DNA Multi-Marker Genotyping and CIAS Morphometric Phenotyping of Fasciola gigantica-Sized Flukes from Ecuador, with an Analysis of the Radix Absence in the New World and the Evolutionary Lymnaeid Snail Vector Filter
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
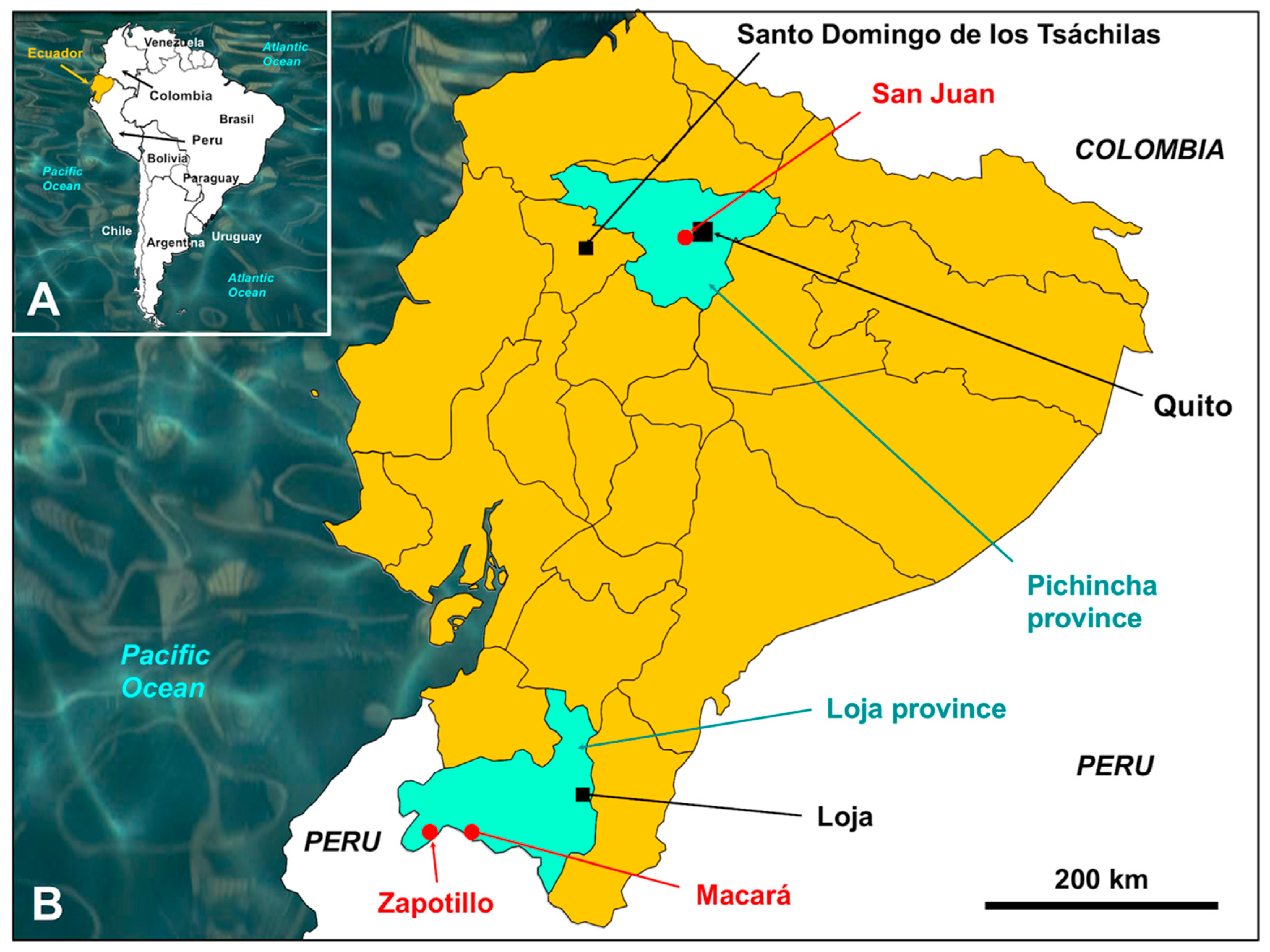
2.1. Parasites
2.2. Phenotyping Analyses
2.2.1. Natural and Experimental Liver Fluke Materials Used for Phenotyping Comparison
- 47 adults of F. hepatica from 5 sheep (range: 3–21/sheep) from Rodicio, Cajamarca Valley, at an altitude of 2726 m a.s.l., northern Peru (nCaj);
- 130 adults of F. hepatica from 8 sheep (range: 2–32/sheep) from Huayucachi, Pachacayo and Huancayo, in the Mantaro Valley, at an altitude of 3231–3989 m a.s.l., Peru (nMan);
- 201 adults of F. hepatica collected in 12 sheep (range: 6–30 worms per sheep) from Batallas, Northern Bolivian Altiplano, at an altitude of 3858 m a.s.l., Bolivia (nAlt);
- 37 adults of F. hepatica from 5 sheep (range: 2–10/sheep) from Massamagrell, Valencia, Comunidad Valenciana, at an altitude of 20 m a.s.l., Spain (nSp);
- 31 adults of F. gigantica from 2 sheep (range: 6–32/sheep) from Giza, Giza Governorate, at an altitude of 40 m a.s.l., Egypt (nEg).
- 127 adults of F. hepatica from 4 experimentally infected sheep (range: 33–66/sheep; 24-week-old) from Spain (expSp);
- 100 adults of F. gigantica from 4 experimentally infected sheep (range: 17–69/sheep; 24-week-old) from Egypt (expEg);
- 70 adults of F. gigantica from 7 experimentally infected sheep (range: 2–69/sheep; 52-week-old) from Vietnam (expVi).
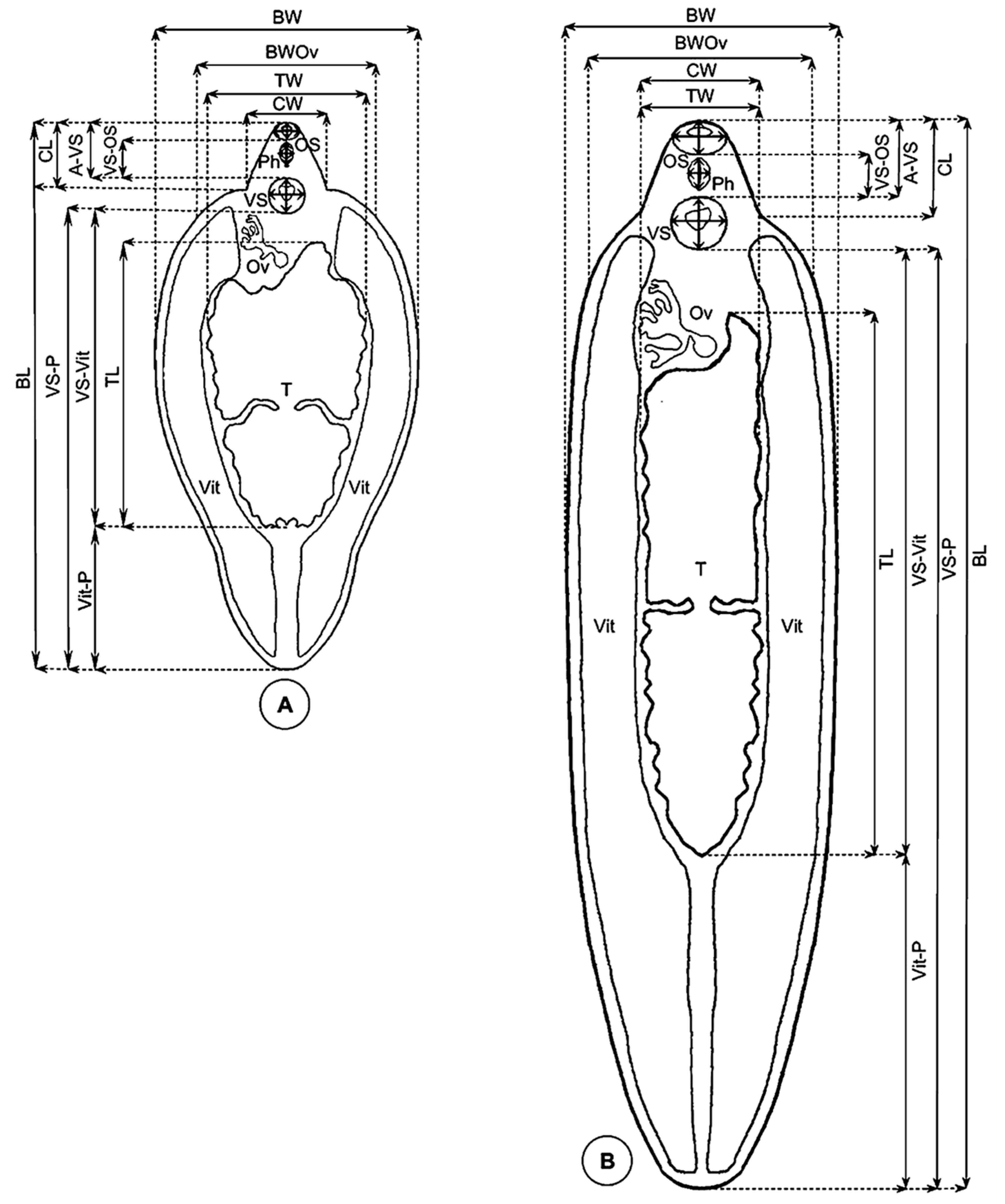
2.2.2. Measurement Techniques
- Lineal biometric characters: body length (BL), maximum body width (BW), body width at ovary level (BWOv), body perimeter (BP), body roundness (BR), cone length (CL), cone width (CW), maximum diameter of oral sucker (OS max), minimum diameter of oral sucker (OS min), maximum diameter of ventral sucker (VS max), minimum diameter of ventral sucker (VS min), distance between the anterior end of the body and the ventral sucker (A-VS), distance between the oral sucker and the ventral sucker (OS-VS), distance between the ventral sucker and the union of the vitelline glands (VSVit), distance between the union of the vitelline glands and the posterior end of the body (Vit-P), distance between the ventral sucker and the posterior end of the body (VS-P), pharynx length (PhL), pharynx width (PhW), testicular length (TL), testicular width (TW), testicular perimeter (TP);
- Areas: body area (BA), oral sucker area (OSA), ventral sucker area (VSA), pharynx area (PhA), and testicle area (taking both testes together, TA);
- Ratios: body length over body width (BL/BW), body width at ovary level over cone width (BWOv/CW), oral sucker area over ventral sucker area (OSA/VSA), and body length over the distance between the ventral sucker and the posterior end of the body (BL/VS-P).
