Simple Summary
Fatness-related traits are economically very important in sheep production and are associated with serious diseases in humans. Using a denser set of SNP markers and a variety of statistical approaches, our results were able to refine the regions associated with fat deposition and to suggest new insights into molecular aspects of fat tail selection. These results may provide a strong foundation for studying the regulation of fat deposition in sheep and do offer hope that the causal mutations and the mode of inheritance of this trait will soon be discovered by further investigation.
Abstract
The fat tail is a phenotype that divides indigenous Iranian sheep genetic resources into two major groups. The objective of the present study is to refine the map location of candidate regions associated with fat deposition, obtained via two separate whole genome scans contrasting thin and fat tail breeds, and to determine the nature of the selection occurring in these regions using a hitchhiking approach. Zel (thin tail) and Lori-Bakhtiari (fat tail) breed samples that had previously been run on the Illumina Ovine 50 k BeadChip, were genotyped with a denser set of SNPs in the three candidate regions using a Sequenom Mass ARRAY platform. Statistical tests were then performed using different and complementary methods based on either site frequency (FST and Median homozygosity) or haplotype (iHS and XP-EHH). The results from candidate regions on chromosome 5 and X revealed clear evidence of selection with the derived haplotypes that was consistent with selection to near fixation for the haplotypes affecting fat tail size in the fat tail breed. An analysis of the candidate region on chromosome 7 indicated that selection differentiated the beneficial alleles between breeds and homozygosity has increased in the thin tail breed which also had the ancestral haplotype. These results enabled us to confirm the signature of selection in these regions and refine the critical intervals from 113 kb, 201 kb, and 2831 kb to 28 kb, 142 kb, and 1006 kb on chromosome 5, 7, and X respectively. These regions contain several genes associated with fat metabolism or developmental processes consisting of TCF7 and PPP2CA (OAR5), PTGDR and NID2 (OAR7), AR, EBP, CACNA1F, HSD17B10, SLC35A2, BMP15, WDR13, and RBM3 (OAR X), and each of which could potentially be the actual target of selection. The study of core haplotypes alleles in our regions of interest also supported the hypothesis that the first domesticated sheep were thin tailed, and that fat tail animals were developed later. Overall, our results provide a comprehensive assessment of how and where selection has affected the patterns of variation in candidate regions associated with fat deposition in thin and fat tail sheep breeds.
1. Introduction
Identifying the regions of the genome that have experienced substantial selective pressure can provide a powerful tool into the location of functionally important polymorphisms and can help prioritize targets for association mapping [1,2]. Fat tailed breeds comprise approximately 25% of the world sheep population [3] and are grazed in a wide range of countries. The major biological role of the fat tail is to serve as an energy store against periodic food scarcity, such as in drought and winter [4]. It has also been used as a source of fat (or ghee) for human consumption for millennia [5]. Historically, the climatic conditions in these areas, as well as the nutritional and religious food requirements of the people, encouraged sheep producers to select for higher fat tail weight [6]. Although fat-tailed sheep breeds are still preferred under local pastoral, desert, or semi-desert conditions in several societies [7], most of the advantages of a large fat tail have recently reduced in importance due to the improved forage availability and decreased price for the product, especially in genetic improvement programs [4,8].
Various genomic-based studies have been conducted to understand the genetic basis and genomic architecture of sheep tails and to find the specific causal genomic variant(s) contributing to sheep tail pattern making [3,9,10,11,12,13,14,15]. A brief overview on genomic outcomes, including proposed potential genes generated from investigations on the sheep tail phenotype has been reported by Kalds et al. [7]. Additionally, transcriptomic analyses were performed, providing sets of potential genes that contribute to the formation and the biological emergence of sheep tails [16,17,18,19]. Taken together, these research efforts have revealed several high-ranking candidate genes with no current consistency or solid opinion about their variant causalities and expression nature. Therefore, to date the genetic mechanism underlying this particular trait has not been fully elucidated and finding the regions associated with fat deposition and the nature of the selection occurring in these locations are two of the most important and challenging areas of research in the countries grazing these breeds.
Genome scan approaches, such as quantitative trait locus (QTL) and hitchhiking mapping, provide the opportunity for investigating the genetic regions associated with different traits. QTL mapping studies have been applied to examine the genetic basis of various economically important traits, e.g., [20,21]. However, QTL mapping basically relies on detecting correlations between genetic markers and phenotypic traits in a segregating population [22]. Also, many traits underlying adaptive divergence are not always easily detectable at the phenotypic level and not well suited to QTL mapping. In this case, one approach for discovering the potential genetic basis of different traits is to use hitchhiking mapping [23,24]. Hitchhiking mapping is a population genetics approach use to identify genomic regions influenced by selection [25,26]. A great advantage of this approach is that it can be performed using molecular markers alone [27,28]. This approach is especially suitable for the traits such as sheep tail pattern where all sheep breeds can be easily classified in different classes (e.g., thin and fat tailed sheep groups) and then, offer ideal materials for comparative analysis of their genetic basis.
Hitchhiking mapping starts with a genome scan using an approximately uniformly spaced set of molecular markers followed by a fine scale analysis of the candidate regions [23]. For example, a marker-based analysis of chromosome 1 in rats from warfarin-resistant populations revealed a 0.5 centimorgan (cM) region that was the likely locus for this trait [29]. This information was used later to identify the warfarin resistance gene through more traditional candidate gene approaches and association mapping techniques [30].
Genomic scans for finding candidate regions associated with fat deposition in thin and fat tailed breeds have been described previously [3]. In brief, two independent experiments, including Iranian and ovine HapMap genotyping data contrasting thin and fat tailed breeds were analyzed, and using different statistics, especially FST, three regions on chromosomes 5 (between 47,149–47,263 kb), 7 (between 46,642–46,843 kb) and X (between 58,621–61,452 kb) were confirmed in both data sets. Interestingly, these regions have also been supported in recently published studies using two different ways consisting of different breeds of thin and fat tailed sheep breeds through a genome-wide analysis of transcriptomic data [17,31] and selective sweeps [32,33], and also using same the breeds but different animals [34].
Unlike FST that has been used in the previous study [3], tests based on linkage disequilibrium (LD) like iHS [35] and XP-EHH [36], are multi-marker tests. These tests are commonly used for human SNP data, where there are now millions of SNPs available (approximately one marker per 1 kb), and that which depend on SNP spacing and frequency [37]. Densely spaced SNPs give greater power when using statistical tests that rely on LD, as signals of selection are less likely to be lost [38]. The Illumina Ovine SNP50k BeadChip, while providing uniform genome wide coverage, has a marker about every 56 kb [3]. Fine mapping, where more SNPs are genotyped in an area of interest, improves the ability to localize causal variants.
The allele frequency spectrum (FST and median homozygosity) and haplotype based (iHS and XP-EHH) statistics used in the current study have been shown by previous power analyses to be largely complementary [39]. FST and median homozygosity are single-marker tests and detect highly differentiated alleles between populations, where positive selection in one area causes larger frequency differences compared to neutrally evolving alleles. iHS, a measure of within breed evidence for selection, has a suitable power to detect partial selective sweeps. However, a disadvantage of this approach is that it loses power when the beneficial allele is close to fixation, because fixation will eliminate variation at and near the selected site. In contrast, XP-EHH can detect selected alleles that have risen to near fixation in one but not another population. Grossman et al. [40] showed that the FST and XP-EHH signals peaked more narrowly around the causal variant, making them useful for spatial localization, while iHS is better to distinguish causal variants and contributed little to spatial resolution. These tests were relatively uncorrelated in neutral regions, and only weakly correlated for neutral variants within selected regions [40].
In this study, three regions recognized as candidate regions associated with fat deposition in the tail were chosen for further analysis. This was achieved through fine-mapping with additional SNPs, using Sequenom technology. The aim of this study was to confirm the signature of selection in the candidate loci, narrow down the chromosomal region showing the selective imprint and to determine the nature of the selection process occurred in these regions. Our analyses focused on two site frequency (FST and Median homozygosity) and haplotype based (iHS and XP-EHH) statistics, and using this variety of selection sweep tests we were able to obtain some novel results associated with fat deposition in these breeds.
