Differentially Expressed miRNAs and mRNAs in Regenerated Scales of Rainbow Trout (Oncorhynchus mykiss) under Salinity Acclimation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. RNA Extraction, Library Construction, mRNA and Small RNA Sequencing, and Analysis
3. Results
3.1. General Description of the Small RNA-Seq Data and mRNA-Seq Data in Rainbow Trout
3.2. Differentially Expressed miRNAs (DE miRNAs), Differentially Expressed Genes (DEGs), and Differentially Expressed Target Genes (DETGs) of the DE miRNAs in Regenerated Scales of SW Group
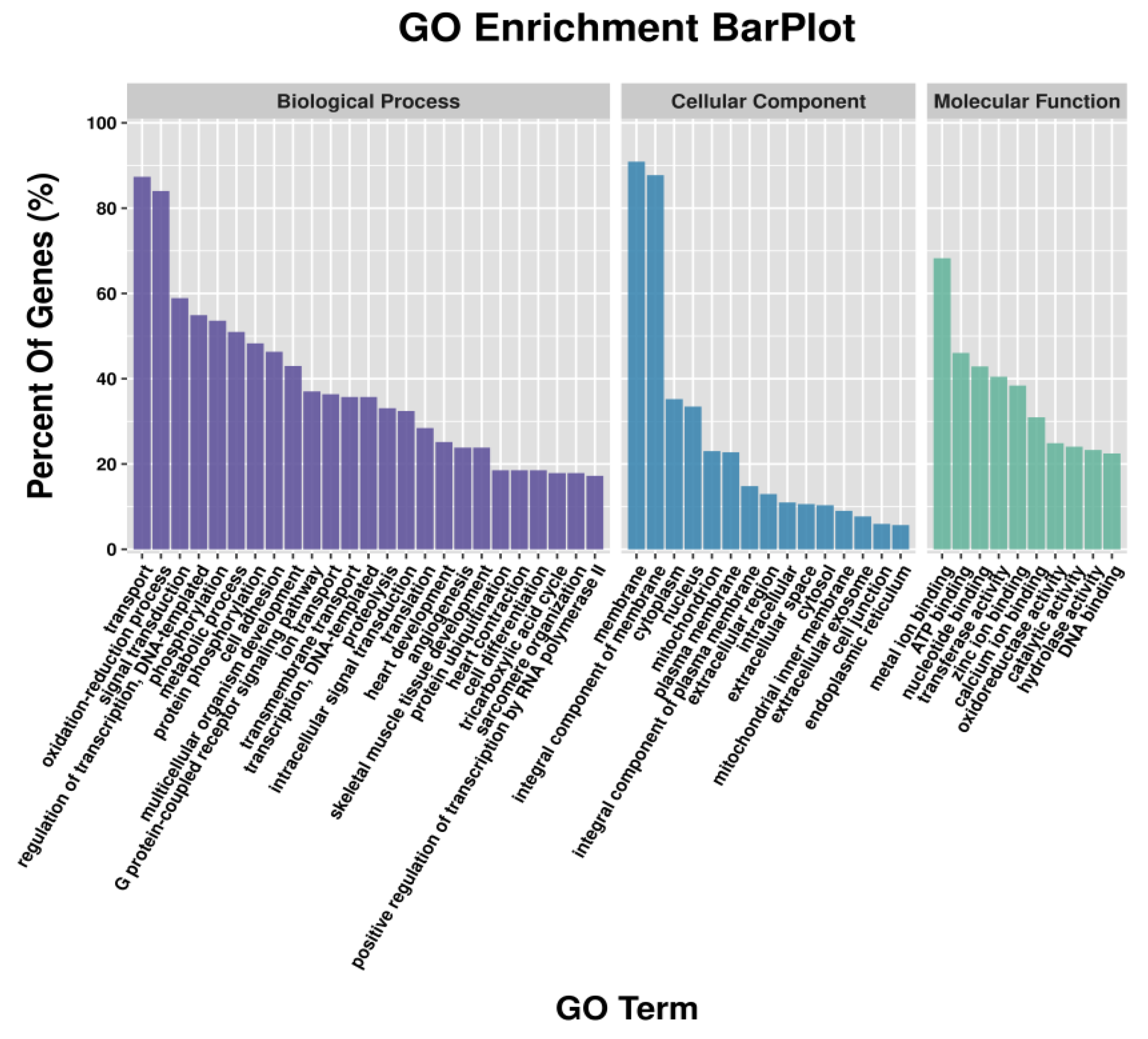
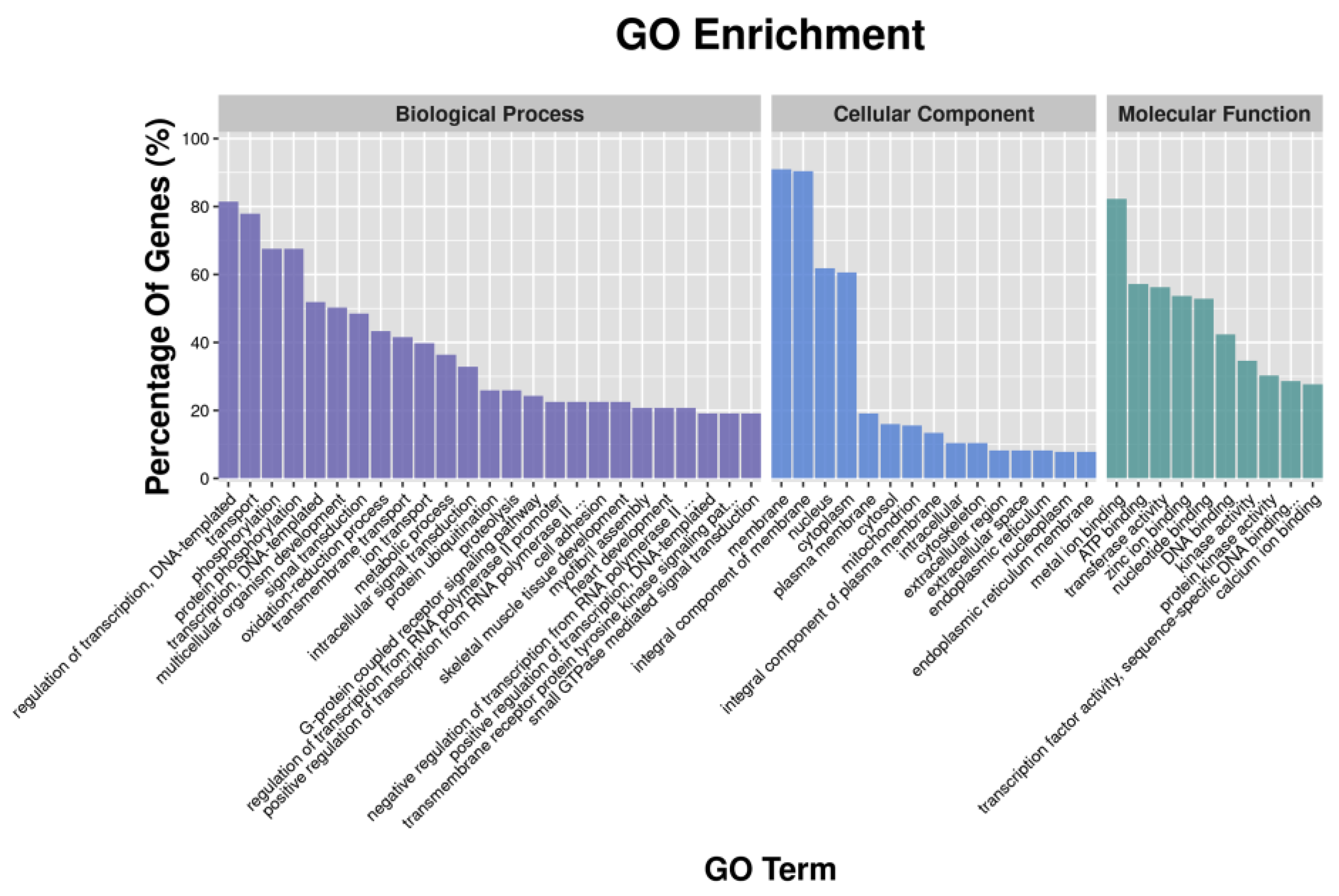
3.3. GO Functional Enrichment Analysis of DEGs and DETGs
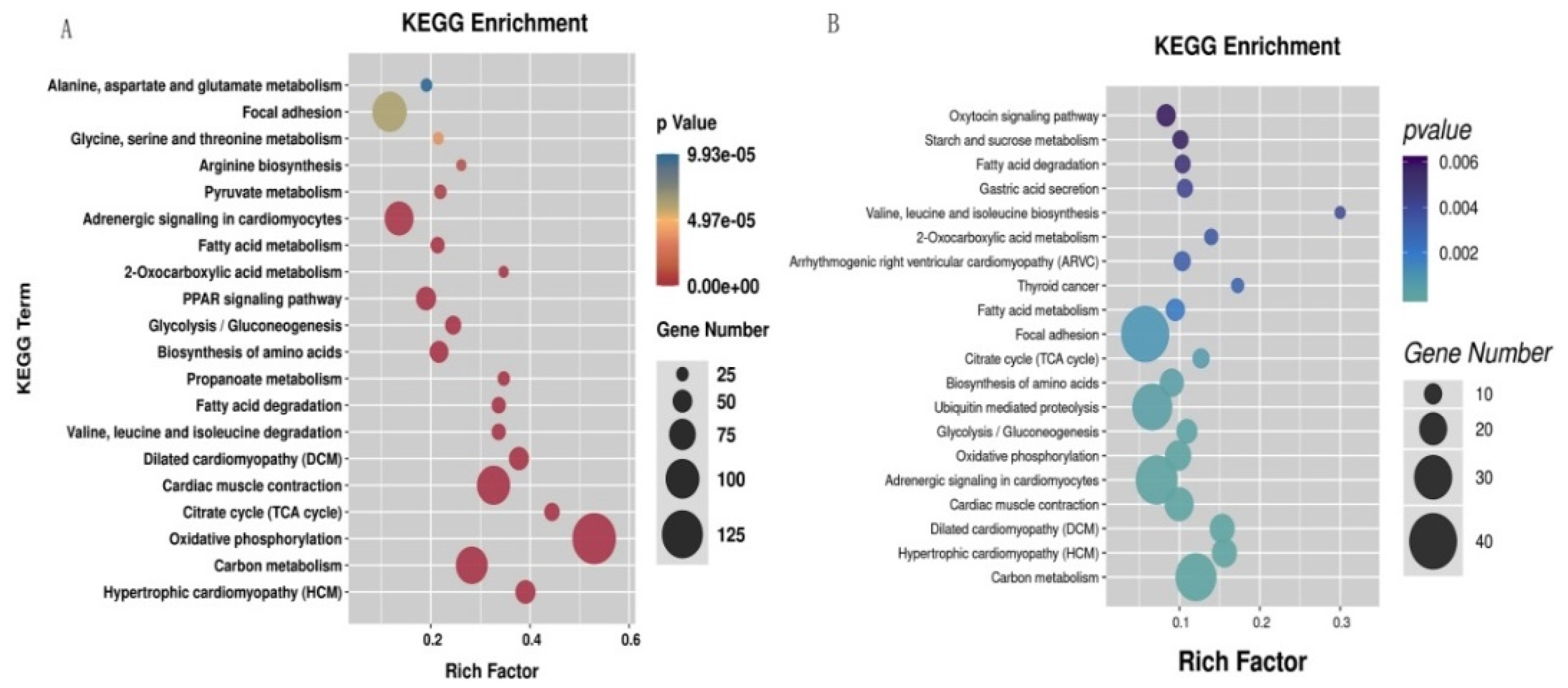
3.4. KEGG Pathway Annotation of DEGs and DETGs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shimada, A.; Kawanishi, T.; Kaneko, T.; Yoshihara, H.; Yano, T.; Inohaya, K.; Kinoshita, M.; Kamei, Y.; Tamura, K.; Takeda, H. Trunk exoskeleton in teleosts is mesodermal in origin. Nat. Commun. 2013, 4, 1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metz, J.; De Vrieze, E.; Lock, E.J.; Schulten, I.; Flik, G. Elasmoid scales of fishes as model in biomedical bone research. J. Appl. Ichthyol. 2012, 28, 382–387. [Google Scholar] [CrossRef]
- Kendall, N.W.; McMillan, J.R.; Sloat, M.R.; Buehrens, T.W.; Quinn, T.P.; Pess, G.R.; Kuzishchin, K.V.; McClure, M.M.; Zabel, R.W. Anadromy and residency in steelhead and rainbow trout (Oncorhynchus mykiss): A review of the processes and patterns. Can. J. Fish. Aquat. Sci. 2015, 72, 319–342. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Wang, L.; Zhao, X.; Yang, Y.; Ma, Q.; Chen, G. Effects of Salinity Acclimation on Histological Characteristics and miRNA Expression Profiles of Scales in Juvenile Rainbow Trout (Oncorhynchus Mykiss). BMC Genom. 2022, 23, 300. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, S.; Ebert, M.S.; Zheng, G.X.; Tsang, J.S.; Sharp, P.A.; van Oudenaarden, A. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011, 43, 854–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, J.W.; Rissland, O.S.; Koppstein, D.; Abreu-Goodger, C.; Jan, C.H.; Agarwal, V.; Yildirim, M.A.; Rodriguez, A.; Bartel, D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell. 2014, 53, 1031–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Torstensen, B.E.; Tocher, D.R.; Dick, J.R.; Henderson, R.J.; Bell, J.G. Environmental and dietary influences on highly unsaturated fatty acid biosynthesis and expression of fatty acyl desaturase and elongase genes in liver of Atlantic salmon (Salmo salar). Biochim. Biophys. Acta-Mol. Cell Res. 2005, 1734, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Wang, X.; Dong, S.; Wang, F.; Yang, J.; Zhou, Y. Comparisons of salinity adaptation in terms of growth, body composition, and energy budget in juveniles of rainbow and steelhead trouts (Oncorhynchus mykiss). J. Ocean Univ. China 2019, 18, 509–518. [Google Scholar] [CrossRef]
- Xiong, Y.; Dong, S.; Huang, M.; Li, Y.; Wang, X.; Wang, F.; Ma, S.; Zhou, Y. Growth, osmoregulatory response, adenine nucleotide contents, and liver transcriptome analysis of steelhead trout (Oncorhynchus mykiss) under different salinity acclimation methods. Aquaculture 2020, 520, 734937. [Google Scholar] [CrossRef]