2.2.3. Numerical Methods
2.3. Genotyping Analyses
2.3.1. DNA Markers
2.3.2. DNA Sequencing
2.3.3. Sequence Analyses
2.3.4. DNA Haplotype Nomenclature
3. Results
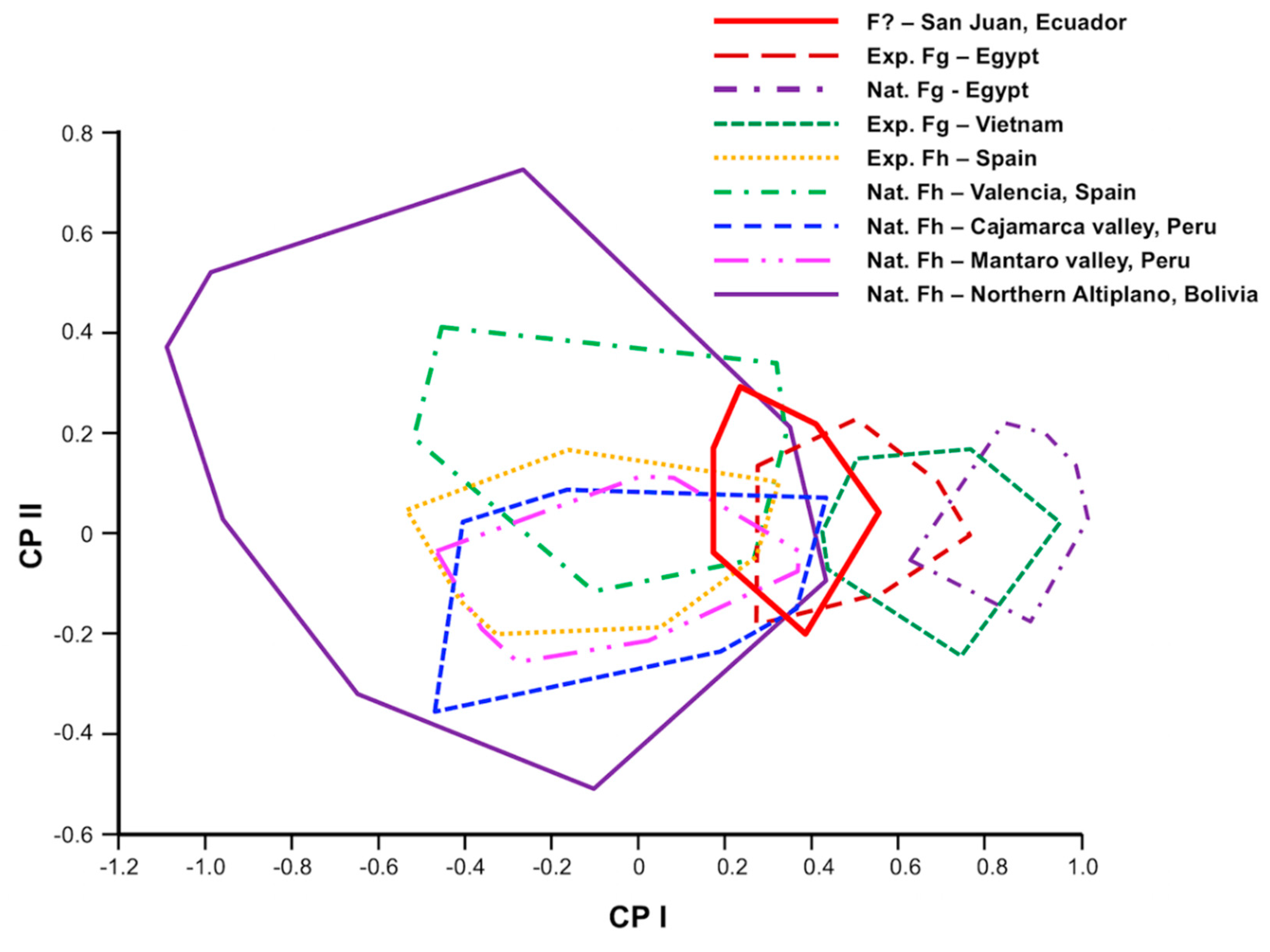
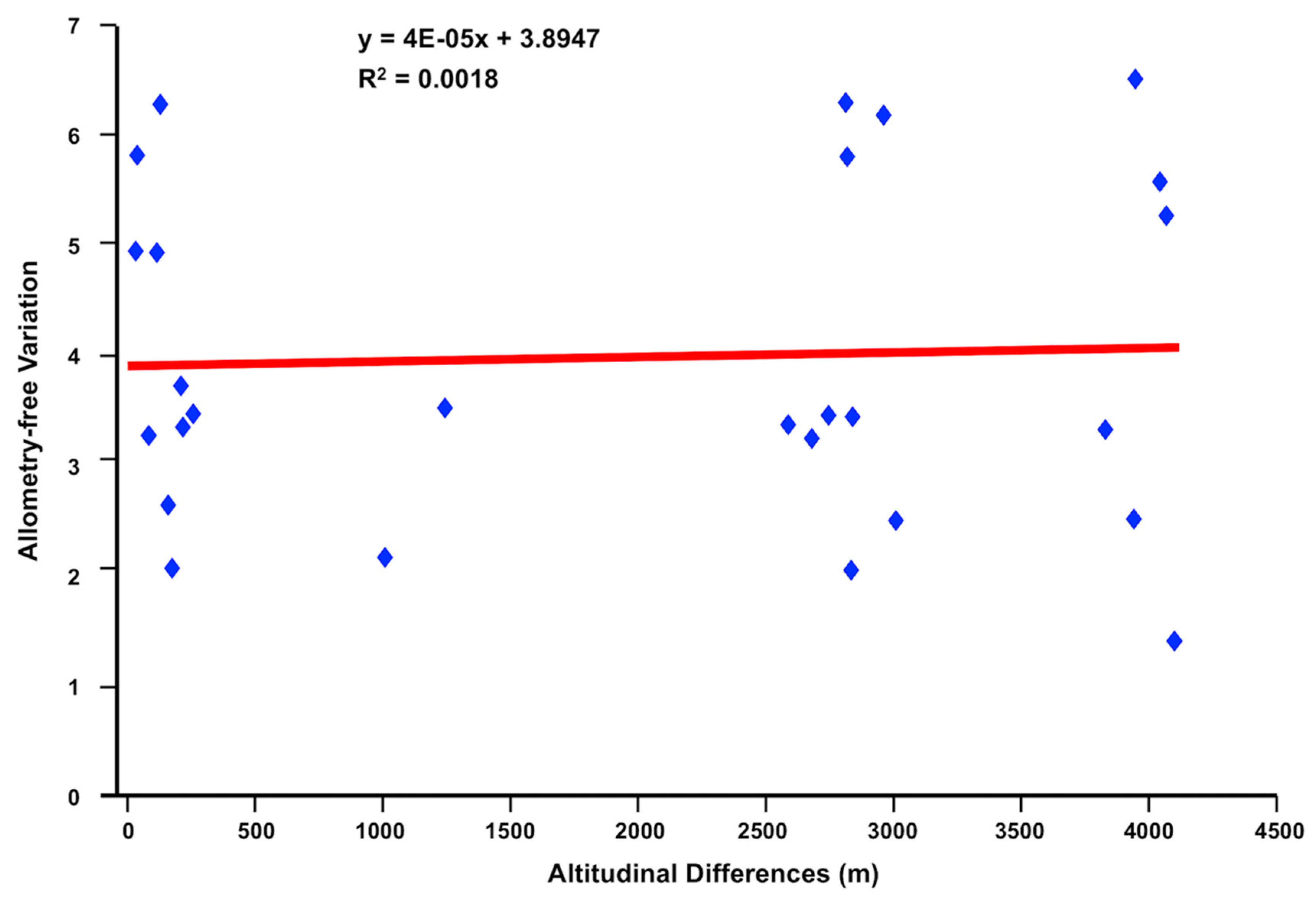
3.1. Morphometric Analyses of Sheep Liver Flukes
3.2. DNA Sequence Analyses
4. Discussion
4.1. Phenotypic Analysis
4.1.1. Fasciola hepatica from Highland and Lowland Sheep
4.1.2. Liver Flukes from Ecuadorian Sheep Compared to F. hepatica and F. gigantica
4.2. Molecular Analysis
4.2.1. DNA Sequence Characterization of Fasciolids from Ecuador
- The colonial period: Three introduction routes were involved. (i) The maritime route through the Pacific coast was the initial entry for livestock from Central America, at the beginning of the colonization of South America. (ii) The southern terrestrial route from Peru started early thereafter, as soon as the European colonizers learned about the interest of Quito for the old Incas. (iii) The northern route through the border with present day Colombia was implemented later, with an exchange of silver transport from the Bolivian Potosi mine up to the Venezuelan haven of Cartagena. The latter silver transport route might have been the most important concerning the probability of introduction of F. hepatica haplotypes from both southern and northern countries into Ecuador.
- The post-colonial period: The original European livestock evidenced problems of adaptation to the Ecuadorian environment both throughout the tropical lowlands along the coast as well as in the Andean highlands. During the XIXth and XXth centuries, Ecuador became a great livestock importer, mainly to improve the local cattle breeds. Importations of many thousands of animals were performed from the XIXth century, but mainly during the first two thirds of the XXth century. These importations were mainly from other countries of South America, but sporadically also from Europe and Asia, as well as North and Central America [90].