2. Materials and Methods
2.1. Animal Sampling and Genotyping
Two independent data sets consisting of Zel-Lori Bakhtiari and Ovine HapMap samples were previously used for a genome-wide scan analysis of selective sweeps in thin and fat tail breeds [3]. Because the latter project did not formally phenotype the individuals concerned and we did not have access to their DNA samples, in this research further analysis was performed for the Zel-Lori Bakhtiari data. This data set consisted of 45 samples each of the thin (Zel) and fat tailed (Lori Bakhtiari) breeds (37 Females and 8 Males per each breed). The animals were collected to be as unrelated as possible. Three regions (Table 1), identified as being under selection based on FST and median homozygosity results from the first genome scan, were chosen for further analysis.

Table 1.
Regions chosen for fine mapping in this study and the number of SNPs that were previously genotyped in these regions using the Ovine SNP50k BeadChip.
Once the regions of interest were defined, all known SNPs in each region were examined for suitability for genotyping. These included SNPs discovered on both the Solexa and 454 platforms (http://www.sheephapmap.org/genseq.php, accessed on 31 May 2017). A total of 156, 89, and 78 putative SNPs flanked by 150 bp on each side (301 bp total) were submitted to Sequenom’s primer design software (Mass ARRAY Assay Design 3.1) for the regions on chromosomes 5, 7, and X, respectively, and primers and probes were designed in multiplex format. Of these SNPs, assays were successfully designed for 140 (90%), 75 (84%), and 66 (84%) of all SNPs initially selected for study, and were grouped into 6, 3, and 4 multiplex assays for the three regions respectively. The remaining SNPs failed primer design, primarily due to high repeated content. The majority of these multiplex assays were located in the first and second plex which contained 71, 62, and 57 SNPs for chromosomes 5, 7, and X respectively. These two SNP assays were used for genotyping. Selected SNPs, their locations, PCR primers, unextended primer, and its extension masses, are represented in Supplementary materials Table S1. All SNPs were genotyped by use of the mass-spectrometry based iPlex Mass Array platform provided by Sequenom (Sequenom, San Diego, CA, USA. http://www.sequenom.com/, accessed on 31 May 2017). Experiments were conducted following the Sequenom iPLEX Assay application note [41].
2.2. Quality Control Filters
All samples with more than 30% missing data and subsequently all loci with more than 15% missing data were excluded. These rejection thresholds were chosen by plotting numbers of animals or loci against percent missing data and the cutoff point was determined as the curve inflection point, where the rate of change in the number of excluded loci, became linear with every increased percent of missing data [42]. For the remaining SNPs, those with a minor allele frequency (MAF) of less than 10% over all samples, and an outlier departure from Hardy—Weinberg equilibrium over all animals of a breed (p-value < 0.0002), were excluded [3,43]. We combined the SNP genotyping results from both the Ovine SNP50k BeadChip (in the regions of interest) and the Sequenom Mass ARRAY platform for final analysis. Across the three regions, 25 SNPs were genotyped on both Sequenom and Illumina platforms with a mean allelic concordance rate of 96% (range 92–99%). For the few SNPs with different genotypes, the Ovine SNP50k Bead Chip results were used due to their greater accuracy.
2.3. Estimates of FST and Median Homozygosity
To determine the pattern of positive selection, the basic form of Wright’s fixation index (FST) was calculated as described by MacEachern et al. [44]. The value of FST can theoretically range from zero (showing no differentiation) to one (indicating complete differentiation, i.e., populations are fixed for different alleles). For each set of five adjacent SNPs, the average of FST values was calculated and termed windowed FST. This is an approximate method of looking for regions where selection is apparent over multiple markers, rather than one-off high values. A window of five markers was chosen as it appeared to provide the better signal compare to other arbitrary window sizes [3]. The windowed FST values were then plotted against genome location.
All analyses presented in this work were also performed using Weir and Cockerham [45] and Hudson [46] methods. The results were almost identical for all FST estimators (r ≥ 99%), so that we have only presented FST results based on Wright’s estimator in the present study for an easier comparison to previous reported results [3]. All scripts for estimating Wright (FST), Weir and Cockerham, and Hudson’s fixation indices were written and performed in R v 4.0.2.
The selection for a new beneficial mutation increases the level of homozygosity in the selected allele and neighboring regions due to the hitchhiking effect. Therefore, one method of looking for the region where selection has taken place is to compare median runs of homozygosity between breeds. To do this, the median run of homozygosity for each SNP was calculated following Moradi et al. [3] in each animal. The length of a run of homozygosity, that is the number of consecutive homozygous SNPs including the one being considered, was calculated (this would be zero if the SNP being considered was heterozygous). For each marker the median length, over the breed, of the run of homozygosity was calculated and plotted against genomic position in the candidate regions (25 SNPs on each side).
It should be noted that since the historical effective population sizes of males and females are not the same for sexual and autosomal chromosomes [47], all analyses presented in this paper for chromosome X were performed by using only females (37 animals per each breed), although the analysis for both sexes produced similar results.
2.4. Determining of Ancestral Alleles
To calculate iHS and XP-EHH, the ancestral allele state of each SNP must be specified. Ancestral alleles for the ovine chip SNP were obtained from international sheep hapmap project (ISGC), and then to obtain the ancestral alleles for the additional Sequenom SNPs, 301 base pairs of sequence (1 bp of the alleles plus 150 bases either side of the SNP) were aligned against Bos taurus (cattle), Sus scrofa (pig), Equus caballus (horse), Canis familiaris (dog) and Homo sapiens (human) genomes using BLAST (http://blast.ncbi.nlm.nih.gov/Blast, accessed on 31 May 2017). A cross-species megaBLAST of Sequenom® primers was used to discover ancestral alleles for the remaining SNPs [37]. The ancestral allele was taken as the base in the genome sequence at the resulting SNP position. For loci where only one SNP allele was represented in the other species, that allele was determined as ancestral. For other SNPs, as an additional tool in determining the ancestral allele, a phylogenetic tree of the five species was used [48]. This provided a crude tree, which could be used in decision making; for example, if all the animals had the same allele (C) apart from humans (T), then the ancestral was more likely to be C, as humans are more distantly related than the other species. In this research, we were able to determine the ancestral status of 36, 28, and 36 from the 41, 30, and 36 SNPs which passed quality control in the regions on chromosome 5, 7, and X respectively.
2.5. Reconstruction of Haplotypes
A pair of haplotypes was reconstructed for each animal in the sample using fastPHASE version 1.2.3 [49]. This software implements an Expectation—Maximization strategy for estimating missing genotypes and for reconstructing haplotypes from unphased SNP genotypes data of unrelated individuals [49].
2.6. Calculation of Integrated Haplotype Score (iHS) and Cross-Population EHH (XP-EHH)
iHS was calculated as in Voight et al. [35] and XP-EHH as in Sabeti et al. [36]. These statistical tests were calculated using the rehh package [50] in R v4.0.2 and the candidate genomic regions under selection were obtained.
Briefly and following Voight et al. [35], the iHS was computed for every SNP with ancestral state information and MAF above 10% (Supplementary Table S2). This test is based on the extended haplotype homozygosity (EHH) statistic [39], which measures the decay of identity, as a function of distance, of haplotypes that carry a specified core allele at one end. The integral of the observed decay of EHH with distance from the core allele is calculated until EHH reaches 0.05. This integrated EHH (iHH) (summed over both directions away from the core SNP) is denoted as iHHA or iHHD, depending on whether it is computed for the ancestral or derived core allele. The unstandardized iHS (uiHS) was then calculated as ln (iHHA/iHHD). The uiHS is thus adjusted so that the final statistic has a mean of 0 and a variance of 1, regardless of allele frequency at the core SNP. To do this, the results for each breed were split into 18 equally sized allele frequency bins, from which a mean and standard deviation were calculated. These were used in the following equation [35]:
where the expectation and standard deviation are calculated using SNPs from the same bin (p). Due to the different demographic histories of the X chromosome and the autosomes (e.g., due to smaller effective population size), we normalized the iHS scores of this chromosome separately from those of the other chromosomes. Results of iHS are presented here as |iHS| for a window of 10 SNPs and then plotted against genome location. We chose a window of 10 SNPs because of the longer extent of LD and SNP spacing in sheep compared to humans, in which the window length used is commonly around 40 SNPs [35]. Large positive and negative values of iHS indicate unusually long haplotypes carrying the ancestral and derived allele, respectively; by taking the absolute iHS value, interesting variants will be shown by large positive values.
