Long-Term Recovery of the Fecal Microbiome and Metabolome of Dogs with Steroid-Responsive Enteropathy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbiome Analysis
2.1.1. 16S rRNA Gene Sequencing
2.1.2. Dysbiosis Index
2.2. Untargeted Metabolomics
2.3. Statistical Analysis
3. Results
3.1. Clinical Data
3.2. Microbiome
3.2.1. 16S rRNA Gene Sequencing
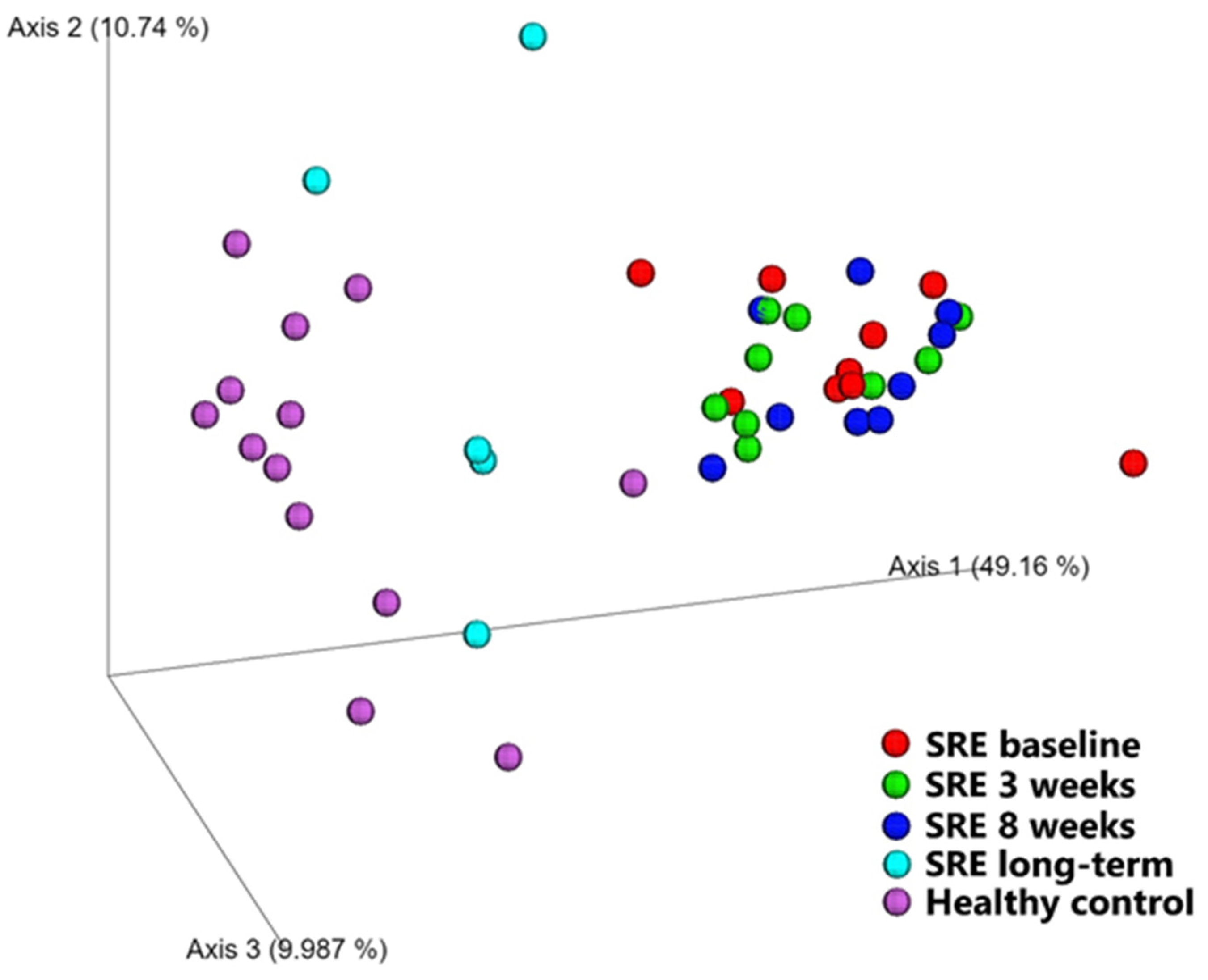
Diversity between Samples (β-Diversity)
Diversity within Samples (α-Diversity)
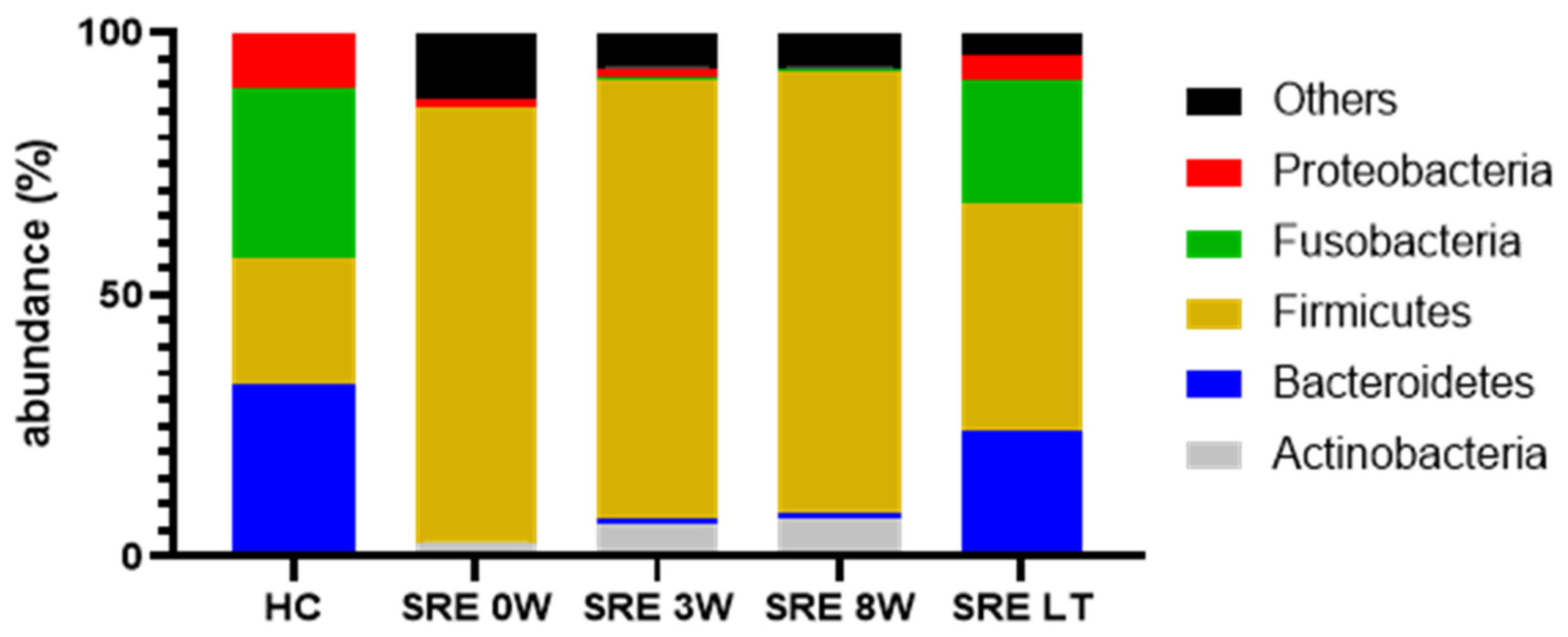
Individual Bacterial Taxa
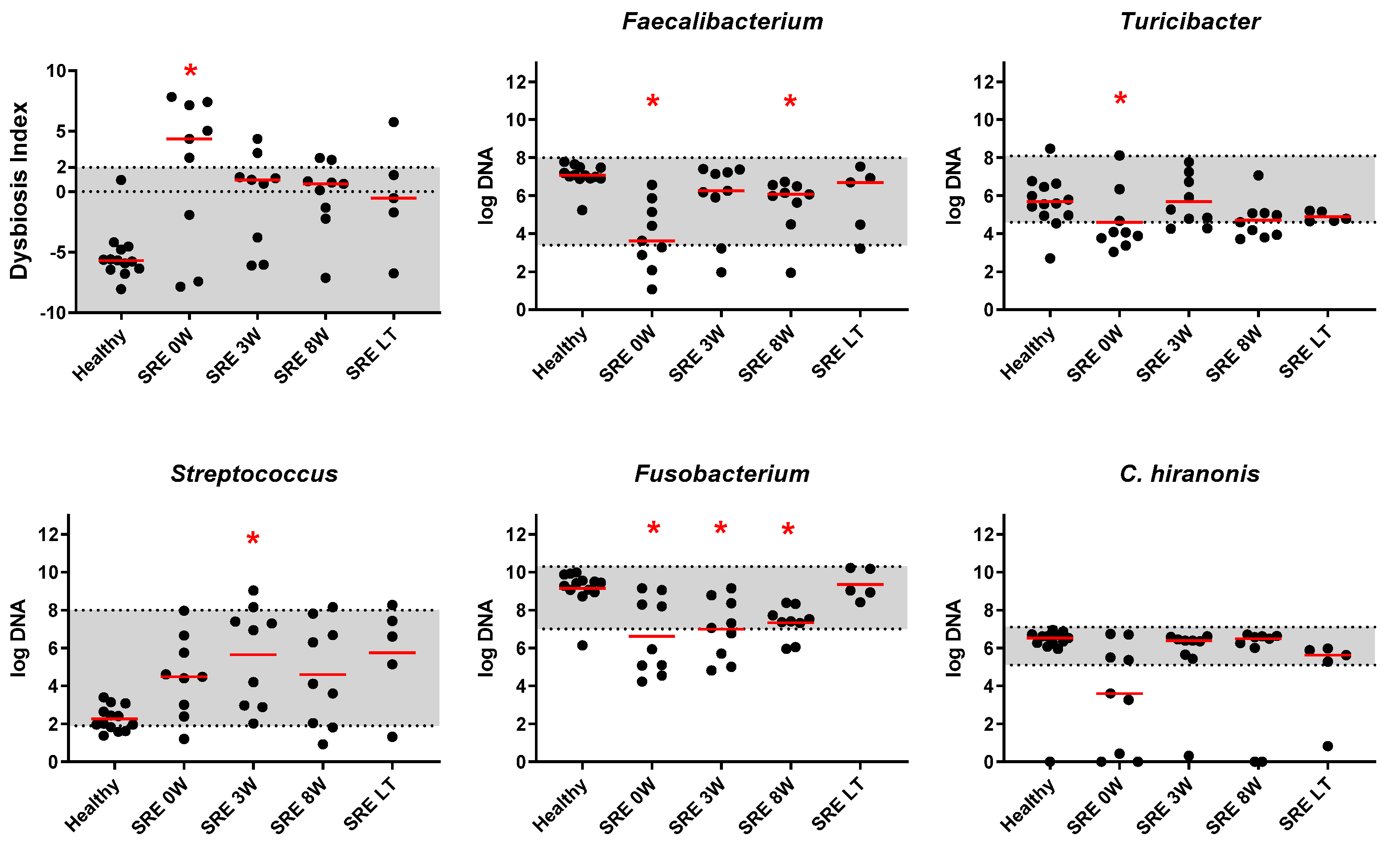
3.2.2. qPCR and DI
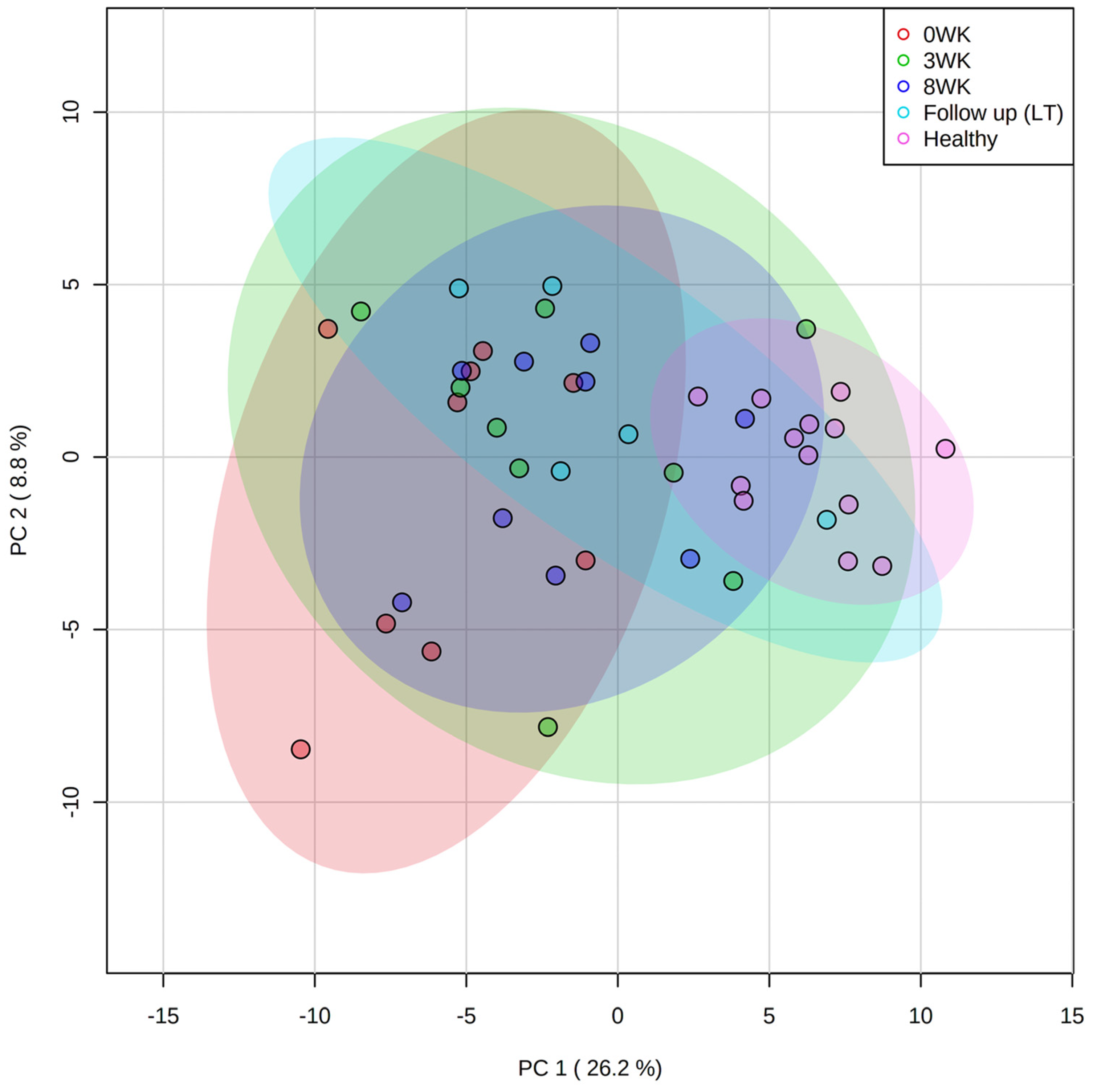
3.3. Metabolomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suchodolski, J.S.; Markel, M.E.; Garcia-Mazcorro, J.F.; Unterer, S.; Heilmann, R.M.; Dowd, S.E.; Kachroo, P.; Ivanov, I.; Minamoto, Y.; Dillman, E.M.; et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS ONE 2012, 7, e51907. [Google Scholar] [CrossRef] [Green Version]
- Zeng, M.Y.; Inohara, N.; Nunez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017, 10, 18–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guard, B.C.; Barr, J.W.; Reddivari, L.; Klemashevich, C.; Jayaraman, A.; Steiner, J.M.; Vanamala, J.; Suchodolski, J.S. Characterization of microbial dysbiosis and metabolomic changes in dogs with acute diarrhea. PLoS ONE 2015, 10, e0127259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziese, A.L.; Suchodolski, J.S.; Hartmann, K.; Busch, K.; Anderson, A.; Sarwar, F.; Sindern, N.; Unterer, S. Effect of probiotic treatment on the clinical course, intestinal microbiome, and toxigenic Clostridium perfringens in dogs with acute hemorrhagic diarrhea. PLoS ONE 2018, 13, e0204691. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Baeza, Y.; Hyde, E.R.; Suchodolski, J.S.; Knight, R. Dog and human inflammatory bowel disease rely on overlapping yet distinct dysbiosis networks. Nat. Microbiol. 2016, 1, 16177. [Google Scholar] [CrossRef] [PubMed]
- Suchodolski, J.S.; Dowd, S.E.; Wilke, V.; Steiner, J.M.; Jergens, A.E. 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS ONE 2012, 7, e39333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minamoto, Y.; Otoni, C.C.; Steelman, S.M.; Büyükleblebici, O.; Steiner, J.M.; Jergens, A.E.; Suchodolski, J.S. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes 2015, 6, 33–47. [Google Scholar] [CrossRef] [Green Version]
- Suchodolski, J.S.; Dowd, S.E.; Westermarck, E.; Steiner, J.M.; Wolcott, R.D.; Spillmann, T.; Harmoinen, J.A. The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rRNA gene sequencing. BMC Microbiol. 2009, 9, 210. [Google Scholar] [CrossRef] [Green Version]