4.2.2. American Big-Sized F. hepatica Introduction Routes
4.3. Radix Absence and the Evolutionary Snail Vector Filter
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Sustaining the Drive to Overcome the Global Impact of Neglected Tropical Diseases; Department of Control of Neglected Tropical Diseases, World Health Organization: Geneva, Switzerland, 2013; pp. 1–138. [Google Scholar]
- World Health Organization. Ending the Neglect to Attain the Sustainable Development Goals. A Road Map for Neglected Tropical Diseases 2021–2030; World Health Organization: Geneva, Switzerland, 2020; pp. 1–47. Available online: https://www.who.int/neglected_diseases/Ending-the-neglect-to-attain-the-SDGs--NTD-Roadmap.pdf (accessed on 4 April 2021).
- Chen, M.G.; Mott, K.E. Progress in assessment of morbidity due to Fasciola hepatica infection: A review of recent literature. Trop. Dis. Bull. 1990, 87, R1–R38. [Google Scholar]
- Mas-Coma, S.; Agramunt, V.H.; Valero, M.A. Neurological and ocular fascioliasis in humans. Adv. Parasitol. 2014, 84, 27–149. [Google Scholar] [PubMed]
- Gonzalez-Miguel, J.; Valero, M.A.; Reguera-Gomez, M.; Mas-Bargues, C.; Bargues, M.D.; Simon-Martin, F.; Mas-Coma, S. Numerous Fasciola plasminogen-binding proteins may underlie blood-brain barrier leakage and explain neurological disorder complexity and heterogeneity in the acute and chronic phases of human fascioliasis. Parasitology 2019, 146, 284–298. [Google Scholar] [CrossRef] [Green Version]
- Mas-Coma, S.; Bargues, M.D.; Valero, M.A. Diagnosis of human fascioliasis by stool and blood techniques: Update for the present global scenario. Parasitology 2014, 141, 1918–1946. [Google Scholar] [CrossRef]
- De, N.V.; Le, T.H.; Agramunt, V.H.; Mas-Coma, S. Early postnatal and preschool age infection by Fasciola spp.: Report of five cases from Vietnam and worldwide review. Am. J. Trop. Med. Hyg. 2020, 103, 1578–1589. [Google Scholar] [CrossRef] [PubMed]
- Valero, M.A.; Santana, M.; Morales, M.; Hernandez, J.L.; Mas-Coma, S. Risk of gallstone disease in advanced chronic phase of fascioliasis: An experimental study in a rat model. J. Infect. Dis. 2003, 188, 787–793. [Google Scholar] [CrossRef] [Green Version]
- Valero, M.A.; Navarro, M.; Garcia-Bodelon, M.A.; Marcilla, A.; Morales, M.; Garcia, J.E.; Hernandez, J.L.; Mas-Coma, S. High risk of bacterobilia in advanced experimental chronic fasciolosis. Acta Trop. 2006, 100, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Valero, M.A.; Girones, N.; Garcia-Bodelon, M.A.; Periago, M.V.; Chico-Calero, I.; Khoubbane, M.; Fresno, M.; Mas-Coma, S. Anaemia in advanced chronic fasciolosis. Acta Trop. 2008, 108, 35–43. [Google Scholar] [CrossRef]
- Rondelaud, D.; Dreyfuss, G.; Vignoles, P. Clinical and biological abnormalities in patients after fasciolosis treatment. Med. Mal. Infect. 2006, 36, 466–468. [Google Scholar] [CrossRef] [PubMed]
- Esteban, J.G.; Flores, A.; Angles, R.; Strauss, W.; Aguirre, C.; Mas-Coma, S. A population-based coprological study of human fascioliasis in a hyperendemic area of the Bolivian Altiplano. Trop. Med. Int. Health 1997, 2, 695–699. [Google Scholar] [CrossRef]
- Esteban, J.G.; Gonzalez, C.; Bargues, M.D.; Angles, R.; Sanchez, C.; Naquira, C.; Mas-Coma, S. High fascioliasis infection in children linked to a man-made irrigation zone in Peru. Trop. Med. Int. Health 2002, 7, 339–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteban, J.G.; Gonzalez, C.; Curtale, F.; Muñoz-Antoli, C.; Valero, M.A.; Bargues, M.D.; El Sayed, M.; El Wakeel, A.; Abdel-Wahab, Y.; Montresor, A.; et al. Hyperendemic fascioliasis associated with schistosomiasis in villages in the Nile Delta of Egypt. Am. J. Trop. Med. Hyg. 2003, 69, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.C.; Esteban, J.G.; Bargues, M.D.; Valero, M.A.; Ortiz, P.; Naquira, C.; Mas-Coma, S. Hyperendemic human fascioliasis in Andean valleys: An altitudinal transect analysis in children of Cajamarca province, Peru. Acta Trop. 2011, 120, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Mas-Coma, S.; Bargues, M.D.; Valero, M.A. Human fascioliasis infection sources, their diversity, incidence factors, analytical methods and prevention measures. Parasitology 2018, 145, 1665–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mas-Coma, S.; Valero, M.A.; Bargues, M.D. Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv. Parasitol. 2009, 69, 41–146. [Google Scholar] [PubMed]
- Ollerenshaw, C.B.; Rowlands, W.T. A method of forecasting the incidence of fascioliasis in Anglesey. Vet. Rec. 1959, 71, 591–598. [Google Scholar]
- Armour, J. The epidemiology and control of bovine fascioliasis. Vet. Rec. 1975, 96, 198–201. [Google Scholar] [CrossRef]
- Dinnik, J.A.; Dinnik, N.N. Effects of seasonal variations of temperature on development of Fasciola gigantica in the snail host in the Kenyan highlands. Bull. Epizoot. Dis. Afr. 1963, 11, 197–207. [Google Scholar]
- Afshan, K.; Fortes-Lima, C.A.; Artigas, P.; Valero, M.A.; Qayyum, M.; Mas-Coma, S. Impact of climate change and man-made irrigation systems on the transmission risk, long-term trend and seasonality of human and animal fascioliasis in Pakistan. Geospat. Health 2014, 8, 317–334. [Google Scholar] [CrossRef]
- Schillhorn Van Veen, T.W. Dynamics of Lymnaea natalensis population in the Zaria area (Nigeria) and the relation to Fasciola gigantica infections. Acta Trop. 1980, 37, 183–194. [Google Scholar]
- Fuentes, M.V.; Valero, M.A.; Bargues, M.D.; Esteban, J.G.; Angles, R.; Mas-Coma, S. Analysis of climatic data and forecast indices for human fascioliasis at very high altitude. Ann. Trop. Med. Parasitol. 1999, 93, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Mas-Coma, S.; Valero, M.A.; Bargues, M.D. Climate change effects on trematodiases, with emphasis on zoonotic fascioliasis and schistosomiasis. Vet. Parasitol. 2009, 163, 264–280. [Google Scholar] [CrossRef]
- Bargues, M.D.; Artigas, P.; Angles, R.; Osca, D.; Duran, P.; Buchon, P.; Gonzalez-Pomar, R.K.; Pinto-Mendieta, J.; Mas-Coma, S. Genetic uniformity, geographical spread and anthropogenic habitat modifications of lymnaeid vectors found in a One Health initiative in the highest human fascioliasis hyperendemic of the Bolivian Altiplano. Parasit Vectors 2020, 13, 171. [Google Scholar] [CrossRef] [PubMed]
- Chougar, L.; Mas-Coma, S.; Artigas, P.; Harhoura, K.; Aissi, M.; Agramunt, V.H.; Bargues, M.D. Genetically "pure" Fasciola gigantica discovered in Algeria: DNA multimarker characterization, trans-Saharan introduction from a Sahel origin and spreading risk into northwestern Maghreb countries. Transb. Emerg. Dis. 2020, 67, 2190–2205. [Google Scholar] [CrossRef]
- Stift, M.; Michel, E.; Sitnikova, T.Y.; Mamonova, E.Y.; Sherbakov, D.Y. Palaearctic gastropod gains a foothold in the dominion of endemics: Range expansion and morphological change of Lymnaea (Radix) auricularia in Lake Baikal. Hydrobiologia 2004, 513, 101–108. [Google Scholar] [CrossRef]
- Valero, M.A.; Bargues, M.D.; Khoubbane, M.; Artigas, P.; Quesada, C.; Berinde, L.; Ubeira, F.M.; Mezo, M.; Hernandez, J.L.; Agramunt, V.H.; et al. Higher physiopathogenicity by Fasciola gigantica than by the genetically close F. hepatica: Experimental long-term follow-up of biochemical markers. Trans. Roy. Soc. Trop. Med. Hyg. 2016, 110, 55–66. [Google Scholar] [PubMed]
- Stemmermann, G.N. Human infestation with Fasciola gigantica. Am. J. Pathol. 1953, 29, 731–759. [Google Scholar]
- Janssens, P.G.; Fain, A.; Limbos, P.; De Muynck, A.; Biemans, R.; Van Meirvenne, N.; De Mulder, P. Trois cas de distomatose hépatique à Fasciola gigantica contractés en Afrique centrale. Ann. Soc. Belg. Med. Trop. 1968, 48, 637–650. [Google Scholar]