XP-EHH compares haplotypes between populations to control for local variation in recombination rates [36]. Briefly, XP-EHH is defined relative to a given SNP i in two populations, A and B. In each population, the expected haplotype homozygosity (EHH) [39] was integrated with respect to genetic distance in both directions from i. The log of the ratio of these integrals, ln(IA/IB), is the abnormal XP-EHH (for more details see [36]). XP-EHH must also be normalized for genome-wide differences in haplotype length between populations, so that there is a mean of zero and unit variance. This was done by subtracting the mean and dividing by the standard deviation of all scores. In this study, we defined the XP-EHH test with respect to thin and fat tail breeds and an unusually positive value suggests selection in fat tailed population and a negative value selection in thin tailed.
2.7. Core SNP Alleles and Haplotype Frequencies in Candidate Regions
After the aforementioned filtering process and reconstruction of haplotypes for candidate regions using PHASE 2.1 [51], the haplotypes were fed into SWEEP v.1.1 [39] to detect core regions based on the EHH statistic in candidate regions, which is fully described by Sabeti et al. [39]. PHASE was chosen here over fastPHASE as haplotype estimates are slightly more accurate using PHASE (http://stephenslab.uchicago.edu/software.html, accessed on 1 February 2021), although substantially slower to compute. For selection of core regions and study of haplotype frequencies in selected area of our interested regions, the results of FST, iHS and XP-EHH tests were also used as additional information. Finally, the pattern of haplotype blocks in the regions of interest were constructed and the decay of LD (pairwise r2) was visualized using Haploview [52].
2.8. Study of Identified Genes in Candidate Regions
Genes located in the genomic regions significantly differentiated between sheep breeds were acquired by the use of the data mining tool Biomart (http://asia.ensembl.org/biomart/martview, accessed on 1 February 2021), with the reference assembly of the O. aries genome OAR v3.1 [53]. The regions of interest in O. aries were also compared to the corresponding areas in B. taurus as its genome is better annotated. Regions chosen for fine mapping onto OARv3.1 and their orthologous coordinates in B. taurus (ARS-UCD1.2, Bostau9) is shown in Supplementary Table S3. It should be noted that, due to the easier comparison of coordinates obtained by the current study with the previous article [3], the coordinates have been presented for different statistics in this study, based on OAR v1.0. However, the coordinate of candidate regions associated with fat deposition reported in Moradi et al. [3] and the fine mapped results in this study have been also shown based on different OAR versions in Supplementary Table S4. Nearby genes within a flanking distance of 500 kbs from each region were acquired. This distance has been selected as previously considered by Moradi et al. [3] for sheep and by Do et al. [54] for Holstein dairy cattle. To determine the biological functions of each gene, Biological Process (BP) and Molecular Functions (MF) of all identified genes in candidate regions were studied using DAVID annotation [55]. Furthermore, a comprehensive literature review was conducted to verify whether these genes have some relevance with fat deposition or developmental process in sheep or other mammals.
3. Results
3.1. SNP Genotyping and Data Mining
A total of 32, 26, and 29 SNPs passed the filtering criteria in our regions of interest on chromosome 5, 7 and X respectively (Table 2). Most of these SNPs that used for further analysis were not available on the Illumina Ovine SNP50k BeadChip due to the lower expected minor allele frequency (MAF) and reliability of the SNPs, discovered on both the Solexa and 454 platforms (http://www.sheephapmap.org/genseq.php, accessed on 31 May 2017).

Table 2.
Summary of SNP characteristics for different regions, genotyped using the Sequenom assay, before and after data cleaning and their combination with Ovine SNP50k BeadChip SNPs, for follow up analysis.
Table 2 presents a descriptive summary of the characteristics of the SNPs used in the final analysis. The distribution of SNPs varied among the regions, especially regarding to those used in statistical tests (Supplementary Table S2); however, the average SNP intervals were relatively consistent and the overall average distance between adjacent SNPs was about 23 kb, 17 kb and 82 kb for candidate regions on chromosome 5, 7 and X respectively, compared to about 60 kb and 115 kb for autosomes and chromosome X on the Illumina Ovine SNP50k BeadChip respectively.
3.2. Distribution of FST and Median Homozygosity
To identify loci that have been targets of selection across thin and fat tailed breeds, the windowed FST was plotted against genomic location for candidate regions (Figure 1a). Variants with unusually large FST values are typically interpreted as being the targets of local selective pressures due to the hitch-hiking effect [25].
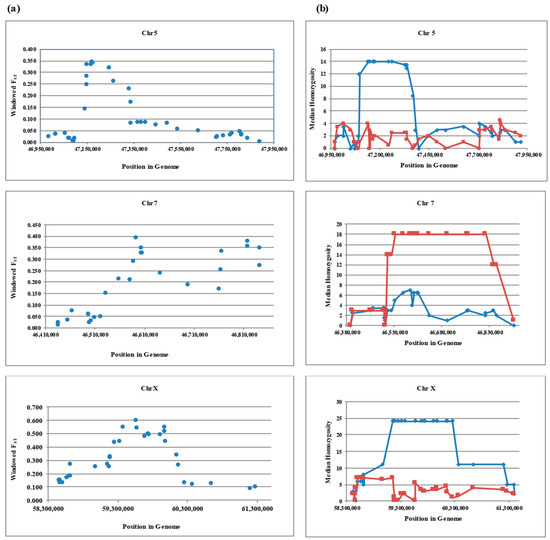
Figure 1.
Plots of windowed FST (a) and run of median homozygosity (b) in relation to genomic position for thin and fat tail breeds in candidate regions: SNP positions in the genome (bp) are shown on the X-axis, and windowed FST or median homozygosity are plotted on the Y-axis. Fat and thin tailed breeds are shown by blue diamonds and red squares respectively on median homozygosity plot.
The average of differentiation between Zel (thin tail) and Lori Bakhtiari (fat tail) for the whole genome was 0.024 (SD = 0.036), while this parameter was 0.093 (SD = 0.136), 0.172 (SD = 0.184) and 0.279 (SD = 0.217) at the areas of interest on chromosome 5, 7 and X respectively. As shown in Figure 1, in these regions we found evidence of selection across relatively short distances with windowed FST values > 0.30 on chromosomes 5 (between 47,149,400–47,245,841 bp), 7 (between 46,587,943–46,843,356 bp) and values > 0.40 on chromosome X (between 59,257,971–59,984,949 bp). The score of 0.30 is in the 99.9 percentile of autosomal SNPs (n = 44,558) and a score of 0.40 is in the 99.0 percentile of chromosome X SNPs (n = 1126).
When there is a selection for a causal mutation in one breed and not the other, the breed under selection shows high homozygosity in a genomic interval while the other does not [39]. To further test this hypothesis, median homozygosity was calculated and plotted against genomic position for candidate regions (Figure 1b). The results indicate that homozygosity increased over the areas of interest on chromosome 5 and X for the fat tailed and at the candidate region on chromosome 7 for the thin tailed breed. The largest differences of median homozygosity were for chromosome X and homozygosity was present for a longer distance as well, whereas these parameters are less on Chromosome 5.
3.3. Calculation of Integrated Haplotype Score (iHS)
iHS detects signatures of strong selection in favor of alleles that have not yet reached fixation [35]. The results revealed that regions on chromosomes 5 and X had no iHS peak (Supplementary Figures S1 and S2). This observation may suggest that selected alleles have already been fixed in these locations. However, analysis of the region on chromosome 7 displayed an obvious peak in both thin and fat tail breeds (Figure 2). For most locations of the region the values are higher in the thin tail breed, however, there is also some even stronger indication of selection in fat tail breed especially at the end of the region.