- Bu, X.; Zhu, J.; Liu, S.; Wang, C.; Xiao, S.; Lu, M.; Li, E.; Wang, X.; Qin, J.G.; Chen, L. Growth, osmotic response and transcriptome response of the euryhaline teleost, Oreochromis mossambicus fed different myo-inositol levels under long-term salinity stress. Aquaculture 2021, 534, 736294. [Google Scholar] [CrossRef]
- Lu, Z.; Huang, W.; Wang, S.; Shan, X.; Ji, C.; Wu, H. Liver transcriptome analysis reveals the molecular responses to low-salinity in large yellow croaker Larimichthys crocea. Aquaculture 2020, 517, 734827. [Google Scholar] [CrossRef]
- Hu, Y.C.; Kang, C.K.; Tang, C.H.; Lee, T.H. Transcriptomic analysis of metabolic pathways in milkfish that respond to salinity and temperature changes. PLoS ONE 2015, 10, e0134959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juanchich, A.; Bardou, P.; Rue, O.; Gabillard, J.C.; Gaspin, C.; Bobe, J.; Guiguen, Y. Characterization of an extensive rainbow trout miRNA transcriptome by next generation sequencing. BMC Genom. 2016, 17, 164. [Google Scholar] [CrossRef] [PubMed]
- Tiago, D.M.; Marques, C.L.; Roberto, V.P.; Cancela, M.L.; Laizé, V. Mir-20a regulates in vitro mineralization and BMP signaling pathway by targeting BMP-2 transcript in fish. Arch. Biochem. Biophys. 2014, 543, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.M.; Yi, S.K.; Zhong, J.; Nie, C.H.; Guan, N.N.; Zhang, W.Z.; Gao, Z.X. Dynamic mRNA and miRNA expression analysis in response to intermuscular bone development of blunt snout bream (Megalobrama amblycephala). Sci. Rep. 2016, 6, 31050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Ren, K.; Yao, T.; Zhu, H.; Xu, Y.; Ye, H.; Chen, Z.; Lv, J.; Shen, S.; Ma, J. MicroRNA-25-3p regulates osteoclasts through nuclear factor I X. Biochem. Biophys. Res. Commun. 