- Curtale, F.; Hassanein, Y.A.W.; Barduagni, P.; Yousef, M.M.; El Wakeel, A.; Hallaj, Z.; Mas-Coma, S. Human fascioliasis infection: Gender difference within school-age children from endemic areas of the Nile Delta, Egypt. Trans. Roy. Soc. Trop. Med. Hyg. 2007, 101, 155–160. [Google Scholar] [CrossRef]
- Fentie, T.; Erqou, S.; Gedefaw, M.; Desta, A. Epidemiology of human fascioliasis and intestinal parasitosis among schoolchildren in Lake Tana Basin, northwest Ethiopia. Trans. R. Soc. Trop. Med. Hyg. 2013, 107, 480–486. [Google Scholar] [CrossRef]
- Ashrafi, K.; Valero, M.A.; Peixoto, R.V.; Artigas, P.; Panova, M.; Mas-Coma, S. Distribution of Fasciola hepatica and F. gigantica in the endemic area of Guilan, Iran: Relationships between zonal overlap and phenotypic traits. Infect. Genet. Evol. 2015, 31, 95–109. [Google Scholar] [CrossRef] [PubMed]
- De, N.V.; Murrell, K.D.; Cong, L.D.; Cam, P.D.; Chau, L.V.; Toan, N.D.; Dalsgaard, A. The food-borne trematode zoonoses of Vietnam. Southeast. Asian J. Trop. Med. Publ. Health 2003, 34, S12–S34. [Google Scholar]
- Chen, J.X.; Chen, M.X.; Ai, L.; Xu, X.N.; Jiao, J.M.; Zhu, T.J.; Su, H.Y.; Zang, W.; Luo, J.J.; Guo, Y.H.; et al. An outbreak of human Fascioliasis gigantica in Southwest China. PLoS ONE 2013, 8, e71520. [Google Scholar]
- Price, E.W. The fluke situation in North American ruminants. J. Parasitol. 1953, 39, 119–134. [Google Scholar] [CrossRef]
- Gomez-Agudelo, T.; Perez-Reyes, R.; Zeron-Bravo, F. Fasciolosis en Mexico. Estado actual y huespedes intermediarios. Rev. Latinoamer. Microbiol. 1978, 120, 121–127. [Google Scholar]
- Carrada-Bravo, T. Fascioliasis. Diagnóstico, epidemiología y tratamientos. Rev. Gastroenterol. México 2003, 68, 135–142. [Google Scholar]
- Robles-Pérez, D.; García-García, P.; Martínez-Pérez, J.M.; Rojo-Vázquez, F.A.; Martínez-Valladares, M. Analysis of genetic variability of Fasciola hepatica populations from different geographical locations by ISSR-PCR. Parasitology 2015, 142, 527–533. [Google Scholar] [CrossRef] [Green Version]
- Valero, M.A.; Bargues, M.D.; Calderon, L.; Artigas, P.; Mas-Coma, S. First phenotypic and genotypic description of Fasciola hepatica infecting highland cattle in the State of Mexico, Mexico. Infect. Genet. Evol. 2018, 64, 231–240. [Google Scholar] [CrossRef]
- Cornelissen, J.B.W.J.; Gaasenbeek, C.P.H.; Borgsteede, F.H.M.; Holland, W.G.; Harmsen, M.M.; Boersma, W.J.A. Early immunodiagnosis of fasciolosis in ruminants using recombinant Fasciola hepatica cathepsin L-like protease. Int. J. Parasitol. 2001, 31, 728–737. [Google Scholar] [CrossRef]
- Villavicencio, A.F.; Bargues, M.D.; Artigas, P.; Guaman, R.; Ulloa, S.M.; Romero, J.; Osca, D.; Mas-Coma, S. Lymnaeid snail vectors of fascioliasis, including the first finding of Lymnaea neotropica in Ecuador, assessed by ribosomal DNA sequencing in the southern zone close to the Peru border. Acta Parasitol. 2019, 64, 839–849. [Google Scholar] [CrossRef]
- Bargues, M.D.; Gayo, V.; Sanchis, J.; Artigas, P.; Khoubbane, M.; BirrieI, S.; Mas-Coma, S. DNA multigene characterization of Fasciola hepatica and Lymnaea neotropica and its fascioliasis transmission capacity in Uruguay, with historical correlation, human report review and infection risk analysis. PLoS Negl. Trop. Dis. 2017, 11, e0005352. [Google Scholar] [CrossRef] [PubMed]
- Bargues, M.D.; Artigas, P.; Khoubbane, M.; Ortiz, P.; Naquira, C.; Mas-Coma, S. Molecular characterisation of Galba truncatula, Lymnaea neotropica and L. schirazensis from Cajamarca, Peru and their potential role in transmission of human and animal fascioliasis. Parasit. Vectors 2012, 5, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bargues, M.D.; Malandrini, J.B.; Artigas, P.; Soria, C.C.; Velasquez, J.N.; Carnevale, S.; Mateo, L.; Khoubbane, M.; Mas-Coma, S. Human fascioliasis endemic areas in Argentina: Multigene characterisation of the lymnaeid vectors and climatic-environmental assessment of the transmission pattern. Parasit. Vectors 2016, 9, 306. [Google Scholar] [CrossRef] [Green Version]
- Valero, M.A.; Darce, N.A.; Panova, M.; Mas-Coma, S. Relationship between host species and morphometric patterns in Fasciola hepatica adults and eggs from the northern Bolivian Altiplano hyperendemic region. Vet. Parasitol. 2001, 102, 85–100. [Google Scholar] [CrossRef]
- Valero, M.A.; Panova, M.; Comes, A.M.; Fons, R.; Mas-Coma, S. Patterns in size and shedding of Fasciola hepatica eggs by naturally and experimentally infected murid rodents. J. Parasitol. 2002, 88, 308–313. [Google Scholar] [CrossRef]
- Valero, M.A.; Perez-Crespo, I.; Periago, M.V.; Khoubbane, M.; Mas-Coma, S. Fluke egg characteristics for the diagnosis of human and animal fascioliasis by Fasciola hepatica and F. gigantica. Acta Trop. 2009, 111, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Valero, M.A.; Panova, M.; Mas-Coma, S. Phenotypic analysis of adults and eggs of Fasciola hepatica by computer image analysis system. J. Helminthol. 2005, 79, 217–225. [Google Scholar] [CrossRef]
- Boray, J.C. Experimental fascioliasis in Australia. Adv. Parasitol. 1969, 8, 95–210. [Google Scholar]
- Valero, M.A.; De Renzi, M.; Panova, M.; García-Bodelon, M.A.; Periago, M.V.; Ordoñez, D.; Mas-Coma, S. Crowding effect on adult growth, pre-patent period and egg shedding of Fasciola hepatica. Parasitology 2006, 133, 453–463. [Google Scholar] [CrossRef]
- Periago, M.V.; Valero, M.A.; Panova, M.; Mas-Coma, S. Phenotypic comparison of allopatric populations of Fasciola hepatica and Fasciola gigantica from European and African bovines using a computer image analysis system (CIAS). Parasitol. Res. 2006, 99, 368–378. [Google Scholar] [CrossRef]
- Periago, M.V.; Valero, M.A.; El Sayed, M.; Ashrafi, K.; El Wakeel, A.; Mohamed, M.Y.; Desquesnes, M.; Curtale, F.; Mas-Coma, S. First phenotypic description of Fasciola hepatica/Fasciola gigantica intermediate forms from the human endemic area of the Nile Delta, Egypt. Infect. Genet. Evol. 2008, 8, 51–58. [Google Scholar] [CrossRef]
- Anonymous. The Proven Solution for Image Analysis. Image-Pro1 Plus, Version 4.5 for WindowsTM, Start-Up Guide; Media Cybernetics, Inc.: Silver Spring, MD, USA, 2001; pp. 8/1–8/16. [Google Scholar]
- Ohnuma, K.; Yomo, T.; Asashima, M.; Kaneko, K. Sorting of cells of the same size, shape, and cell cycle stage for a single cell level assay without staining. BMC Cell Biol. 2006, 7, 25. [Google Scholar] [CrossRef] [Green Version]
- Rohlf, F.J.; Marcus, L.F. A revolution in morphometrics. TREE 1993, 8, 129–132. [Google Scholar]
- Bookstein, F.L. Size and shape: A comment on semantics. Syst. Zool. 1989, 38, 173–180. [Google Scholar] [CrossRef]
- Dujardin, J.-P. Morphometrics applied to medical entomology. Infect. Genet. Evol. 2008, 8, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, C.P. Multivariate allometry. In Advances in Morphometrics, Proceedings of the 1993 NATO-ASI on Morphometrics, Ciocco, Tuscany, Italy, 18–30 July 1993; NATO ASI; Series A Life Sciences; Marcus, L.F., Corti, M., Loy, A., Naylor, G.J.P., Slice, D., Eds.; Plenum Publishers: New York, NY, USA, 1996; pp. 23–49. 587p. [Google Scholar]
- Dujardin, J.-P.; Le Pont, F. Geographical variation of metric properties within the neotropical sandflies. Infect. Genet. Evol. 2004, 4, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Dos Reis, S.F.; Pessoa, L.M.; Strauss, R.E. Application of size-free canonical discriminant analysis to studies of geographic differentation. Braz. J. Gen. 1990, 13, 509–520. [Google Scholar]
- Dujardin, J.-P. BAC Software; Institute de Reserche pour le Développement—IRD: Montpellier, France, 2002; Available online: http://www.fsf.org/copyleft/gpl.html (accessed on 14 March 2012).