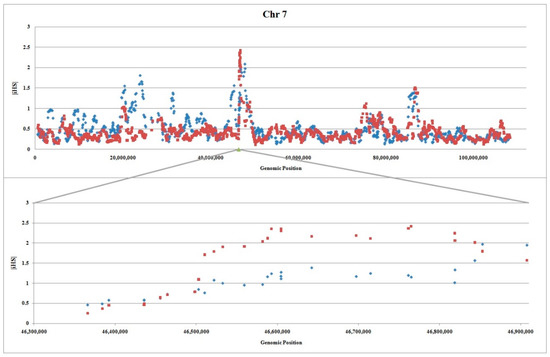
Figure 2.
Plot of |iHS| in relation to the genomic position (bp) for thin and fat tail breeds on chromosome 7 (Upper) and our candidate region (lower): Fat and thin tail breeds are shown by blue diamonds and red squares respectively, and |iHS| statistic averaged over 10 SNPs. The information for calculating |iHS| on whole chromosome was obtained from Moradi et al. [3].
3.4. Cross-Population EHH (XP-EHH)
To identify selective sweeps in which the selected allele has approached or achieved fixation in a subpopulation, but remains polymorphic in the population as a whole, the standardized cross population extent of haplotype homozygosity scores (XP-EHH) were plotted against genomic locations (Figure 3). The results revealed clear peaks in all of our regions of interest, suggesting that selected alleles approached fixation or have risen to near fixation in favor of fat tail breed on chromosome 5 and X while on chromosome 7 the frequency of alleles have risen close to fixation in thin tail breed. These results are based on linkage disequilibrium around given SNP and are in agreement with the results of median homozygosity plots.
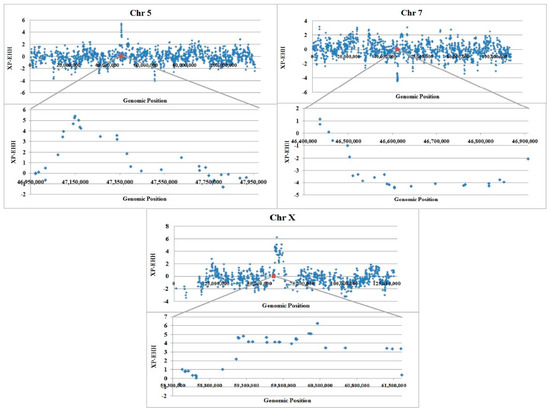
Figure 3.
Plot of XP-EHH relation to genomic position (bp) for thin and fat tailed breeds on whole chromosome (upper) and our candidate region (lower) in different chromosomes: High positive values suggest selection in fat tailed population and negative values selection in thin tailed population. The genotyping information required for the presentation of the entire chromosomes were obtained from Moradi et al. [3].
Sabeti et al. [36] considered a region as a candidate for selection in the human HapMap Phase 2 dataset when the two population XP-EHH was above 4.34. This score is in the 99.9 percentile for thin versus fat tail breeds. As shown in Figure 3, we found evidence of selection with |XP-EHH| value > 4.34 on chromosomes 5 (47,141,229–47,171,110 bp), 7 (46,604,500–46,642,359 bp) and X (59,187,456–60,264,325 bp). These regions are almost identical to the positions with highest FST.
3.5. Study of Core Haplotypes and Their Ancestral Status in Candidate Regions
To evaluate the haplotype frequencies and the status of selected alleles (derived or ancestral), the core SNP alleles, and their haplotype frequencies were investigated in the selected regions (Table 3).

Table 3.
Core SNP alleles and haplotype frequencies in candidate regions for fat tail (Lori) and thin tail (Zel) breeds: The haplotypes with higher frequency in fat and thin tailed breeds have been highlighted.
For chromosome 5 (Table 3), we defined a core region of 26 k where both FST and XP-EHH statistics were at their highest. There are 5 genotyped SNPs in this region. The SNPs defined 14 core haplotypes (denoted haplotype 1 to 14) in thin tailed but only 6 core haplotypes in fat tailed sheep. As shown in Table 3, there is a common haplotype (haplotype 1) with a frequency of 90% in fat tailed sheep whereas its frequency for thin tailed sheep is 15%. In contrast, the common haplotype in thin tail breed is haplotype 8 with a frequency of 31%, while it is almost absent (2%) in the fat tail breed. The interesting result in this region is that all the SNPs in the common haplotype for the fat tail breed are derived SNPs whereas all SNPs in the common haplotype for the thin tail breed are ancestral.
For chromosome 7 (Table 3) we defined a core region of 37 k in the selected area with 4 genotyped SNPs, where all statistics showed strong evidence of selection. There were only 4 core haplotypes (denoted haplotype 1 to 4) in thin tailed and 6 haplotypes in fat tailed sheep. The common haplotypes of fat and thin tail breeds in this region were haplotype 5 and 1 with frequency of 31% (2% in thin tailed) and 80% (6% in fat tailed), respectively. Once again, while the common haplotype in the fat tail breed had all derived alleles, the common haplotype in the thin tail breed were almost all ancestral alleles (3 out of 4 SNPs). However, it is notable that in this region the haplotypes appear more polymorphic in the fat tail breed and the frequency of the common haplotype of the thin tail breed is not as high as for the other candidate regions.
Subsequently, this approach was performed in the candidate region on chromosome X. We defined a core region of 242 k corresponding to 6 genotyped SNPs. The longer length of the core region, in this case is due to the wider SNP spacing on chromosome X. The genotyped SNPs defined seven core haplotypes in the thin tail breed and only two haplotypes in the fat tail breed (Table 3). The common haplotype in the fat tail breed was haplotype 1 with a frequency of 89%, whereas its frequency in thin tailed was 12%. In this region the common haplotype in the thin tail breed (54%) was absent in the fat tail breed. The results in this region were similar to the previous regions in that the derived alleles were more prevalent in the common haplotype of fat tailed (4 out of 6 core SNPs) with the same result for ancestral alleles in the common haplotype of the thin tail breed. Together, these results could suggest that ancestral alleles have been under selection in the thin tail breed while the contrary happened for the fat tail breed where derived alleles have undergone selection.
3.6. Study of the Identified Genes Associated with Fat Metabolisms in Candidate Regions
The regions of interest were investigated to determine if any genes related to fat deposition could be identified in sheep or their corresponding areas of the cow genome. The genes obtained in O. aries or by orthology with B. taurus and their functions are presented in Table 4.

Table 4.
Fat metabolism related genes located within the candidate regions in O. aries and their orthologous area in B. taurus.
There is also the hypothesis that some genes selected for, in fat tail sheep breeds are likely to be also associated with developmental defects or ectopic expression of organs [56,57]. Our results revealed that these regions contain many genes, having some known biological functions associated with the developmental process in O. aries (Table 5) and their orthologous coordinates in B. taurus (Supplementary Table S5).

Table 5.
Developmental process or gene expression related genes, located within the candidate regions in O. aries.
4. Discussion
An increasing number of studies have been conducted to detect signals of recent positive selection on a genome-wide scale in different domestic animals [58,59,60]; however, there are relatively few genomic regions identified that have been subject to selection for a specific mutation underlying evolutionary shifts in a trait. In this paper, in order to fine map and get more insight into the genomic basis of fat deposition in thin and fat tail breeds, we have investigated three candidate regions using this approach. These candidate regions were analyzed using a variety of statistics to clarify the signals of selection observed in these regions. Our analyses focused on two allele frequency spectrum (FST and median homozygosity) and haplotype based (|iHS| and XP-EHH) statistics. These tests were chosen because previous power analyses suggested that these are largely complementary [39].
The study of the candidate regions on chromosome 5 and X revealed obvious evidence of the selection using an FST, median homozygosity, and XP-EHH test in a relatively narrow region; while an examination of these regions identified no particular |iHS| peak. With a hypothesis that historically different selection pressures operated in thin and fat tail breeds and somehow selection acted on a variant that was advantageous only in one breed, these results suggest that selection in these regions occurred for mutations affecting fat tail size as the beneficial mutations have risen to near fixation in fat tailed breeds. This suggestion is also supported by the core haplotype frequencies observed in candidate regions on these chromosomes as the common core haplotypes in fat tail breeds were near fixation (Table 3).