- Manchester, A.C.; Webb, C.B.; Blake, A.B.; Sarwar, F.; Lidbury, J.A.; Steiner, J.M.; Suchodolski, J.S. Long-term impact of tylosin on fecal microbiota and fecal bile acids of healthy dogs. J. Vet. Intern. Med. 2019, 33, 2605–2617. [Google Scholar] [CrossRef]
- Pilla, R.; Gaschen, F.P.; Barr, J.W.; Olson, E.; Honneffer, J.; Guard, B.C.; Blake, A.B.; Villanueva, D.; Khattab, M.R.; AlShawaqfeh, M.K.; et al. Effects of metronidazole on the fecal microbiome and metabolome in healthy dogs. J. Vet. Intern. Med. 2020, 34, 1853–1866. [Google Scholar] [CrossRef]
- Chaitman, J.; Ziese, A.-L.; Pilla, R.; Minamoto, Y.; Blake, A.B.; Guard, B.C.; Isaiah, A.; Lidbury, J.A.; Steiner, J.M.; Unterer, S. Fecal microbial and metabolic profiles in dogs with acute diarrhea receiving either fecal microbiota transplantation or oral metronidazole. Front. Vet. Sci. 2020, 7, 192. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Chavez, F.; Lopez, C.A.; Baumler, A.J. Oxygen as a driver of gut dysbiosis. Free Radic. Biol. Med. 2017, 105, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Minamoto, Y.; Minamoto, T.; Isaiah, A.; Sattasathuchana, P.; Buono, A.; Rangachari, V.R.; McNeely, I.H.; Lidbury, J.; Steiner, J.M.; Suchodolski, J.S. Fecal short-chain fatty acid concentrations and dysbiosis in dogs with chronic enteropathy. J. Vet. Intern. Med. 2019, 33, 1608–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guard, B.C.; Honneffer, J.B.; Jonika, M.M.; Lidbury, J.A.; Steiner, J.M.; Jergens, A.; Suchodolski, J.S. Longitudinal characterization of dysbiosis and unconjugated bile acid profiles in the feces of dogs with inflammatory bowel disease. Gastroenterology 2017, 152, S992. [Google Scholar] [CrossRef]
- Blake, A.B.; Guard, B.C.; Honneffer, J.B.; Lidbury, J.A.; Steiner, J.M.; Suchodolski, J.S. Altered microbiota, fecal lactate, and fecal bile acids in dogs with gastrointestinal disease. PLoS ONE 2019, 14, e0224454. [Google Scholar] [CrossRef] [Green Version]
- Kitahara, M.; Takamine, F.; Imamura, T.; Benno, Y. Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7alpha-dehydroxylating activity. Int. J. Syst. Evol. Microbiol. 2001, 51, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Martins, R.; Sullivan, M.C.; Friedman, E.S.; Misic, A.M.; El-Fahmawi, A.; De Martinis, E.C.P.; O’Brien, K.; Chen, Y.; Bradley, C.; et al. Diet-induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. Microbiome 2019, 7, 126. [Google Scholar] [CrossRef] [Green Version]
- Kathrani, A.; Allenspach, K.; Fascetti, A.J.; Larsen, J.A.; Hall, E.J. Alterations in serum amino acid concentrations in dogs with protein-losing enteropathy. J. Vet. Intern. Med. 2018, 32, 1026–1032. [Google Scholar] [CrossRef] [Green Version]
- Benvenuti, E.; Pierini, A.; Gori, E.; Bartoli, F.; Erba, P.; Ruggiero, P.; Marchetti, V. Serum amino acid profile in 51 dogs with immunosuppressant-responsive enteropathy (IRE): A pilot study on clinical aspects and outcomes. BMC Vet. Res. 2020, 16, 117. [Google Scholar] [CrossRef] [Green Version]