- Dujardin, J.-P.; Kaba, D.; Henry, A.B. The exchangeability of shape. BMC Res. Notes 2010, 3, 266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahalanobis, P.C. On the generalized distance in statistics. Proc. Natl. Inst. Sci. India 1936, 2, 49–55. [Google Scholar]
- Mas-Coma, S.; Funatsu, R.; Bargues, M.D. Fasciola hepatica and lymnaeid snails occurring at very high altitude in South America. Parasitology 2001, 123, S115–S127. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.G.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Sys. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Glez-Peña, D.; Gómez-Blanco, D.; Reboiro-Jato, M.; Fdez-Riverola, F.; Posada, D. ALTER: Program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010, 38, W14–W18. [Google Scholar] [CrossRef] [PubMed]
- Bargues, M.D.; Mas-Coma, S. Reviewing lymnaeid vectors of fascioliasis by ribosomal DNA sequence analyses. J. Helminthol. 2005, 79, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Dujardin, J.-P.; Costa, J.; Bustamante, D.; Jaramillo, N.; Catalã, S. Deciphering morphology in Triatominae: The evolutionary signals. Acta Trop. 2009, 110, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.R.; Mayhew, T.M.; Haas, J.D. The volumetric composition of human term placentae: Altitudinal, ethnic and sex differences in Bolivia. J. Anat. 1987, 52, 173–187. [Google Scholar]
- Frisancho, D.; Frisancho, O. Fisiología y patología digestiva en gran altitud. Rev. Gastroenterol. Peru 1992, 12, 155–158. [Google Scholar] [PubMed]
- Valero, M.A.; Panova, M.; Mas-Coma, S. Development differences in the uterus of Fasciola hepatica between livestock liver fluke populations from Bolivian highland and European lowlands. Parasitol. Res. 2001, 87, 337–342. [Google Scholar] [CrossRef]
- Valero, M.A.; Perez-Crespo, I.; Khoubbane, M.; Artigas, P.; Panova, M.; Ortiz, P.; Maco, V.; Espinoza, J.R.; Mas-Coma, S. Fasciola hepatica phenotypic characterisation in Andean human endemic areas: Valley versus altiplanic patterns analysed in liver flukes from sheep from Cajamarca and Mantaro, Peru. Infect. Genet. Evol. 2012, 12, 403–410. [Google Scholar] [CrossRef]
- Valero, M.A.; Marcos, M.D.; Comes, A.M.; Sendra, M.; Mas-Coma, S. Comparison of adult liver flukes from highland and lowland populations of Bolivian and Spanish sheep. J. Helminthol. 1999, 73, 341–345. [Google Scholar] [CrossRef]
- Mas-Coma, S.; Buchon, P.; Funatsu, I.R.; Angles, R.; Artigas, P.; Valero, M.A.; Bargues, M.D. Sheep and cattle reservoirs in the highest human fascioliasis hyperendemic area: Experimental transmission capacity, field epidemiology, and control within a One Health initiative in Bolivia. Front. Vet. Sci. 2020, 7, 583204. [Google Scholar] [CrossRef] [PubMed]
- Mansour, T.E. The effect of serotonin on phenol oxidase from the liver fluke Fasciola hepatica, and from other sources. Biochim. Biophys. Acta 1958, 30, 492–500. [Google Scholar] [CrossRef]
- Bjorkman, N.; Thorsell, W. On the fine morphology of the egg-shell globules in the vitelline glands of the liver fluke (F. hepatica). Exp. Cell. Res. 1963, 32, 153–156. [Google Scholar] [CrossRef]
- McGonigle, S.; Dalton, J.P. Isolation of Fasciola hepatica haemoglobin. Parasitology 1995, 111, 209–215. [Google Scholar] [CrossRef]
- Bargues, M.D.; Angles, R.; Coello, J.; Artigas, P.; Funatsu, I.R.; Cuervo, P.F.; Buchon, P.; Mas-Coma, S. One Health initiative in the Bolivian Altiplano human fascioliasis hyperendemic area: Lymnaeid biology, population dynamics, microecology and climatic factor influences. Braz. J. Vet. Parasitol. 2021, 30, e025620. [Google Scholar] [CrossRef] [PubMed]
- Valero, M.A.; Marcos, M.D.; Mas-Coma, S. A mathematical model for the ontogeny of Fasciola hepatica in the definitive host. Res. Rev. Parasitol. 1996, 56, 13–20. [Google Scholar]
- Valero, M.A.; Marcos, M.D.; Fons, R.; Mas-Coma, S. Fasciola hepatica development in experimentally infected black rat, Rattus rattus. Parasitol. Res. 1998, 84, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, K.; Valero, M.A.; Panova, M.; Periago, M.V.; Massoud, J.; Mas-Coma, S. Phenotypic analysis of adults of Fasciola hepatica, Fasciola gigantica and intermediate forms from the endemic region of Gilan, Iran. Parasitol. Int. 2006, 55, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Afshan, K.; Valero, M.A.; Qayyum, M.; Peixoto, R.V.; Magraner, A.; Mas-Coma, S. Phenotypes of intermediate forms of Fasciola hepatica and Fasciola gigantica in buffaloes from Central Punjab, Pakistan. J. Helminthol. 2014, 88, 417–426. [Google Scholar] [CrossRef]
- Ahasan, S.A.; Valero, M.A.; Chowdhury, E.H.; Islam, M.T.; Islam, M.R.; Mondal, M.M.H.; Peixoto, R.V.; Berinde, L.; Panova, M.; Mas-Coma, S. CIAS detection of Fasciola hepatica/F. gigantica intermediate forms in bovines from Bangladesh. Acta Parasitol. 2016, 61, 267–277. [Google Scholar] [CrossRef] [Green Version]
- Mas-Coma, S.; Bargues, M.D. Populations, hybrids and the systematic concepts of species and subspecies in Chagas disease triatomine vectors inferred from nuclear ribosomal and mitochondrial DNA. Acta Trop. 2009, 110, 112–136. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, S.; Ohari, Y.; Jin, S.; Calvopina, M.; Takagi, H.; Sugiyama, H.; Itagaki, T. Molecular characterization revealed Fasciola specimens in Ecuador are all Fasciola hepatica, none at all of Fasciola gigantica or parthenogenic Fasciola species. Parasitol. Int. 2021, 80, 102215. [Google Scholar] [CrossRef]
- Semyenova, S.K.; Morozova, E.V.; Chrisanfova, G.C.; Gorokhov, V.V.; Arkhipov, I.A.; Moskvin, A.S.; Movsessian, S.O.; Ryskov, A.P. Genetic differentiation in eastern European and western Asian populations of the liver fluke, Fasciola hepatica, as revealed by mitochondrial nad1 and cox1 genes. J. Parasitol. 2006, 92, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Saijuntha, W.; Tantrawatpan, C.; Agatsuma, T.; Wang, C.; Intapane, P.M.; Maleewong, W.c; Petney, T.N. Revealing genetic hybridization and DNA recombination of Fasciola hepatica and Fasciola gigantica in nuclear introns of the hybrid Fasciola flukes. Mol. Biochem. Parasitol. 2018, 223, 31–36. [Google Scholar] [CrossRef]
- Vizcarra, R.; Tapia, D.; Lasso, R.; Jimenez, M. La Leche del Ecuador—Historia de la Lechería Ecuatoriana; Centro de la Industria Láctea del Ecuador: Quito, Ecuador, 2015; 183p. [Google Scholar]
- Valero, M.A.; Mas-Coma, S. Comparative infectivity of Fasciola hepatica metacercariae from isolates of the main and secondary reservoir animal host species in the Bolivian Altiplano high human endemic region. Folia Parasitol. 2000, 47, 17–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valero, M.A.; Perez-Crespo, I.; Chillon-Marinas, C.; Khoubbane, M.; Quesada, C.; Reguera-Gomez, M.; Mas-Coma, S.; Fresno, M.; Girones, N. Fasciola hepatica reinfection potentiates a mixed Th1/Th2/Th17/Treg response and correlates with the clinical phenotypes of anemia. PLoS ONE 2017, 12, e0173456. [Google Scholar] [CrossRef]
- Valero, M.A.; Girones, N.; Reguera-Gomez, M.; Perez-Crespo, I.; Lopez-Garcia, M.P.; Quesada, C.; Bargues, M.D.; Fresno, M.; Mas-Coma, S. Impact of fascioliasis reinfection on Fasciola hepatica egg shedding: Relationship with the immune-regulatory response. Acta Trop. 2020, 209, 105518. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.O. History and development of zebu cattle in the United States. J. Anim. Sci. 1980, 50, 1188–1200. [Google Scholar] [CrossRef]
- Meirelles, F.V.; Rosa, A.J.M.; Lobo, R.B.; García, J.M.; Smith, L.C.; Duarte, F.A.M. Is the American zebu really Bos indicus? Genet. Mol. Biol. 1999, 22, 543–546. [Google Scholar] [CrossRef] [Green Version]
- Mirol, M.; Giovambattista, G.; Liron, J.P.; Dulout, F.N. African and European mitochondrial haplotypes in South American Creole cattle. Heredity 2003, 91, 248–254. [Google Scholar] [CrossRef] [Green Version]
- Sinitsin, D.F. Studien über die Phylogenie der Trematoden. VI. The life histories of some American liver flukes. Z. Parasitenk. 1933, 6, 170–191. [Google Scholar] [CrossRef]
- Borchart de Moreno, C. Llamas y ovejas: El desarrollo del ganado lanar en la Audiencia de Quito, S. XVI. In La Audiencia de Quito. Aspectos Económicos y Sociales (Siglos XVI–XVIII); Colección Pendoneros, Ediciones del Banco Central de Ecuador y Abya-Yala: Quito, Ecuador, 1998; Volume 23, pp. 15–46. [Google Scholar]
- Mendoza, B.; Peña, L. Situación actual y perspectivas de los ovinos en Ecuador. In Biodiversidad Ovina Iberoamericana. Caracterización y Uso Sostenible; Delgado Bermejo, J.V., Nogales Baena, S., Eds.; Córdoba, Argentina, 2009; pp. 433–446. [Google Scholar]
- Edifarm. Vademécum Veterinario. Vol. 1; Grupo Latino Editores, Edifarm, División de Publicaciones Técnicas: Quito, Ecuador, 2006; pp. 1–189. [Google Scholar]
- Anonymous. Ecuador importará 12000 cabezas de ganado este año. Rev. Líderes Lect. 2015, 2588, 1–2. Available online: https://www.revistalideres.ec/lideres/ecuador-importacion-ganado.html (accessed on 24 March 2021).