Analysis of the region on chromosome 7 indicated strong evidence of selection using all selective sweep statistics. The results of FST and median homozygosity suggested that the selection differentiated the beneficial alleles between breeds and that homozygosity has been increased in favor of thin tailed in this region. However, |iHS| and XP-EHH revealed additional information. |iHS| provided evidence of partial selection in both breeds, while the XP-EHH results showed that the selected alleles have approached fixation in the thin tail breed.
As discussed earlier, the power of the |iHS| statistics to detect selective sweeps is greatest at a moderate allele frequency (~40–60%), while XP-EHH test is more powerful for detecting selective sweeps close to fixation (>80%) [35,61]. However, both of these methods do have power outside their optimal ranges. Sabeti et al. [36] demonstrated that the iHS statistic could detect signals over the range of 20–80% and XP-EHH do have power between 60–100%. Overall, these results suggest that while the frequency of selected alleles has been raised to fixation in thin tail breed, its frequency should be ~60–80% to be picked up by both methods. Simultaneously, the favorable allele should be increased to mid-range frequency in the fat tail breed. The results of core haplotype frequencies in this region (Table 3) are in consistent to this point as the common haplotype in thin tail breed has a frequency of 80%, whereas all haplotypes observed in this region appears polymorphic in the fat tail breed. One inference is that there has been an ongoing infusion of fat tailed haplotypes into this breed, but also selection for the thin-tailed phenotype.
To further test this hypothesis, we constructed the pattern of haplotype blocks in this region and the decay of LD (pairwise r2) was visualized using Haploview [52]. The effect of a selective sweep on patterns of variation is expected to decline with time (due to recombination) and if a selective sweep is still ongoing in a subpopulation, the hitchhiking haplotype is expected to be rather long [39,57]. Our results (Supplementary Figure S3) revealed that although there were haplotype blocks in both breeds, they extended for longer distances in the fat tailed compared to the thin tail breed. This suggests that as the prevalence of selected alleles increased in thin tail breed, the LD around variants decayed due to recombination, while the selection in the fat tail breed is younger (longer haplotype blocks). These results confirm our observations for evidence of selection in this region with both |iHS| and XP-EHH tests.
The earliest known depiction of a fat tail sheep is on an Uruk III stone vessel about 5000 years before present, approximately 4000 years after initial domestication [62]. Given that fat tailed breeds are now prevalent in the Fertile Crescent, where sheep were originally domesticated, while thin tailed sheep breeds are predominant in peripheral areas and that the wild ancestor of sheep is thin tail, it has been assumed that the first domesticated sheep were thin tailed and fat tail was developed later [62,63]. We have investigated this hypothesis through the classification of selected alleles in the core haplotypes of our regions of interest as ancestral or derived. Our results provide the preliminary molecular evidence to confirm this assumption since, we observed that in almost all cases, derived alleles have been under selection pressure in the fat tail breed, and this is consistent with the selection of a new mutation in these breeds (Table 3).
Population demographic history can also cause similar patterns on DNA sequence variation and could be a source of error in making inferences on genomic targets of selection. This caveat can be avoided by screening a large number of markers (as has previously been performed in this research) spaced across whole genome [3], as selection will result in regional patterns compared to the genome-wide effects of population history and demographic events [23,64]. Similarly conducting this type of fine-scale analysis at the candidate genomic regions using a dense set of markers and multiple statistical tests, reduces the chance that a signature of selection will be a false positive if it is detected in more than one marker locus and statistical test [40].
The candidate regions of interest have previously been studied using OAR v1.0 in O. aries and their corresponding area of B. taurus and no particular candidate genes associated with fat deposition were identified [3]. In this study, investigation of these regions using the newly available sheep genome OAR v3.1 [53], defined some genes associated with fat metabolism in O. aries or their orthologous areas of B. taurus (Table 4). Protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform (PPP2CA) has a variety of roles in different biological process such as cellular lipid metabolic, membrane lipid metabolic and sphingolipid metabolic process, while hydroxysteroid (17-beta) dehydrogenase 10 (HSD17B10) and emopamil binding protein (EBP) genes play some roles in lipid metabolic process and androgen receptor (Ar) and synaptophysin (Syp) genes get participate in lipid binding [65]. Interestingly, most of the genes (or their gene families) identified here, have been recently reported as candidate genes associated with lipid metabolisms using various molecular techniques in sheep. Yuan et al. [66] implemented differential expression analysis using RNA-seq technology in longissimus dorsi muscle tissue (MUT), perirenal adipose tissue (PAT) and tail adipose tissue (TAT) of different Chines short and fat tailed sheep breeds and revealed that PPP1CA is highly expressed in TAT. Also, PPP1CA was identified as plausible genes associated with the fat-tailed or fat-rumped phenotype by comparing copy number variations (CNVs) with different tail types [67]. Moreover, protein phosphatase 1, catalytic subunit, gamma isozyme (PPP1CC) have been under selection signature in Chinese thin and fat-tailed sheep breed and reported to be associated with tail type [68]. HSD17B12 has also been reported to be associated with fat tail metabolism in thin and fat tailed sheep breeds using deep transcriptome analysis with RNA-Seq data [69]. It is reported that HSD17B12 (act as elongates) are important genes for controlling the overall balance of fatty acid composition [69]. Therefore, it seems the gene families and isoforms of Protein phosphatase (PPP) and hydroxysteroid (HSD) have important roles in molecular regulations of fat deposition in tail.
To confirm the results of our study, the exon 1 of PPP2CA gene was amplified and its variation patterns were sequenced in an independent study on Zel and Lori-Bakhtiari sheep breeds [34]. Two patterns were identified and the results of sequencing showed that in Lori-Bakhtiari, Del/Del genotype resulted in heavier fat tail than T/T genotype (5.20 ± 0.21 kg vs. 3.28 ± 0.12 kg) (p < 0.05) while, in Zel, the effect of genotypes on carcass fat percentage and triglyceride was significant, so that the T/T genotype had more carcass fat percentage comparing to Del/Del genotypes (p < 0.05). Overall, it seems as the annotation of the ovine genome becomes more complete, all genes located in the candidate regions will be identified and promising targets can then be verified by further experimentation.
A result which is irrelevant to the inheritance of the trait, but provides an insight into a possible mechanism of fat deposition in this organ, are the results of Gokdal et al. [56] who examined the effects of docking in fat tail breeds. The carcasses of the docked group contained more kidney, pelvic and internal fat than the intact lambs as well as a higher percentage of subcutaneous and intramuscular fat. The weights of the different carcass cut of the docked lambs were also heavier than those of the intact group. However, there was little change in overall carcass composition, suggesting that the genes affecting the fat tail phenotype are associated with the localization of fat stores to a regional depot rather than control of the overall level of fat deposition. This observation also may provide support to the suggestion that some genes selected for in fat tail sheep breeds in these regions are likely to be also associated with developmental defects or ectopic expression of organs. Our results revealed that these regions contain many genes, having some known biological functions associated with developmental process (Table 5 and Table S5). Several earlier studies provide evidence for this issue. For example, the transcription factor (TCF) genes have been among highly differentially expressed genes in perirenal adipose tissue (PAT) and identified as being the most likely to account for the fat-tailed phenotype of sheep [66]. TCF7 is involved in the Wnt/β-catenin signaling pathway, and this pathway plays a critical role in regulating sheep [69] and porcine [70] adipogenesis genes expression. Zhu et al. [71] studied copy number variations (CNVs) and selection signatures on the X chromosome of Chinese indigenous sheep with different tail types and revealed that the regions harboring CNVs and selective sweeps in different sheep breeds overlapped with calcium channel, voltage-dependent, L type, alpha 1F subunit (CACNA1F) gene that could be as associated with tail type in these breeds [43]. In addition, HSD17B10 [69], SLC35A2 [68], AR and TIMP1 [31], have been identified as candidate genes that affect fat tail development. Moreover, it is important to note that our regions of interest overlapped with some genes, for example BMP15, WDR13 and RBM3 that belong to the gene families that their closely related genes consisting BMP2 [9,32,68,72,73], WDR92 [17,68] and RBM11 [6] have recently reported to be associated with fat tail formation and adipose tissue gene expression in sheep.