- Sakai, K.; Maeda, S.; Yonezawa, T.; Matsuki, N. Decreased plasma amino acid concentrations in cats with chronic gastrointestinal diseases and their possible contribution in the inflammatory response. Vet. Immunol. Immunopathol. 2018, 195, 1–6. [Google Scholar] [CrossRef]
- Gupta, N.K.; Thaker, A.I.; Kanuri, N.; Riehl, T.E.; Rowley, C.W.; Stenson, W.F.; Ciorba, M.A. Serum analysis of tryptophan catabolism pathway: Correlation with Crohn’s disease activity. Inflamm. Bowel Dis. 2012, 18, 1214–1220. [Google Scholar] [CrossRef] [Green Version]
- Burgener, I.A.; Konig, A.; Allenspach, K.; Sauter, S.N.; Boisclair, J.; Doherr, M.G.; Jungi, T.W. Upregulation of toll-like receptors in chronic enteropathies in dogs. J. Vet. Intern. Med. 2008, 22, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sancho, M.; Rodriguez-Franco, F.; Sainz, A.; Mancho, C.; Rodriguez, A. Evaluation of clinical, macroscopic, and histopathologic response to treatment in nonhypoproteinemic dogs with lymphocytic-plasmacytic enteritis. J. Vet. Intern. Med. 2007, 21, 11–17. [Google Scholar] [CrossRef]
- Schreiner, N.M.S.; Gaschen, F.; Grone, A.; Sauter, S.N.; Allenspach, K. Clinical signs, histology, and CD3-positive cells before and after treatment of dogs with chronic enteropathies. J. Vet. Intern. Med. 2008, 22, 1079–1083. [Google Scholar] [CrossRef] [PubMed]
- White, R.; Atherly, T.; Guard, B.; Rossi, G.; Wang, C.; Mosher, C.; Webb, C.; Hill, S.; Ackermann, M.; Sciabarra, P.; et al. Randomized, controlled trial evaluating the effect of multi-strain probiotic on the mucosal microbiota in canine idiopathic inflammatory bowel disease. Gut Microbes 2017, 8, 451–466. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Mazcorro, J.F.; Dowd, S.E.; Poulsen, J.; Steiner, J.M.; Suchodolski, J.S. Abundance and short-term temporal variability of fecal microbiota in healthy dogs. Microbiologyopen 2012, 1, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Honneffer, J.B.; Steiner, J.M.; Lidbury, J.A.; Suchodolski, J.S. Variation of the microbiota and metabolome along the canine gastrointestinal tract. Metabolomics 2017, 13, 1–20. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- AlShawaqfeh, M.K.; Wajid, B.; Minamoto, Y.; Markel, M.; Lidbury, J.A.; Steiner, J.M.; Serpedin, E.; Suchodolski, J.S. A dysbiosis index to assess microbial changes in fecal samples of dogs with chronic inflammatory enteropathy. FEMS Microbiol. Ecol. 2017, 93, fix136. [Google Scholar] [CrossRef] [Green Version]