- Ortiz Bonilla, M.J. Consolidación del Turismo de Aventura en la Ruta Quito-Chiriboga. Trabajo de Titulación, Previo a la Obtención del Título de Ingeniero en Administración Turística y Hotelera; Departamento de Ciencias Económicas Administrativas y de Comercio, Universidad de las Fuerzas Armadas: Sangolqui, Ecuador, 2017; pp. 1–185. [Google Scholar]
- Bargues, M.D.; Vigo, M.; Horak, P.; Dvorak, J.; Patzner, R.A.; Pointier, J.P.; Jackiewicz, M.; Meier-Brook, C.; Mas-Coma, S. European Lymnaeidae (Mollusca: Gastropoda), intermediate hosts of trematodiases, based on nuclear ribosomal DNA ITS-2 sequences. Infect. Genet. Evol. 2001, 1, 85–107. [Google Scholar] [CrossRef]
- Weider, L.J.; Elser, J.J.; Crease, T.J.; Mateos, M.; Cotner, J.B.; Markow, T.A. The functional significance of ribosomal (r)DNA Variation: Impacts on the evolutionary ecology of organisms. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 219–242. [Google Scholar] [CrossRef] [Green Version]
- White, D.J.; Wolff, J.N.; Pierson, M.; Gemmell, N.J. Revealing the hidden complexities of mtDNA inheritance. Mol. Ecol. 2008, 17, 4925–4942. [Google Scholar] [CrossRef] [PubMed]
- Paraense, W.L. Lymnaea viatrix and Lymnaea columella in the Neotropical region: A distributional outline. Mem. Inst. Osw. Cruz 1982, 77, 181–188. [Google Scholar] [CrossRef]
- Paraense, W.L. Planorbidae, Lymnaeidae and Physidae of Ecuador (Mollusca: Basommatophora). Mem. Inst. Osw. Cruz 2004, 99, 357–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bargues, M.D.; Artigas, P.; Khoubbane, M.; Mas-Coma, S. DNA sequence characterisation and phylogeography of Lymnaea cousini and related species, vectors of fascioliasis in northern Andean countries, with description of L. meridensis n. sp. (Gastropoda: Lymnaeidae). Parasit. Vectors 2011, 4, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bargues, M.D.; Artigas, P.; Khoubbane, M.; Flores, R.; Glöer, P.; Rojas-Garcia, R.; Ashrafi, K.; Falkner, G.; Mas-Coma, S. Lymnaea schirazensis, an overlooked snail distorting fascioliasis data: Genotype, phenotype, ecology, worldwide spread, susceptibility, applicability. PLoS ONE 2011, 6, e24567. [Google Scholar] [CrossRef]
- Narvaez, A.O.; Aroca, J.M.; Alda, P.; Macías Castro, V.; Lounnas, M.; Hurtrez-Bousses, S.; Noya, O.; Martini Robles, L.; Pointier, J.P. Primer reporte de Galba cubensis (Gastropoda: Lymnaeidae) en el Ecuador, hospedador potencial de Fasclola hepatica en arrozales de la cosla ecuatoriana. Misionero Agro 2017, 13, 36–47. [Google Scholar]
- Dreyfuss, G.; Correa, A.C.; Djuikwo-Teukeng, F.F.; Novobilsky, A.; Höglund, J.; Pankrac, J.; Kasny, M.; Vignoles, P.; Hurtrez-Bousses, S.; Pointier, J.P.; et al. Differences in the compatibility of infection between the liver flukes Fascioloides magna and Fasciola hepatica in a Colombian population of the snail Galba sp. J. Helminthol. 2015, 89, 720–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganley, A.R.D.; Kobayashi, T. Monitoring the rate and dynamics of concerted evolution in the ribosomal DNA repeats of Saccharomyces cerevisiae using experimental evolution. Mol. Biol. Evol. 2011, 28, 2883–2891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbons, J.G.; Branco, A.T.; Godinho, S.A.; Yu, S.; Lemos, B. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc. Natl. Acad. Sci. USA 2015, 112, 2485–2490. [Google Scholar] [CrossRef] [Green Version]
- Reblanova, M.; Spakulova, M.; Orosova, M.; Kralova-Hromadova, I.; Bazsalovicsova, E.; Rajsky, D. A comparative study of karyotypes and chromosomal location of rDNA genes in important liver flukes Fasciola hepatica and Fascioloides magna (Trematoda: Fasciolidae). Parasitol. Res. 2011, 109, 1021–1028. [Google Scholar] [CrossRef]

| Adult Measurements (mm) | Altiplano Bolivia n = 201 nAlt | Mantaro Peru n = 47 nMan | Cajamarca Peru n = 130 nCaj | San Juan Ecuador n = 42 nEc |
|---|---|---|---|---|
| Body area, BA | 31.11–236.14 | 79.89–250.58 | 47.34–283.95 | 185,21–352.28 |
| 106.39 ± 3.35 | 149.30 ± 5.64 | 135.87 ± 3.73 | 266.88 ± 4.78 | |
| Body length, BL | 9.64–31.04 | 13.41–27.75 | 13.48–30.97 | 21.49–32.78 |
| 18.08 ± 0.31 | 19.70 ± 0.40 | 18.86 ± 0.31 | 28.38 ± 0.41 | |
| Body width, BW | 4.23–13.41 | 7.60–13.93 | 5.06–14.23 | 11.74–15.25 |
| 8.26 ± 0.14 | 10.88 ± 0.23 | 10.25 ± 0.14 | 13.31 ± 0.14 | |
| BL/BW ratio | 1.41–3.74 | 1.30–2.46 | 1.31–3.73 | 1.68–2.62 |
| 2.22 ± 0.03 | 1.83 ± 0.04 | 1.86 ± 0.03 | 2.14 ± 0.04 | |
| BW at ovary level, BWOv | 3.51–11.71 | 5.48–11.31 | 4.23–11.97 | 8.20–11.63 |
| 6.71 ± 0.12 | 8.50 ± 0.19 | 8.30 ± 0.11 | 9.54 ± 0.12 | |
| Body perimeter, BP | 26.89–68.35 | 33.71–64.51 | 30.20–71.11 | 44.36–77.30 |
| 47.51 ± 0.72 | 48.15 ± 0.98 | 45.70 ± 0.66 | 65.84 ± 0.92 | |
| Body roundness, BR | 1.11–2.12 | 1.13–1.48 | 1.09–1.94 | 1.16–1.54 |
| 1.53 ± 0.01 | 1.26 ± 0.01 | 1.25 ± 0.01 | 1.33 ± 0.01 | |
| Cone length, CL | 1.32–3.04 | 1.36–2.59 | 1.32–2.98 | 1.88–2.81 |
| 2.12 ± 0.02 | 1.89 ± 0.04 | 2.11 ± 0.02 | 2.26 ± 0.03 | |
| Cone width, CW | 1.78–3.92 | 2.30–4.21 | 2.12–4.41 | 2.77–4.44 |
| 2.65 ± 0.03 | 3.04 ± 0.06 | 3.17 ± 0.04 | 3.54 ± 0.06 | |
| BWOv/CW ratio | 1.53–3.83 | 1.86–3.94 | 1.79–3.72 | 2.19–4.19 |
| 2.53 ± 0.03 | 2.82 ± 0.07 | 2.64 ± 0.03 | 2.72 ± 0.06 | |
| Oral sucker area, OSA | 0.21–0.66 | 0.19–0.72 | 0.20–0.67 | 0.29–0.66 |
| 0.38 ± 0.01 | 0.42 ± 0.01 | 0.46 ± 0.01 | 0.49 ± 0.01 | |
| Maximum diameter of the oral sucker, OSmax | 0.53–1.06 | 0.61–1.02 | 0.63–1.14 | 0.79–0.98 |
| 0.76 ± 0.01 | 0.81 ± 0.01 | 0.84 ± 0.01 | 0.88 ± 0.01 | |
| Minimum diameter of the oral sucker, OSmin | 0.42–0.86 0.63 ± 0.01 | 0.29–0.90 0.62 ± 0.02 | 0.36–0.85 0.66 ± 0.01 | 0.35–0.88 0.67 ± 0.02 |
| Ventral sucker area, VSA | 0.44–1.23 | 0.49–1.26 | 0.53–1.22 | 0.82–1.20 |
| 0.78 ± 0.01 | 0.82 ± 0.02 | 0.89 ± 0.01 | 1.01 ± 0.02 | |
| Maximum diameter of the ventral sucker, VSmax | 0.75–1.25 | 0.83–1.31 | 0.89–1.29 | 1.07–1.35 |
| 1.00 ± 0.01 | 1.08 ± 0.01 | 1.12 ± 0.01 | 1.18 ± 0.01 | |
| Minimum diameter of the ventral sucker, VSmin | 0.67–1.29 | 0.72–1.22 | 0.77–1.21 | 0.95–1.19 |
| 0.98 ± 0.01 | 0.95 ± 0.01 | 0.99 ± 0.01 | 1.07 ± 0.01 | |
| OSA/VSA ratio | 0.31–0.71 | 0.31–0.69 | 0.23–0.76 | 0.28–0.64 |
| 0.49 ± 0.01 | 0.51 ± 0.01 | 0.52 ± 0.01 | 0.49 ± 0.01 | |
| Distance between the anterior body end and ventral sucker, A-VS | 1.52–3.35 | 1.75–2.88 | 1.61–3.26 | 2.19–3.22 |
| 2.24 ± 0.02 | 2.29 ± 0.04 | 2.48 ± 0.02 | 2.82 ± 0.03 | |
| Distance between oral sucker and ventral sucker, OS-VS | 0.87–2.56 | 1.16–2.15 | 1.19–2.40 | 1.68–2.52 |
| 1.60 ± 0.02 | 1.66 ± 0.03 | 1.81 ± 0.02 | 2.13 ± 0.03 | |
| Distance between ventral sucker and vitelline gland union, VS-Vit | 4.49–19.82 | 8.11–17.36 | 6.78–20.60 | 14.14–23.60 |
| 11.32 ± 0.22 | 12.30 ± 0.30 | 11.56 ± 0.21 | 19.40 ± 0.42 | |
| Distance between vitelline gland union and posterior body end, Vit-P | 0.49–9.83 | 2.64–8.09 | 2.69–9.47 | 3.65–10.44 |