Finally, fine mapping of candidate regions using different sweep statistical tests has enabled us to confirm the signature of selection in these chromosomal regions and better refine the critical regions from 113 kb (47,149,400–47,263,230) to 28 kb (47,146,931–47,175,489) on chromosome 5, from 201 kb (46,642,359–46,843,356) to 142 kb (consisting to shorter intervals: 46,587,943–46,642,359 and 46,765,080–46,852,870) on chromosome 7 and from 2831 kb (58,621,412–61,452,816) to 1006 kb (59,257,971–60,264,325) on chromosome X. These regions were refined considering all statistical test results (Supplementary Figure S4). Acquiring of the genes located within the regions of interest after fine mapping revealed some genes consisting TCF7 on chromosome 5, PTGDR and NID2 on chromosome 7 and finally AR on chromosome X that have a multiple effect on lipid metabolisms, macromolecule metabolic process, organ/gland development or associated with ectopic expression of organs simultaneously.
Recently published study on the origin of European sheep as revealed by the diversity of the Balkan breeds and by optimizing population-genetic analysis tools [74] using a variety of sheep breed samples from Southwest-Asian, Mediterranean, Central-European and North-European showed that the thin-tailed Zel sheep is found to be in the same genetic cluster as the fat-tailed Iranian sheep, whereas the fat-tailed Italian Laticauda is related to other breeds in central Italy. This may imply that the tail phenotype is encoded by a limited number of genes. By combining information of the present study, previously reported and annotated biological functional genes, we suggest PPP2CA and TCF7 (OAR5), PTGDR and NID2 (OAR7), AR, EBP, CACNA1F, HSD17B10, SLC35A2, BMP15, WDR13 and RBM3 (OAR X) as the most promising candidate genes for type of tail traits.
It is obvious that understanding the mechanisms that underlie fat tail inheritance in sheep is difficult to verify solely by selective sweep profiling. While this study has now refined three regions located on chromosome 5, 7 and X, associated with fat tail sheep breeds, it is likely that other genomic regions may also be involved. Some other studies have identified a region on Chromosome 15 for example, close to PDGFD as associated with the fat tail phenotype [12,13]. Definitive studies of the actions of these regions will require larger-scale designs in segregating populations where trait measurements are recorded. Also, these regions may still be too large to efficiently implement technologies such as marker assisted selection or positional cloning. More detailed and larger scale experiments from these and other thin and fat tailed breeds may allow us to refine the location of the causal mutations. Likewise, it does not exclude contemporaneous selection for other traits, and any regions identified still need to be tested for a functional genetic relationship via trait measurement in contrasting genotypes or phenotypes. Specifically, future studies should be conducted in reciprocal F2 crosses to provide independent and causal evidence and verify the mode of inheritance. If these areas are shown to have a significant effect, then further work sequencing either side of the regions can help the search for causal variants.
5. Conclusions
Our results provide a comprehensive assessment of how and where selection has affected the patterns of variation in candidate regions associated with fat deposition in thin and fat tail sheep breeds. These results enabled us to confirm the signature of selection in these regions, refine the critical intervals regions, and to identify the most promising candidate genes associated with fat deposition in thin and fat tail sheep. These results may provide a strong foundation for studying the regulation of fat deposition in sheep and do offer hope that the causal mutations and the mode of inheritance of this trait will soon be discovered by further experimentation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ani12111423/s1, Table S1: Selected SNPs for genotyping via Sequenom iPlex Mass ARRAY platform and their locations, PCR primers, unextended primer and its extension masses. (XLS Format), Table S2: The number of animals, distances and number of SNPs used in this research for different statistical tests in Zel (thin tail) and Lori-Bakhtiari (fat tail) breeds, Table S3: Regions chosen for gene mapping onto O. aries (OAR v3.1) and their orthologous coordinates in B. taurus (ARS-UCD1.2, Bostau9), Table S4: The coordinate of candidate regions associated with fat deposition reported in article Moradi et al. [3] and the fine mapped results in this study based on different OAR versions, Table S5: Developmental process or gene expression related genes, located in/close the orthologous area of the regions of interest in B. taurus, Supplementary Figure S1: Plot of iHS relation to genomic position (bp) for thin and fat tailed breeds in candidate region on Chromosome 5, Figure S2: Plot of iHS relation to genomic position (bp) for thin and fat tailed breeds in candidate region on Chromosome X, Figure S3: A graphical representation of pairwise r2 for the candidate region on chromosome 7 in thin and fat tail breeds calculated and visualized using Haploview, Figure S4: Refinement of candidate regions associated with fat deposition in thin and fat tail breeds using different sweep statistical tests: refined candidate regions has been shown by green line and the gene located within each region of interest highlighted by pink colours. XP-EHH and |iHS| values have had 4 and 2 subtracted respectively, so the values fit on the same scale.
Author Contributions
M.H.M. planned and performed the analyses, and drafted the manuscript, A.N.-J. coordinated the study and supervised the analysis, M.M.-S. coordinated the study and sample collection, K.G.D. and R.B. provided statistical and analysis support and J.C.M. supervised the analysis and participated in the design of the study. All authors have contributed to the editing of the article, and approved the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by Animal Science Research Institute of Iran, Mobarakandish Institute and AgResearch, New Zealand, Project number: PRJ-2016/11547. The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Institutional Review Board Statement
Sampling was carried out by trained veterinarians within the frame of vaccination campaigns, hence no permission from the animal research ethics committee was necessary. Veterinarians adhered to standard procedures and relevant national guidelines to ensure appropriate animal care.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this manuscript and its supplementary information files. In addition, more detail data are available from the corresponding author on reasonable request.
Acknowledgments
The authors gratefully acknowledge the International Sheep Genomics Consortium for access to the Ovine HapMap genotypes and the Animal Breeding Center of Iran (ABCI) for access to the records and animals of the Iranian breeds. Thanks to the staff of the University of Tehran, Animal Science Research Institute of Iran and AgResearch, especially Tim Manley, Rayna Anderson and Kathryn McRae who helped and supported this research. The authors also acknowledge the financial contributions of Animal Science Research Institute of Iran, Mobarakandish Institute and AgResearch, New Zealand. We thank Pardis Sabeti and Benjamin Voight for their invaluable advises during this research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sabeti, P.C.; Schaffner, S.F.; Fry, B.; Lohmueller, J.; Varilly, P.; Shamovsky, O.; Palma, A.; Mikkelsen, T.S.; Altshuler, D.; Lander, E.S. Positive natural selection in the human lineage. Science 2006, 312, 1614–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fariello, M.-I.; Servin, B.; Tosser-Klopp, G.; Rupp, R.; Moreno, C.; Consortium, I.S.G.; Cristobal, M.S.; Boitard, S. Selection signatures in worldwide sheep populations. PLoS ONE 2014, 9, e103813. [Google Scholar] [CrossRef] [Green Version]