- Blake, A.B.; Cigarroa, A.; Klein, H.L.; Khattab, M.R.; Keating, T.; Van De Coevering, P.; Lidbury, J.A.; Steiner, J.M.; Suchodolski, J.S. Developmental stages in microbiota, bile acids, and clostridial species in healthy puppies. J. Vet. Intern. Med. 2020, 34, 2345–2356. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Lee, D.Y.; Lu, Y.; Moon, S.; Nikolau, B. Quality control for plant metabolomics: Reporting MSI-compliant studies. Plant J. 2008, 53, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigottier-Gois, L. Dysbiosis in inflammatory bowel diseases: The oxygen hypothesis. ISME J. 2013, 7, 1256–1261. [Google Scholar] [CrossRef] [PubMed]
- Guard, B.C.; Honneffer, J.B.; Jergens, A.E.; Jonika, M.M.; Toresson, L.; Lawrence, Y.A.; Webb, C.B.; Hill, S.; Lidbury, J.A.; Steiner, J.M.; et al. Longitudinal assessment of microbial dysbiosis, fecal unconjugated bile acid concentrations, and disease activity in dogs with steroid-responsive chronic inflammatory enteropathy. J. Vet. Intern. Med. 2019, 33, 1295–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilla, R.; Suchodolski, J.S. The role of the canine gut microbiome and metabolome in health and gastrointestinal disease. Front. Vet. Sci. 2020, 6, 498. [Google Scholar] [CrossRef] [Green Version]
- Bresciani, F.; Minamoto, Y.; Suchodolski, J.S.; Galiazzo, G.; Vecchiato, C.G.; Pinna, C.; Biagi, G.; Pietra, M. Effect of an extruded animal protein-free diet on fecal microbiota of dogs with food-responsive enteropathy. J. Vet. Intern. Med. 2018, 32, 1903–1910. [Google Scholar] [CrossRef] [Green Version]
- Honneffer, J.B.; Minamoto, Y.; Suchodolski, J.S. Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J. Gastroenterol. 2014, 20, 16489–16497. [Google Scholar] [CrossRef]
- Xenoulis, P.G.; Palculict, B.; Allenspach, K.; Steiner, J.M.; Van House, A.M.; Suchodolski, J.S. Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol. Ecol. 2008, 66, 579–589. [Google Scholar] [CrossRef] [Green Version]
- Park, M.J.; Pilla, R.; Panta, A.; Pandey, S.; Sarawichitr, B.; Suchodolski, J.; Sohrabji, F. Reproductive senescence and ischemic stroke remodel the gut microbiome and modulate the effects of estrogen treatment in female rats. Transl. Stroke Res. 2019, 11, 812–830. [Google Scholar] [CrossRef]
- Jeffery, N.D.; Barker, A.K.; Alcott, C.J.; Levine, J.M.; Meren, I.; Wengert, J.; Jergens, A.E.; Suchodolski, J.S. The association of specific constituents of the fecal microbiota with immune-mediated brain disease in dogs. PLoS ONE 2017, 12, e0170589. [Google Scholar] [CrossRef] [Green Version]
- Marchesi, J.R.; Holmes, E.; Khan, F.; Kochhar, S.; Scanlan, P.; Shanahan, F.; Wilson, I.D.; Wang, Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 2007, 6, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Allenspach, K.; Steiner, J.M.; Shah, B.N.; Berghoff, N.; Ruaux, C.; Williams, D.A.; Blum, J.W.; Gaschen, F. Evaluation of gastrointestinal permeability and mucosal absorptive capacity in dogs with chronic enteropathy. Am. J. Vet. Res. 2006, 67, 479–483. [Google Scholar] [CrossRef]