| 3.76 ± 0.14 | 5.20 ± 0.19 | 4.91 ± 0.12 | 6.56 ± 0.23 | |
| Distance between ventral sucker and posterior body end, VS-P | 7.11–27.39 | 11.27–25.36 | 11.39–28.37 | 18.78–30.47 |
| 15.07 ± 0.31 | 17.50 ± 0.43 | 16.47 ± 0.30 | 25.96 ± 0.43 | |
| BL/VS-P ratio | 0.88–1.42 | 1.09–1.19 | 1.08–1.22 | 1.06–1.19 |
| 1.22 ± 0.01 | 1.13 ± 0.004 | 1.15 ± 0.003 | 1.09 ± 0.004 | |
| Pharynx area, PhA | 0.05–0.34 | 0.13–0.31 | 0.12–0.35 | 0.11–0.30 |
| 0.17 ± 0.003 | 0.20 ± 0.01 | 0.21 ± 0.004 | 0.20 ± 0.01 | |
| Pharynx length, PhL | 0.37–0.93 | 0.58–0.91 | 0.55–0.96 | 0.60–1.06 |
| 0.68 ± 0.01 | 0.75 ± 0.01 | 0.78 ± 0.01 | 0.85 ± 0.02 | |
| Pharynx width, PhW | 0.18–0.50 | 0.25–0.48 | 0.24–0.55 | 0.52–0.19 |
| 0.34 ± 0.004 | 0.36 ± 0.01 | 0.36 ± 0.005 | 0.33 ± 0.01 | |
| Testes area, TA | 9.31–89.48 | 22.54–71.71 | 13.10–112.85 | 57.36–121.39 |
| 37.43 ± 1.35 | 48.33 ± 1.92 | 46.25 ± 1.32 | 96.11 ± 2.22 | |
| Testes length, TL | 3.12–14.62 | 5.56–12.70 | 4.54–23.53 | 7.85–18.77 |
| 8.12 ± 0.17 | 9.05 ± 0.26 | 8.86 ± 0.20 | 14.06 ± 0.35 | |
| Testes width, TW | 2.84–8.97 | 4.84–9.87 | 3.12–9.07 | 7.69–10.33 |
| 5.69 ± 0.10 | 7.01 ± 0.16 | 6.84 ± 0.09 | 8.93 ± 0.10 | |
| Testes perimeter, TP | 13.06–48.53 | 20.08–41.44 | 16.76–48.75 | 33.35–51.66 |
| 26.94 ± 0.56 | 30.68 ± 0.75 | 28.71 ± 0.44 | 44.55 ± 0.60 | |
| Testes roundness, TR | 1.26–2.50 | 1.22–2.07 | 1.19–1.94 | 1.36–1.99 |
| 1.57 ± 0.01 | 1.58 ± 0.03 | 1.45 ± 0.01 | 1.66 ± 0.02 |
| Adult Measurements (mm) | Experimental Spanish Isolate n = 127 expSp | Natural, Valencia, Spain n = 37 nSp | ||
|---|---|---|---|---|
| Extreme Values | Mean ± SD | Extreme Values | Mean ± SD | |
| Body area, BA | 68.09–227.11 | 129.78 ± 3.17 | 75.09–239.13 | 142.75 ± 6.22 |
| Body length, BL | 12.45–26.68 | 18.52 ± 0.29 | 14.21–31.17 | 20.82 ± 0.64 |
| Body width, BW | 6.88–12.74 | 10.19 ± 0.10 | 7.49–12.76 | 9.75 ± 0.16 |
| BL/BW ratio | 1.30–2.62 | 1.82 ± 0.02 | 1.70–2.89 | 2.14 ± 0.05 |
| BW at ovary level, BWOv | 5.69–10.17 | 7.98 ± 0.08 | 6.45–10.57 | 8.13 ± 0.17 |
| Body perimeter, BP | 33.22–66.06 | 47.91 ± 0.64 | 39.85–71.41 | 52.90 ± 1.33 |
| Body roundness, BR | 1.23–1.73 | 1.43 ± 0.01 | 1.31–1.76 | 1.46 ± 0.02 |
| Cone length, CL | 1.10–3.07 | 2.01 ± 0,03 | 1.55–2.98 | 2.10 ± 0.06 |
| Cone width, CW | 2.08–4.19 | 3.27 ± 0.03 | 2.46–4.12 | 3.25 ± 0.06 |
| BWOv/CW ratio | 1.86–3.81 | 2.46 ± 0.03 | 2.03–3.90 | 2.51 ± 0–05 |
| Oral sucker area, OSA | 0.24–0.53 | 0.39 ± 0.01 | 0.33–0.66 | 0.44 ± 0.01 |
| Maximum diameter of the oral sucker, OSmax | 0.70–1.01 | 0.86 ± 0.01 | 0.63–1.00 | 0.84 ± 0.01 |
| Minimum diameter of the oral sucker, OSmin | 0.33–0.74 | 0.57 ± 0.1 | 0.52–0.86 | 0.67 ± 0.01 |
| Ventral sucker area, VSA | 0.56–1.31 | 0.98 ± 0.01 | 0.95–1.35 | 1.13 ± 0.01 |
| Maximum diameter of the ventral sucker, VSmax | 0.89–1.40 | 1.19 ± 0.01 | 0.92–1.40 | 1.19 ± 0.02 |
| Minimum diameter of the ventral sucker, VSmin | 0.79–1.23 | 1.04 ± 0.01 | 0.68–1.48 | 1.06 ± 0.03 |
| OSA/VSA ratio | 0.24–0.58 | 0.41 ± 0.01 | 0.27–0.51 | 0.39 ± 0.01 |
| Distance between anterior body end and ventral sucker, A-VS | 1.16–3.09 | 2.13 ± 0.03 | 1.66–3.15 | 2.32 ± 0.05 |
| Distance between oral sucker and ventral sucker, OS-VS | 0.64–2.50 | 1.56 ± 0.03 | 1.13–2.49 | 1.65 ± 0.05 |
| Distance between ventral sucker and vitelline gland union, VS-Vit | 7.71–19.75 | 12.33 ± 0.23 | 9.65–22.97 | 13.84 ± 0.47 |
| Distance between vitelline gland union and posterior body end, Vit-P | 1.99–7.02 | 4.35 ± 0.09 | 1.12–6.89 | 3.45 ± 0.20 |
| Distance between ventral sucker and posterior body end, VS-P | 10.43–24.66 | 16.68 ± 0.29 | 11.21–26.84 | 17.29 ± 0.58 |
| BL/VS-P ratio | 1.04–1.20 | 1.11 ± 0.002 | 1.15–1.27 | 1.21 ± 0.004 |
| Pharynx area, PhA | 0.11–0.34 | 0.20 ± 0.003 | 0.15–0.31 | 0.22 ± 0.01 |
| Pharynx length, PhL | 0.51–1.07 | 0.77 ± 0.01 | 0.59–0.89 | 0.74 ± 0.01 |
| Pharynx width, PhW | 0.26–0.56 | 0.36 ± 0.004 | 0.30–0.51 | 0.40 ± 0.01 |
| Testes area, TA | 24.12–89.10 | 50.99 ± 1.29 | 29.90–87.37 | 46.27 ± 2.35 |
| Testes length, TL | 5.33–14.50 | 8.92 ± 0.17 | 6.03–14.53 | 9.39 ± 0.32 |
| Testes width, TW | 4.66–9.16 | 7.37 ± 0.07 | 5.08–9.38 | 6.42 ± 0.16 |
| Testes perimeter, TP | 22.88–47.93 | 33.13 ± 0.49 | 20.81–44.36 | 30.93 ± 0.82 |
| Testes roundness, TR | 1.41–2.25 | 1.74 ± 0.02 | 1.30–2.22 | 1.69 ± 0.04 |
| Adult Measurements (mm) | Experimental Egyptian Isolate n = 100 expEg | Natural Giza, Egypt n = 31 nEg | Experimental Vietnam Isolate n = 70 expVi |
|---|---|---|---|
| Body area, BA | 156.61–305.35 | 385.29–556.58 | 438.46–474.11 |
| 219.64 ± 2.87 | 470.0 ± 26.35 | 460.97 ± 9.58 | |
| Body length, BL | 24.41–40.51 | 40.10–54.98 | 26.88–46.85 |
| 31.47 ± 0.30 | 48.39 ± 3.36 | 36.17 ± 4.05 | |
| Body width, BW | 8.28–12.04 | 8.73–12.45 | 9.82–16.30 |
| 9.97 ± 0.09 | 10.63 ± 0.91 | 12.89 ± 1.27 | |
| BL/BW ratio | 2.23–4.31 | 3.59–5.45 | 1.84–4.59 |
| 3.18 ± 0.04 | 4.58 ± 0.47 | 2.83 ± 0.41 | |
| BW at ovary level, BWOv | 6.33–9.98 | 9.05–11.53 | 6.33–9.98 |
| 8.06 ± 0.07 | 10.07 1.02 | 8.06 ± 0.07 | |
| Body perimeter, BP | 57.98–105.24 | 57.63–114.17 | 96.70–104.11 |
| 74.41 ± 0.07 | 101.88 ± 10.32 | 100.10 ± 3.73 | |
| Body roundness, BR | 1.53–3.47 | 2.11–2.48 | 1.67–1.81 |
| 2.02 ± 0.02 | 2.28 ± 0.18 | 1.73 ± 0.07 | |
| Cone length, CL | 1.46–3.14 | 3.46–4.24 | 3.22–4.01 |
| 2.42 ± 0.03 | 3.09 ± 0.40 | 2.99 ± 0.40 | |
| Cone width, CW | 3.05–4.71 | 3.05–4.71 | 3.20–4.95 |
| 3.83 ± 0.03 | 3.83 ± 0.03 | 3.99 ± 0.03 | |
| BWOv/CW ratio | 1.62–2.66 | 1.45–2.22 | 1.30–2.25 |
| 2.11 ± 0.02 | 2.01 ± 0.02 | 2.00 ± 0.02 | |
| Oral sucker area, OSA | 0.26–0.78 | 0.28–0.89 | 0.25–0.86 |
| 0.49 ± 0.01 | 0.53 ± 0.01 | 0.55 ± 0.01 | |
| Maximum diameter of the oral sucker, OSmax | 0.79–1.20 | 0.55–1.12 | 0.88–1.23 |
| 1.01 ± 0.01 | 1.01 ± 0.10 | 1.05 ± 0.08 | |
| Minimum diameter of the oral sucker, OSmin | 0.39–0.83 | 0.45–0.98 | 0.55–0.99 |
| 0.61 ± 0.01 | 0.82 ± 0.09 | 0.82 ± 0.08 | |
| Ventral sucker area, VSA | 1.39–2.21 | 1.39–2.21 | 1.40–2.10 |
| 1.83 ± 0.02 | 1.83 ± 0.02 | 1.88 ± 0.02 | |
| Maximum diameter of the ventral sucker, VSmax | 1.36–1.92 | 1.12–1.82 | 1.18–1.75 |
| 1.61 ± 0.01 | 1.57 ± 0.14 | 1.54 012 | |
| Minimum diameter of the ventral sucker, VSmin | 1.01–1.59 | 0.95–1.64 | 0.97–1.60 |
| 1.44 ± 0.01 | 1.41 ± 0.15 | 1.38 ± 0.12 | |
| OSA/VSA ratio | 0.13–0.47 | 0.23–0.57 | 0.27–0.64 |
| 0.27 ± 0.01 | 0.38 ± 0.07 | 0.41 ± 0.06 | |
| Distance between anterior body end and ventral sucker, A-VS | 1.40–3.48 | 2.61–3.87 | 2.39–3.63 |
| 2.37 ± 0.03 | 3.33 ± 0.31 | 3.02 ± 0.27 | |
| Distance between oral sucker and ventral sucker, OS-VS | 0.75–2.67 | 0.85–2.98 | 0.84–2.99 |
| 1.76 ± 0.03 | 1.78 ± 0.03 | 1.79 ± 0.03 | |
| Distance between ventral sucker and vitelline gland union, VS-Vit | 14.24–26.61 | 21.57–38.18 | 16.27–29.09 |