- Moradi, M.H.; Nejati-Javaremi, A.; Moradi-Shahrbabak, M.; Dodds, K.G.; McEwan, J.C. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genet. 2012, 13, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khaldari, M.; Azarfar, A.; Pahlavan, R. The size of fat tail does not have an effect on growth performance and carcass characteristics in Lori-Bakhtiari lambs. Small Rumin. Res. 2020, 187, 106088. [Google Scholar] [CrossRef]
- Negussie, E.; Rottmann, O.J.; Pirchner, F.; Rege, J.E.O. Patterns of growth and partitioning of fat depots in tropical fat-tailed Menz and Horro sheep breeds. Meat Sci. 2003, 64, 491–498. [Google Scholar] [CrossRef]
- Zhang, T.; Gao, H.; Sahana, G.; Zan, Y.; Fan, H.; Liu, J.; Shi, L.; Wang, H.; Du, L.; Wang, L. Genome-wide association studies revealed candidate genes for tail fat deposition and body size in the Hulun Buir sheep. J. Anim. Breed. Genet. 2019, 136, 362–370. [Google Scholar] [CrossRef]
- Kalds, P.; Luo, Q.; Sun, K.; Zhou, S.; Chen, Y.; Wang, X. Trends towards revealing the genetic architecture of sheep tail patterning: Promising genes and investigatory pathways. Anim. Genet. 2021, 52, 799–812. [Google Scholar] [CrossRef]
- Nejati-Javaremi, A.; Izadi, F.; Rahmati, G.H.; Moradi, M.; Izadi, F. Selection in fat-tailed sheep based on two traits of fat-tail and body weight versus single-trait total body weight. Int. J. Agri. Biol. 2007, 9, 645–648. [Google Scholar]
- Moioli, B.; Pilla, F.; Ciani, E. Signatures of selection identify loci associated with fat tail in sheep. J. Anim. Sci. 2015, 93, 4660–4669. [Google Scholar] [CrossRef]
- Wei, C.; Wang, H.; Liu, G.; Wu, M.; Cao, J.; Liu, Z.; Liu, R.; Zhao, F.; Zhang, L.; Lu, J. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genom. 2015, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Pan, Z.; Li, S.; Liu, Q.; Wang, Z.; Zhou, Z.; Di, R.; An, X.; Miao, B.; Wang, X.; Hu, W. Rapid evolution of a retro-transposable hotspot of ovine genome underlies the alteration of BMP2 expression and development of fat tails. BMC Genom. 2019, 20, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Yang, M.; Han, J.; Ma, Q.; Han, J.; Song, Z.; Luosang, C.; Gorkhali, N.A.; Yang, B.; He, X.; et al. Genomic analysis of worldwide sheep breeds reveals PDGFD as a major target of fat-tail selection in sheep. BMC Genom. 2020, 21, 800. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, J.; Shen, M.; Xie, X.-L.; Liu, G.-J.; Xu, Y.-X.; Lv, F.-H.; Yang, H.; Yang, Y.-L.; Liu, C.-B.; et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 2020, 11, 2815. [Google Scholar] [CrossRef]
- Shao, J.; He, S.; Pan, X.; Yang, Z.; Nanaei, H.A.; Chen, L.; Li, R.; Wang, Y.; Gao, S.; Xu, H. Allele-specific expression reveals the phenotypic differences between thin-and fat-tailed sheep. Res. Sq. 2020. [CrossRef]
- Luo, R.; Zhang, X.; Wang, L.; Zhang, L.; Li, G.; Zheng, Z. GLIS1, a potential candidate gene affect fat deposition in sheep tail. Mol. Biol. Rep. 2021, 48, 4925–4931. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, G.; Xu, X.; Geng, R.; Zhou, J.; Yang, Y.; Yang, Z.; Chen, Y. Transcriptome profile analysis of adipose tissues from fat and short-tailed sheep. Gene 2014, 549, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, Z.; Cai, Y.; Xu, H.; Yang, R.; Lan, X. Genetic variants in fat-and short-tailed sheep from high-throughput RNA-sequencing data. Anim. Genet. 2018, 49, 483–487. [Google Scholar] [CrossRef]
- Bakhtiarizadeh, M.R.; Alamouti, A.A. RNA-Seq based genetic variant discovery provides new insights into controlling fat deposition in the tail of sheep. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, M.; Wang, J.; Wang, S.; Wang, X.; Yang, J.; Gao, L.; Gan, S. Comparative transcriptome analysis of key genes and pathways activated in response to fat deposition in two sheep breeds with distinct tail phenotype. Front. Genet. 2021, 12, 1–20. [Google Scholar] [CrossRef]
- Schrooten, C.; Bink, M.; Bovenhuis, H. Whole genome scan to detect chromosomal regions affecting multiple traits in dairy cattle. J. Dairy Sci. 2004, 87, 3550–3560. [Google Scholar] [CrossRef] [Green Version]
- McClure, M.C.; Morsci, N.S.; Schnabel, R.D.; Kim, J.W.; Yao, P.; Rolf, M.M.; McKay, S.D.; Gregg, S.J.; Chapple, R.H.; Northcutt, S.L. A genome scan for quantitative trait loci influencing carcass, post-natal growth and reproductive traits in commercial Angus cattle. Anim. Genet. 2010, 41, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Mackay, T.F.C. The genetic architecture of quantitative traits. Annu. Rev. Genet. 2001, 35, 303–339. [Google Scholar] [CrossRef] [PubMed]
- Makinen, H.S.; Shikano, T.; Cano, J.M.; Merila, J. Hitchhiking mapping reveals a candidate genomic region for natural selection in three-spined stickleback chromosome VIII. Genetics 2008, 178, 453–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moioli, B.; Scatà, M.C.; Steri, R.; Napolitano, F.; Catillo, G. Signatures of selection identify loci associated with milk yield in sheep. BMC Genet. 2013, 14, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.M.; Haigh, J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974, 23, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Moon, S.; Kim, T.-H.; Lee, K.-T.; Kwak, W.; Lee, T.; Lee, S.-W.; Kim, M.-J.; Cho, K.; Kim, N.; Chung, W.-H. A genome-wide scan for signatures of directional selection in domesticated pigs. BMC Genom. 2015, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Akey, J.M. Constructing genomic maps of positive selection in humans: Where do we go from here? Genome Res. 2009, 19, 711–722. [Google Scholar] [CrossRef] [Green Version]
- Shikano, T.; Ramadevi, J.; Merilä, J. Identification of local-and habitat-dependent selection: Scanning functionally important genes in nine-spined sticklebacks (Pungitius pungitius). Mol. Biol. Evol. 2010, 27, 2775–2789. [Google Scholar] [CrossRef] [Green Version]
- Kohn, M.H.; Pelz, H.-J.; Wayne, R.K. Natural selection mapping of the warfarin-resistance gene. Proc. Natl. Acad. Sci. USA 2000, 97, 7911–7915. [Google Scholar] [CrossRef] [Green Version]
- Rost, S.; Fregin, A.; Ivaskevicius, V.; Conzelmann, E.; Hörtnagel, K.; Pelz, H.-J.; Lappegard, K.; Seifried, E.; Scharrer, I.; Tuddenham, E.G.D. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 2004, 427, 537–541. [Google Scholar] [CrossRef]
- Kang, D.; Zhou, G.; Zhou, S.; Zeng, J.; Wang, X.; Jiang, Y.; Yang, Y.; Chen, Y. Comparative transcriptome analysis reveals potentially novel roles of Homeobox genes in adipose deposition in fat-tailed sheep. Sci. Rep. 2017, 7, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastrangelo, S.; Moioli, B.; Ahbara, A.; Latairish, S.; Portolano, B.; Pilla, F.; Ciani, E. Genome-wide scan of fat-tail sheep identifies signals of selection for fat deposition and adaptation. Anim. Prod. Sci. 2018, 59, 835–848. [Google Scholar] [CrossRef]
- Ahbara, A.; Bahbahani, H.; Almathen, F.; Al Abri, M.; Agoub, M.O.; Abeba, A.; Kebede, A.; Musa, H.H.; Mastrangelo, S.; Pilla, F. Genome-wide variation, candidate regions and genes associated with fat deposition and tail morphology in Ethiopian indigenous sheep. Front. Genet. 2019, 9, 699. [Google Scholar] [CrossRef] [PubMed]
- Zare, M.; Sadeghi, M.; Moradi-Shahrbabak, M. Identification of single nucleotide polymorphisms of PPP2CA gene and its effect on carcass, growth and fat storage traits in Lori Bakhtiari and Zel sheep. Anim. Prod. Res. 2016, 5, 70–78. [Google Scholar]
- Voight, B.F.; Kudaravalli, S.; Wen, X.; Pritchard, J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006, 4, e72. [Google Scholar]
- Sabeti, P.C.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; McCarroll, S.A.; Gaudet, R. Genome-wide detection and characterization of positive selection in human populations. Nature 2007, 449, 913–918. [Google Scholar] [CrossRef]