- Wennogle, S.A.; Priestnall, S.L.; Webb, C.B. Histopathologic characteristics of intestinal biopsy samples from dogs with chronic inflammatory enteropathy with and without hypoalbuminemia. J. Vet. Intern. Med. 2017, 31, 371–376. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef] [Green Version]
- Ciorba, M.A. Indoleamine 2,3 dioxygenase in intestinal disease. Curr. Opin. Gastroenterol. 2013, 29, 146–152. [Google Scholar] [CrossRef] [Green Version]
- Foster, J.A.; Rinaman, L.; Cryan, J.F. Stress & the gut-brain axis: Regulation by the microbiome. Neurobiol. Stress 2017, 7, 124–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.J.; Kovacs-Nolan, J.A.; Yang, C.B.; Archbold, T.; Fan, M.Z.; Mine, Y. L-Tryptophan exhibits therapeutic function in a porcine model of dextran sodium sulfate (DSS)-induced colitis. J. Nutr. Biochem. 2010, 21, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Wils-Plotz, E.L.; Jenkins, M.C.; Dilger, R.N. Modulation of the intestinal environment, innate immune response, and barrier function by dietary threonine and purified fiber during a coccidiosis challenge in broiler chicks. Poult. Sci. 2013, 92, 735–745. [Google Scholar] [CrossRef]
- Chen, Y.; Li, D.; Dai, Z.; Piao, X.; Wu, Z.; Wang, B.; Zhu, Y.; Zeng, Z. L-methionine supplementation maintains the integrity and barrier function of the small-intestinal mucosa in post-weaning piglets. Amino Acids 2014, 46, 1131–1142. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Bazer, F.W.; Burghardt, R.C.; Johnson, G.A.; Kim, S.W.; Knabe, D.A.; Li, P.; Li, X.; McKnight, J.R.; Satterfield, M.C.; et al. Proline and hydroxyproline metabolism: Implications for animal and human nutrition. Amino Acids 2011, 40, 1053–1063. [Google Scholar] [CrossRef] [Green Version]
- Kang, P.; Zhang, L.; Hou, Y.; Ding, B.; Yi, D.; Wang, L.; Zhu, H.; Liu, Y.; Yin, Y.; Wu, G. Effects of L-proline on the growth performance, and blood parameters in weaned lipopolysaccharide (LPS)-challenged pigs. Asian Australas J. Anim. Sci. 2014, 27, 1150–1156. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wu, C.; Li, P.; Li, N.; Zhang, D.; Zhu, Q.; Ren, W.; Peng, Y. Functions and signaling pathways of amino acids in intestinal inflammation. Biomed. Res. Int. 2018, 2018, 9171905. [Google Scholar] [CrossRef]
- Yoshii, K.; Hosomi, K.; Sawane, K.; Kunisawa, J. Metabolism of dietary and microbial vitamin B family in the regulation of host immunity. Front. Nutr. 2019, 6, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, B.C.; Randolph, T.W.; Lim, U.; Monroe, K.R.; Cheng, I.; Wilkens, L.R.; Le Marchand, L.; Lampe, J.W.; Hullar, M.A.J. Temporal Variability and Stability of the Fecal Microbiome: The Multiethnic Cohort Study. Cancer Epidemiol. Biomark. Prev. 2019, 28, 154–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Patient and Time Point | Histopathologic Diagnosis | Cellular Infiltrates | Mucosal Architecture | Inflammation Scores |
