human phosphatase-substrate networks. Sci. Signal. 2013, 6, s10. [Google Scholar] [CrossRef]
- Tonks, N.K. Protein tyrosine phosphatases-from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013, 280, 346–378. [Google Scholar] [CrossRef]
- Stanford, S.M.; Bottini, N. Targeting tyrosine phosphatases: Time to end the stigma. Trends Pharm. Sci. 2017, 38, 524–540. [Google Scholar] [CrossRef]
- Alonso, A.; Sasin, J.; Bottini, N.; Friedberg, I.; Friedberg, I.; Osterman, A.; Godzik, A.; Hunter, T.; Dixon, J.; Mustelin, T. Protein tyrosine phosphatases in the human genome. Cell 2004, 117, 699–711. [Google Scholar] [CrossRef]
- Zhang, Z. Drugging the undruggable: Therapeutic potential of targeting protein tyrosine phosphatases. Acc. Chem. Res. 2017, 50, 122–129. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, Z. Regulatory mechanisms and novel therapeutic targeting strategies for protein tyrosine phosphatases. Chem. Rev. 2018, 118, 1069–1091. [Google Scholar] [CrossRef]
- Zong, M.; Yuan, H.; He, X.; Zhou, Z.; Qiu, X.; Yang, J.; Ji, M. Disruption of striatal-enriched protein tyrosine phosphatase signaling might contribute to memory impairment in a mouse model of sepsis-associated encephalopathy. Neurochem. Res. 2019, 44, 2832–2842. [Google Scholar] [CrossRef]
- Mahaman, Y.A.R.; Huang, F.; Embaye, K.S.; Wang, X.; Zhu, F. The implication of STEP in synaptic plasticity and cognitive impairments in alzheimer’s disease and other neurological disorders. Front. Cell Dev. Biol. 2021, 9, 680118. [Google Scholar] [CrossRef]
- Keyse, S.M. Protein phosphatases and the regulation of mitogen-activated protein kinase signalling. Curr. Opin. Cell Biol. 2000, 12, 186–192. [Google Scholar] [CrossRef]
- Palaniappan, M.; Edwards, D.; Creighton, C.J.; Medina, D.; Conneely, O.M. Reprogramming of the estrogen responsive transcriptome contributes to tamoxifen-dependent protection against tumorigenesis in the p53 null mammary epithelial cells. PLoS ONE 2018, 13, e194913. [Google Scholar] [CrossRef]
- Rajagopal, S.; Deb, I.; Poddar, R.; Paul, S. Aging is associated with dimerization and inactivation of the brain-enriched tyrosine phosphatase STEP. Neurobiol. Aging 2016, 41, 25–38. [Google Scholar] [CrossRef][Green Version]
- Rajagopal, S.; Yang, C.; DeMars, K.M.; Poddar, R.; Candelario-Jalil, E.; Paul, S. Regulation of post-ischemic inflammatory response: A novel function of the neuronal tyrosine phosphatase STEP. Brain Behav. Immun. 2021, 93, 141–155. [Google Scholar] [CrossRef]
- Zhang, Y.; Kurup, P.; Xu, J.; Carty, N.; Fernandez, S.M.; Nygaard, H.B.; Pittenger, C.; Greengard, P.; Strittmatter, S.M.; Nairn, A.C.; et al. Genetic reduction of striatal-enriched tyrosine phosphatase (STEP) reverses cognitive and cellular deficits in an Alzheimer’s disease mouse model. Proc. Natl. Acad. Sci. USA 2010, 107, 19014–19019. [Google Scholar] [CrossRef]
- Akhtar, M.; Guo, S.; Guo, Y.F.; Zahoor, A.; Shaukat, A.; Chen, Y.; Umar, T.; Deng, P.G.; Guo, M. Upregulated-gene expression of pro-inflammatory cytokines (TNF-alpha, IL-1beta and IL-6) via TLRs following NF-kappaB and MAPKs in bovine mastitis. Acta Trop. 2020, 207, 105458. [Google Scholar] [CrossRef]