- McRae, K.M.; McEwan, J.C.; Dodds, K.G.; Gemmell, N.J. Signatures of selection in sheep bred for resistance or susceptibility to gastrointestinal nematodes. BMC Genom. 2014, 15, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Qanbari, S. On the extent of linkage disequilibrium in the genome of farm animals. Front. Genet. 2020, 10, 1304. [Google Scholar] [CrossRef] [Green Version]
- Sabeti, P.C.; Reich, D.E.; Higgins, J.M.; Levine, H.Z.P.; Richter, D.J.; Schaffner, S.F.; Gabriel, S.B.; Platko, J.V.; Patterson, N.J.; McDonald, G.J.; et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 2002, 419, 832–837. [Google Scholar] [CrossRef]
- Grossman, S.R.; Shylakhter, I.; Karlsson, E.K.; Byrne, E.H.; Morales, S.; Frieden, G.; Hostetter, E.; Angelino, E.; Garber, M.; Zuk, O. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 2010, 327, 883–886. [Google Scholar] [CrossRef] [Green Version]
- Oeth, P.; Beaulieu, M.; Park, C.; Kosman, D.; del Mistro, G.; van den Boom, D.; Jurinke, C. iPLEX assay: Increased plexing efficiency and flexibility for MassArray system through single base primer extension with mass-modified terminators. Seq. Appl. Note 2005, 27, 1–12. [Google Scholar]
- Barendse, W.; Harrison, B.E.; Bunch, R.J.; Thomas, M.B.; Turner, L.B. Genome wide signatures of positive selection: The comparison of independent samples and the identification of regions associated to traits. BMC Genom. 2009, 10, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Huang, X.; Li, M.; Qin, S.; Fang, S.; Ma, Y. GWAS and post-GWAS to identification of genes associated with sheep tail fat deposition. Preprints 2019, 6, 1–11. [Google Scholar]
- MacEachern, S.; Hayes, B.; McEwan, J.; Goddard, M. An examination of positive selection and changing effective population size in Angus and Holstein cattle populations (Bos taurus) using a high density SNP genotyping platform and the contribution of ancient polymorphism to genomic diversity in Domestic ca. BMC Genom. 2009, 10, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Hudson, R.R.; Slatkin, M.; Maddison, W.P. Estimation of levels of gene flow from DNA sequence data. Genetics 1992, 132, 583–589. [Google Scholar] [CrossRef]
- Sayres, M.A.W. Genetic diversity on the sex chromosomes. Genome Biol. Evol. 2018, 10, 1064. [Google Scholar] [CrossRef]
- McRae, K.M. Signatures of Selective Sweeps in Parasite Selection Flocks; Univeristy of Otago: Mosgiel, New Zealand, 2012. [Google Scholar]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M. The structure of haplotype blocks in the human genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef] [Green Version]
- Gautier, M.; Vitalis, R. rehh: An R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics 2012, 28, 1176–1177. [Google Scholar] [CrossRef]
- Stephens, M.; Smith, N.J.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef] [Green Version]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S. Ensembl 2019. Nucleic Acids Res. 2019, 47, D745–D751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Do, D.N.; Bissonnette, N.; Lacasse, P.; Miglior, F.; Sargolzaei, M.; Zhao, X.; Ibeagha-Awemu, E.M. Genome-wide association analysis and pathways enrichment for lactation persistency in Canadian Holstein cattle. J. Dairy Sci. 2017, 100, 1955–1970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Gokdal, O.; Ulker, H.; Karakus, F.; Cengiz, F.; Temur, C.; Handil, H. Growth, feedlot performance and carcass characteristics of Karakas and crossbred lambs (F1)(Ile de France x Akkaraman (G1) x Karakas) under rural farm conditions in Turkey. S. Afr. J. Anim. Sci. 2004, 34, 223–232. [Google Scholar]
- Qanbari, S.; Gianola, D.; Hayes, B.; Schenkel, F.; Miller, S.; Moore, S.; Thaller, G.; Simianer, H. Application of site and haplotype-frequency based approaches for detecting selection signatures in cattle. BMC Genom. 2011, 12, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Stainton, J.J.; Haley, C.S.; Charlesworth, B.; Kranis, A.; Watson, K.; Wiener, P. Detecting signatures of selection in nine distinct lines of broiler chickens. Anim. Genet. 2015, 46, 37–49. [Google Scholar] [CrossRef]
- Ma, Y.; Wei, J.; Zhang, Q.; Chen, L.; Wang, J.; Liu, J.; Ding, X. A genome scan for selection signatures in pigs. PLoS ONE 2015, 10, e0116850. [Google Scholar] [CrossRef] [Green Version]
- Gurgul, A.; Jasielczuk, I.; Semik-Gurgul, E.; Pawlina-Tyszko, K.; Stefaniuk-Szmukier, M.; Szmatoła, T.; Polak, G.; Tomczyk-Wrona, I.; Bugno-Poniewierska, M. A genome-wide scan for diversifying selection signatures in selected horse breeds. PLoS ONE 2019, 14, e0210751. [Google Scholar] [CrossRef] [Green Version]
- Pickrell, J.K.; Coop, G.; Novembre, J.; Kudaravalli, S.; Li, J.Z.; Absher, D.; Srinivasan, B.S.; Barsh, G.S.; Myers, R.M.; Feldman, M.W. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009, 19, 826–837. [Google Scholar] [CrossRef] [Green Version]
- Davidson, A. The Oxford Companion to Food; Oxford University Press: Cary, NC, USA; ISBN 019104072X.
- Ryder, M.L. Sheep & Man; Duckworth & Co. Press: London, UK, 1983; ISBN 0715636472. [Google Scholar]
- Hayes, B.J.; Chamberlain, A.J.; Maceachern, S.; Savin, K.; McPartlan, H.; MacLeod, I.; Sethuraman, L.; Goddard, M.E. A genome map of divergent artificial selection between Bos taurus dairy cattle and Bos taurus beef cattle. Anim. Genet. 2009, 40, 176–184. [Google Scholar] [CrossRef]
- Huang, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Yuan, Z.; Xiang, R.; Li, W.; Li, F.; Yue, X. Transcriptomic analyses revealed common tailed and perirenal adipose differentially expressed genes in four Chinese indigenous sheep breeds. Livest. Sci. 2019, 230, 103832. [Google Scholar] [CrossRef]
- Zhu, C.; Fan, H.; Yuan, Z.; Hu, S.; Ma, X.; Xuan, J.; Wang, H.; Zhang, L.; Wei, C.; Zhang, Q. Genome-wide detection of CNVs in Chinese indigenous sheep with different types of tails using ovine high-density 600K SNP arrays. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Z.; Liu, E.; Liu, Z.; Kijas, J.W.; Zhu, C.; Hu, S.; Ma, X.; Zhang, L.; Du, L.; Wang, H. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Anim. Genet. 2017, 48, 55–66. [Google Scholar] [CrossRef]
- Bakhtiarizadeh, M.R.; Salehi, A.; Alamouti, A.A.; Abdollahi-Arpanahi, R.; Salami, S.A. Deep transcriptome analysis using RNA-Seq suggests novel insights into molecular aspects of fat-tail metabolism in sheep. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Sun, H.; Zhang, Z.; Zhang, C.-Y.; Zhao, Q.; Xiao, Q.; Olasege, B.S.; Ma, P.-P.; Zhang, X.-Z.; Wang, Q.-S. Selection signature reveals genes associated with susceptibility loci affecting respiratory disease due to pleiotropic and hitchhiking effect in Chinese indigenous pigs. Asian-Australas. J. Anim. Sci. 2020, 33, 187. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Li, M.; Qin, S.; Zhao, F.; Fang, S. Detection of copy number variation and selection signatures on the X chromosome in Chinese indigenous sheep with different types of tail. Asian-Australas. J. Anim. Sci. 2020, 33, 1378. [Google Scholar] [CrossRef]
- Mastrangelo, S.; Bahbahani, H.; Moioli, B.; Ahbara, A.; Al Abri, M.; Almathen, F.; Da Silva, A.; Belabdi, I.; Portolano, B.; Mwacharo, J.M. Novel and known signals of selection for fat deposition in domestic sheep breeds from Africa and Eurasia. PLoS ONE 2019, 14, e0209632. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Li, N.; Cheng, H.; Ma, Y. Genome wide association study for the identification of genes associated with tail fat deposition in Chinese sheep breeds. Biol. Open 2021, 10, bio054932. [Google Scholar] [CrossRef]
- Ciani, E.; Mastrangelo, S.; Da Silva, A.; Marroni, F.; Ferenčaković, M.; Ajmone-Marsan, P.; Baird, H.; Barbato, M.; Colli, L.; Delvento, C. On the origin of European sheep as revealed by the diversity of the Balkan breeds and by optimizing population-genetic analysis tools. Genet. Sel. Evol. 2020, 52, 1–14. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).