|---|---|---|---|---|
| Dog 1 Baseline | Mild-mod LP gastritis; mild-mod LP enteritis | Lymphocytes, plasma cells | No abnormalities | 3 |
| 8 weeks | Mild-mod LP gastritis; mod-severe LP enteritis | Lymphocytes, plasma cells, eosinophils | Multi-focal lacteal dilation | 1 |
| Dog 2 Baseline | Moderate L gastritis; moderate LP enteritis; mild L colitis | Lymphocytes, plasma cells | Lacteal dilation; crypt hyperplasia | 2 |
| 8 weeks | Mild L gastritis; mild LP enteritis; mild LP colitis | Lymphocytes, plasma cells | Lacteal dilation; crypt hyperplasia | 1 |
| Dog 3 Baseline | Moderate LP enteritis; mild L colitis | Lymphocytes, plasma cells | Lacteal dilation; multi-focal crypt abscesses | 1 |
| 8 weeks | Mild-mod LP enteritis; mild LP colitis; intestinal lymphangiectasia | Lymphocytes, plasma cells | Lacteal dilation | 1 |
| Dog 4 Baseline | Severe L gastritis with fibrosis; severe L enteritis; moderate L colitis | Lymphocytes | Gastric fibrosis; multi-focal dilated lacteals; increased IELs; crypt hyperplasia; diffuse villus blunting; increased goblet cells | 1 |
| 8 weeks | Severe LP gastritis; mild-mod LP enteritis; mild LP colitis | Lymphocytes, plasma cells, neutrophils | Tortuous colonic glands | 0 |
| Dog 5 Baseline | Mild LP gastritis; mild-mod LP enteritis | Lymphocytes, plasma cells | Multi-focal dilated lacteals | 1 |
| 8 weeks | Mild LP enteritis; mild LP colitis | Lymphocytes, plasma cells | No abnormalities | 2 |
| Dog 6 Baseline | Moderate LP enteritis | Lymphocytes, plasma cells | No abnormalities | 1 0 |
| 8 weeks | Mild LP gastritis; moderate LP enteritis; moderate LP colitis | Lymphocytes, plasma cells | No abnormalities | |
| Dog 7 Baseline | Mild LP gastritis; mod plasmactyic enteritis; mild LP colitis | Lymphocytes, plasma cells | Gastric edema; multi-focal crypt abscesses; hyperplasia of colonic glandular epithelium | 2 |
| 8 weeks | Mild LP gastritis; mild-mod LP enteritis; mild LP colitis | Lymphocytes, plasma cells | Multifocal lacteal dilation; multi-focal crypt abscesses | 1 |
| Dog 8 Baseline | Mild-mod LP enteritis | Lymphocytes, plasma cells | No abnormalities | 1 |
| 8 weeks | Moderate P enteritis; moderate P colitis with fibrosis | Plasma cells | Multi-focal lacteal dilation; mild colonic fibrosis | 1 |
| Dog 9 Baseline | Moderate LP gastritis; moderate LP enteritis | Lymphocytes, plasma cells | Multifocal lacteal dilation; multifocal crypt abscesses | 2 |
| 8 weeks | Moderate LP gastritis; moderate LP enteritis | Lymphocytes, plasma cells | No abnormalities | 2 |