2020, 29, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Flynt, A.S.; Thatcher, E.J.; Burkewitz, K.; Li, N.; Liu, Y.; Patton, J.G. miR-8 microRNAs regulate the response to osmotic stress in zebrafish embryos. J. Cell Biol. 2009, 185, 115–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, B.; Guo, J.T.; Zhao, L.H.; Zhao, J.L. MiR-30c: A novel regulator of salt tolerance in tilapia. Biochem. Biophys. Res. Commun. 2012, 425, 315–320. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Zhou, Q.; Ma, Q.; Wang, L.; Yang, Y.; Chen, G. Differentially Expressed miRNAs and mRNAs in Regenerated Scales of Rainbow Trout (Oncorhynchus mykiss) under Salinity Acclimation. Animals 2022, 12, 1265. https://doi.org/10.3390/ani12101265
Yang C, Zhou Q, Ma Q, Wang L, Yang Y, Chen G. Differentially Expressed miRNAs and mRNAs in Regenerated Scales of Rainbow Trout (Oncorhynchus mykiss) under Salinity Acclimation. Animals. 2022; 12(10):1265. https://doi.org/10.3390/ani12101265
Chicago/Turabian StyleYang, Changgeng, Qiling Zhou, Qian Ma, Liuyong Wang, Yunsheng Yang, and Gang Chen. 2022. "Differentially Expressed miRNAs and mRNAs in Regenerated Scales of Rainbow Trout (Oncorhynchus mykiss) under Salinity Acclimation" Animals 12, no. 10: 1265. https://doi.org/10.3390/ani12101265
APA StyleYang, C., Zhou, Q., Ma, Q., Wang, L., Yang, Y., & Chen, G. (2022). Differentially Expressed miRNAs and mRNAs in Regenerated Scales of Rainbow Trout (Oncorhynchus mykiss) under Salinity Acclimation. Animals, 12(10), 1265. https://doi.org/10.3390/ani12101265