- Hagiwara, K.; Yamanaka, H.; Hisaeda, K.; Taharaguchi, S.; Kirisawa, R.; Iwai, H. Concentrations of IL-6 in serum and whey from healthy and mastitic cows. Vet. Res. Commun. 2001, 25, 99–108. [Google Scholar] [CrossRef]
| Gene Name | Primer Sequence (5′-3′) | Product Length/bp |
|---|---|---|
| miR-375 | F: CTCTGCTTTTGTTCGTTCGG | 64 |
| R: AGTGCAGGGTCCGAGGTATT | ||
| NR4A1 | F: CGGCTTTGCTGAACTGTCTC | 80 |
| R: CCAGACGGAGGATAAAGAGC | ||
| PTPN5 | F: AGGGCTTCGGCTATCTCAT | 228 |
| R: TGTGAGGGTTGGGAAGGAT | ||
| PADI1 | F: ACCTGTCCTACGCAGTGGC | 139 |
| R: TCAGGCAGGGTCTTGGTG | ||
| IL-6 | F: CACTCCATTCGCTGTCT | 227 |
| R: GTGTCTCCTTGCTGCTT | ||
| RAB39B | F: TTACCAACCGCAGGTCTTTCC | 164 |
| R: ATACGCAGCAGCCAGTTTCTC | ||
| SERPINF1 | F: CGCCAATGTGCTGCTGTCT | 135 |
| R: CCGTGGATGTCTGGGTTACTG | ||
| DHRS3 | F: AATGCCTGAAGGAGACGACG | 224 |
| R: GGTGTTGATGTGCTGGGACTT | ||
| C19H17orf78 | F: TGGTTCTGGGAGCAAAGTGA | 271 |
| R: AGGAGTTACGGAGGTAGTAGTGG | ||
| GADPH | F: GGCATCGTGGAGGGACTTATG | 186 |
| R: GCCAGTGAGCTTCCCGTTGAG | ||
| RPS18 | F: GTGGTGTTGAGGAAAGCAGACA | 79 |
| R: TGATCACACGTTCCACCTCATC |
| Group | Sample | Raw Reads | Error (%) | Q30 (%) | GC (%) | Clean Reads | Clean Bases | Total Mapped |
|---|---|---|---|---|---|---|---|---|
| Inhibitor group | i-375-1 | 43,314,930 | 0.03 | 93.24 | 47.38 | 43,038,262 | 5.95G | 41,300,139 (95.96%) |
| i-375-2 | 42,091,914 | 0.03 | 92.55 | 47.27 | 41,737,124 | 5.77G | 39,883,875 (95.56%) | |
| i-375-3 | 46,367,836 | 0.03 | 93.29 | 47.23 | 46,051,060 | 6.37G | 44,258,208 (96.11%) | |
| Inhibitor NC group | i-NC-1 | 41,433,410 | 0.03 | 93.02 | 47.07 | 41,141,648 | 5.7G | 39,472,120 (95.94%) |
| i-NC-2 | 46,829,314 | 0.03 | 92.6 | 47.08 | 46,481,256 | 6.42G | 44,516,188 (95.77%) | |
| i-NC-3 | 46,103,512 | 0.03 | 93.28 | 47.05 | 45,797,992 | 6.33G | 44,030,956 (96.14%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Hu, Q.; Luoreng, Z.; Yang, J.; Wang, X.; Ma, Y.; Wei, D. Differential mRNA Expression Profiling Reveals the Role of MiR-375 in Inflammation of Bovine Mammary Epithelial Cells. Animals 2022, 12, 1431. https://doi.org/10.3390/ani12111431
Li Y, Hu Q, Luoreng Z, Yang J, Wang X, Ma Y, Wei D. Differential mRNA Expression Profiling Reveals the Role of MiR-375 in Inflammation of Bovine Mammary Epithelial Cells. Animals. 2022; 12(11):1431. https://doi.org/10.3390/ani12111431
Chicago/Turabian StyleLi, Yuhang, Qichao Hu, Zhuoma Luoreng, Jian Yang, Xingping Wang, Yun Ma, and Dawei Wei. 2022. "Differential mRNA Expression Profiling Reveals the Role of MiR-375 in Inflammation of Bovine Mammary Epithelial Cells" Animals 12, no. 11: 1431. https://doi.org/10.3390/ani12111431
APA StyleLi, Y., Hu, Q., Luoreng, Z., Yang, J., Wang, X., Ma, Y., & Wei, D. (2022). Differential mRNA Expression Profiling Reveals the Role of MiR-375 in Inflammation of Bovine Mammary Epithelial Cells. Animals, 12(11), 1431. https://doi.org/10.3390/ani12111431