| Pairwise Comparisons | Unweighted UniFrac | Weighted UniFrac | ||
|---|---|---|---|---|
| R-Value | p-Value | R-Value | p-Value | |
| SRE baseline vs. week 3 | 0.014 | 0.373 | 0.042 | 0.198 |
| SRE baseline vs. week 8 | 0.052 | 0.172 | 0.063 | 0.163 |
| SRE baseline vs. LT | −0.101 | 0.761 | 0.449 | 0.001 |
| Healthy vs. SRE baseline | 0.348 | 0.002 | 0.780 | 0.001 |
| Healthy vs. SRE week 3 | 0.410 | 0.001 | 0.833 | 0.001 |
| Healthy vs. SRE week 8 | 0.457 | 0.001 | 0.886 | 0.001 |
| Healthy vs. SRE LT | 0.168 | 0.119 | 0.169 | 0.141 |
| Compound Name | Compound Type | Change in Relation to HC | Time Points Significantly Different from HC | p-Value | FDR |
|---|---|---|---|---|---|
| alanine | amino acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| asparagine | amino acid | increased | baseline | 0.001 | 0.006 |
| aspartic acid | amino acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| cysteine | amino acid | increased | baseline | 0.009 | 0.029 |
| cystine | amino acid | increased | baseline | 0.001 | 0.003 |
| glutamic acid | amino acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| glycine | amino acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| isoleucine | amino acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| leucine | amino acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| lysine | amino acid | increased | 3 weeks, 8 weeks | 0.012 | 0.037 |
| methionine | amino acid | increased | baseline, 3 weeks, 8 weeks, long term | 0.000 | 0.000 |
| phenylalanine | amino acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.002 |
| proline | amino acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| serine | amino acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| threonine | amino acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| tryptophan | amino acid | increased | baseline, 3 weeks, 8 weeks, long term | 0.000 | 0.001 |
| valine | amino acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| glycyl tyrosine | dipeptide | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| glycyl-proline | dipeptide | increased | baseline | 0.012 | 0.037 |
| homocystine | dipeptide | increased | baseline | 0.018 | 0.048 |
| glucose | glucose | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.002 |
| 6-deoxyglucose | glucose metabolite | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.002 |
| gluconic acid | glucose metabolite | increased | baseline | 0.012 | 0.036 |
| myo-inositol | glucose metabolite | increased | baseline | 0.000 | 0.001 |
| ribose | glucose metabolite | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.002 |
| indole-3-lactate | indole | decreased | 3 weeks, 8 weeks, long term | 0.000 | 0.002 |
| isothreonic acid | metabolite vitamin C | increased | baseline | 0.005 | 0.019 |
| threonic acid | metabolite vitamin C | increased | baseline, long term | 0.003 | 0.014 |
| fructose | monosaccharide | increased | baseline, 8 weeks | 0.000 | 0.003 |
| N-acetyl-D- mannosamine | monosaccharide | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.000 |
| tagatose | monosaccharide | increased | baseline | 0.006 | 0.022 |
| xylulose NIST | monosaccharide | increased | baseline, 3 weeks, 8 weeks | 0.003 | 0.011 |
| arachidonic acid | omega 6 fatty acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.002 |
| arachidic acid | saturated fatty acid | increased | baseline, 3 weeks | 0.002 | 0.009 |
| caprylic acid | saturated fatty acid | increased | baseline, 3 weeks | 0.001 | 0.006 |
| isoheptadecanoic acid NIST | saturated fatty acid | increased | baseline, 8 weeks | 0.006 | 0.022 |
| lauric acid | saturated fatty acid | increased | baseline, 3 weeks, 8 weeks | 0.000 | 0.001 |
| lignoceric acid | saturated fatty acid | increased | baseline | 0.001 | 0.005 |
| myristic acid | saturated fatty acid | increased | 3 weeks | 0.019 | 0.050 |
| palmitic acid | saturated fatty acid | increased | baseline | 0.018 | 0.048 |
| stearic acid | saturated fatty acid | increased | baseline, 3 weeks | 0.000 | 0.001 |
| nicotinic acid | vitamin B3 | increased | 3 weeks, 8 weeks | 0.002 | 0.008 |
| tocopherol delta- NIST | vitamin E | increased | 3 weeks, 8 weeks | 0.012 | 0.036 |
| tocopherol gamma- | vitamin E | increased | 3 weeks, 8 weeks, long term | 0.000 | 0.002 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilla, R.; Guard, B.C.; Blake, A.B.; Ackermann, M.; Webb, C.; Hill, S.; Lidbury, J.A.; Steiner, J.M.; Jergens, A.E.; Suchodolski, J.S. Long-Term Recovery of the Fecal Microbiome and Metabolome of Dogs with Steroid-Responsive Enteropathy. Animals 2021, 11, 2498. https://doi.org/10.3390/ani11092498
Pilla R, Guard BC, Blake AB, Ackermann M, Webb C, Hill S, Lidbury JA, Steiner JM, Jergens AE, Suchodolski JS. Long-Term Recovery of the Fecal Microbiome and Metabolome of Dogs with Steroid-Responsive Enteropathy. Animals. 2021; 11(9):2498. https://doi.org/10.3390/ani11092498
Chicago/Turabian StylePilla, Rachel, Blake C Guard, Amanda B Blake, Mark Ackermann, Craig Webb, Steve Hill, Jonathan A Lidbury, Jörg M Steiner, Albert E. Jergens, and Jan S Suchodolski. 2021. "Long-Term Recovery of the Fecal Microbiome and Metabolome of Dogs with Steroid-Responsive Enteropathy" Animals 11, no. 9: 2498. https://doi.org/10.3390/ani11092498
APA StylePilla, R., Guard, B. C., Blake, A. B., Ackermann, M., Webb, C., Hill, S., Lidbury, J. A., Steiner, J. M., Jergens, A. E., & Suchodolski, J. S. (2021). Long-Term Recovery of the Fecal Microbiome and Metabolome of Dogs with Steroid-Responsive Enteropathy. Animals, 11(9), 2498. https://doi.org/10.3390/ani11092498