| 20.96 ± 0.24 | 30.45 ± 3.76 | 22.53 ± 2.92 | |
| Distance between vitelline gland union and posterior body end, Vit-P | 5.17–12.37 | 9.82–21.37 | 6.69–18.30 |
| 8.52 ± 0.12 | 14.83 ± 2.50 | 11.84 ± 2.04 | |
| Distance between ventral sucker and posterior body end, VS-P | 22.88–38.06 | 37.25–51.23 | 25.6–45.8 |
| 29.48 ± 0.30 | 45.28 ± 3.25 | 34.3 ± 3.7 | |
| BL/VS-P ratio | 1.02–1.27 | 1.04–1.13 | 0.97–1.08 |
| 1.07 ± 0.003 | 1.07 ± 0.02 | 1.05 ± 0.02 | |
| Pharynx area, PhA | 0.14–0.41 | 0.25–0.40 | 0.19–0.39 |
| 0.26 ± 0.01 | 0.32 ± 0.01 | 0.25 ± 0.01 | |
| Pharynx length, PhL | 0.50–1.01 | 0.48–1.04 | 0.71–1.21 |
| 0.77 ± 0.01 | 0.86 ± 0.11 | 0.99 ± 0.10 | |
| Pharynx width, PhW | 0.30–0.66 | 0.27–0.61 | 0.32–0.71 |
| 0.46 ± 0.01 | 0.44 ± 0.07 | 0.48 ± 0.09 | |
| Testes area, TA | 41.51–107.06 | 42.98–125.06 | 42.50–120.02 |
| 75.96 ± 1.29 | 85.09 ± 1.30 | 85.01 ± 1.20 | |
| Testes length, TL | 8.23–18.85 | 15.45–27.60 | 9.78–22.57 |
| 14.32 ± 0.21 | 22.10 ± 3.51 | 15.73 ± 2.57 | |
| Testes width, TW | 5.40–8.72 | 5.02–7.32 | 5.01–7.00 |
| 6.90 ± 0.07 | 6.01 ± 0.06 | 5.99 ± 0.06 | |
| Testes perimeter, TP | 30.22–56.13 | 31.33–57.56 | 31.33–57.56 |
| 44.29 ± 0.51 | 44.45 ± 0.52 | 44.45 ± 0.52 | |
| Testes roundness, TR | 1.48–2.76 | 1.48–2.66 | 1.50–2.69 |
| 2.07 ± 0.03 | 2.10 ± 0.03 | 2.18 ± 0.03 |
| Fasciola spp. and Combined Haplotypes | Polymorphic Sites Intergenic Region (ITS-1, 5.8S, ITS-2) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positions | 24 | 114 | 208 | 286 | 306 | 821 | 834 | 860 | 866 | 874 | 917 | 924 | |
| Polymorphic sites ITS-1 | Polymorphic sites ITS-2 | ||||||||||||
| Positions | 24 | 114 | 208 | 286 | 306 | 234 | 248 | 273 | 279 | 288 | 330 | 337 | |
| F. hepatica 1A | C | A | C | T | C | T | A | C | C | C | T | G | |
| F. hepatica 2A | C | A | C | T | C | T | A | C | C | T | T | G | |
| F. hepatica 1/2A * | C | A | C | T | C | T | A | C | C | C/T | T | G | |
| F. gigantica 1A | T | T | T | A | T | C | A | T | T | C | - | A | |
| F. gigantica 2A | T | T | T | A | T | C | C | T | T | C | - | A | |
| F. gigantica 1/2A ** | T | T | T | A | T | C | C/A | T | T | C | - | A | |
| Fasciola hepatica Haplotypes | cox1 Nucleotide Sequence | COX1 Amino Acid Sequence | Nucleotide Composition (AT%) |
|---|---|---|---|
| Positions | 111 1111111111 222333555 6777999112 2333444445 7145027467 2246023381 4666488992 2935985673 7670670788 0567325561 | 11444 01159 30469 | |
| F. hepatica cox1-1 * cox1-H2-H69 ^ | GGCGGTGAGA TCTCTGCATA GTTCAGTAAC tttttcagag cactcatgcg acctgacttw | LSVSN fpilf | |
| Fh-cox1-16 Fh-cox1-23 Fh-cox1-53 Fh-cox1-54 Fh-cox1-55 Fh-cox1-56 Fh-cox1-57 Fh-cox1-70 (new) Fh-cox1-5 § Fh-cox1-42 § Fh-AF216697 † Fh-M93388 ‡ | .T........ ......T... .......... T......G.. C..TC.TG.. .......... .......... .A....T.C. ....G.C... .......... .AC...T... ....G.C... T......G.. C..TC.TG.. .........T .......... .A....T... ....G.CTT. T......G.. ...TC.TG.. .......... T......... ......T... .......... .......... .A....T... ....G..... ........AG ......T..G .......... T......G.. C.....T... ..CT...... ..TTTCA... ...T.ATG.. AC...A.... | ..... ..... ..... ..... ..... ....F ..... ..... ..... ..... ...L. FPI.. | 62.75 62.56 62.56 62.56 62.62 62.62 62.62 62.75 62.60 62.60 62.60 63.00 |
| Fasciola hepatica Haplotypes | nad1 Nucleotide Sequence | NAD1 Amino Acid Sequence | Nucleotide Composition (AT%) |
|---|---|---|---|
| Positions | 22335667 88 2345097176 07 0663394523 14 | 7 | |
| F. hepatica nad1-1 * nad1-H2-H51 ^ | CACTTTTTTT AT tgtccacccc gc | A v | |
| Fh-nad1-2 Fh-nad1-6 Fh-nad1-14 Fh-nad1-23 Fh-nad1-42 Fh-nad1-43 Fh-nad1-12 § Fh-AF216697 † Fh-M93388 ‡ | ..TC...C.. .. ..T...C... .. .GT....... .. ..T...C..C .. ..TC...C.. .C ..TC..CC.. .C ..T....... .. TGT.C.C.C. G. ..T..AC..C .. | . . . . . . . V . | 65.23 65.34 65.34 65.23 65.12 65.01 65.50 65.01 65.23 |
| DNA Marker | Haplotypes | Ecuador | Other Countries | ||
|---|---|---|---|---|---|
| San Juan | Zapotillo | Macará | |||
| ITS-2 | FhITS2-1 | sheep/cattle | cattle | cattle | Spain, France, Poland, Mexico, Venezuela, |
| Peru, Bolivia, Uruguay, Argentina | |||||
| FhITS2-2 | cattle | Spain, Andorra, Mexico, Bolivia, Uruguay | |||
| FhITS2-1/2 | sheep | ||||
| ITS-1 | FhITS2-A | sheep/cattle | cattle | cattle | Spain, France, Poland, Mexico, Venezuela, |
| Peru, Bolivia, Uruguay, Argentina | |||||
| cox1 | Fhcox1-16 | sheep | cattle | cattle | Venezuela (cattle), Peru (sheep/cattle), |
| Bolivia (sheep/cattle), Uruguay (cattle) | |||||
| Fhcox1-23 | sheep | Mexico (cattle), Peru (cattle), | |||
| Argentina (cattle), Chile (cattle) | |||||
| Fhcox1-53 | cattle | Chile (cattle) | |||
| Fhcox1-54 | cattle | ||||
| Fhcox1-55 | cattle | Mexico (cattle) | |||
| Fhcox1-56 | sheep | ||||
| Fhcox1-57 | sheep | ||||
| Fhcox1-70 new | sheep | ||||
| nad1 | Fhnad1-2 | sheep | Spain (sheep/cattle), Poland (bison), Venezuela (cattle), Peru | ||
| (cattle), Bolivia (cattle), Uruguay (cattle), | |||||
| Argentina (sheep/cattle), Chile (cattle) | |||||
| Fhnad1-6 | cattle | Spain (cattle) | |||
| Fhnad1-14 | sheep | cattle | cattle | Mexico (cattle), Venezuela (cattle), Peru (sheep/cattle), Uruguay (cattle), Argentina (cattle) | |
| Fhnad1-23 | sheep | Mexico (cattle), Peru (cattle), Bolivia (sheep), | |||
| Argentina (cattle), Chile (cattle) | |||||
| Fhnad1-42 | cattle | ||||
| Fhnad1-43 | cattle | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bargues, M.D.; Valero, M.A.; Trueba, G.A.; Fornasini, M.; Villavicencio, A.F.; Guamán, R.; De Elías-Escribano, A.; Pérez-Crespo, I.; Artigas, P.; Mas-Coma, S. DNA Multi-Marker Genotyping and CIAS Morphometric Phenotyping of Fasciola gigantica-Sized Flukes from Ecuador, with an Analysis of the Radix Absence in the New World and the Evolutionary Lymnaeid Snail Vector Filter. Animals 2021, 11, 2495. https://doi.org/10.3390/ani11092495
Bargues MD, Valero MA, Trueba GA, Fornasini M, Villavicencio AF, Guamán R, De Elías-Escribano A, Pérez-Crespo I, Artigas P, Mas-Coma S. DNA Multi-Marker Genotyping and CIAS Morphometric Phenotyping of Fasciola gigantica-Sized Flukes from Ecuador, with an Analysis of the Radix Absence in the New World and the Evolutionary Lymnaeid Snail Vector Filter. Animals. 2021; 11(9):2495. https://doi.org/10.3390/ani11092495
Chicago/Turabian StyleBargues, Maria Dolores, Maria Adela Valero, Gabriel A. Trueba, Marco Fornasini, Angel F. Villavicencio, Rocío Guamán, Alejandra De Elías-Escribano, Ignacio Pérez-Crespo, Patricio Artigas, and Santiago Mas-Coma. 2021. "DNA Multi-Marker Genotyping and CIAS Morphometric Phenotyping of Fasciola gigantica-Sized Flukes from Ecuador, with an Analysis of the Radix Absence in the New World and the Evolutionary Lymnaeid Snail Vector Filter" Animals 11, no. 9: 2495. https://doi.org/10.3390/ani11092495
APA StyleBargues, M. D., Valero, M. A., Trueba, G. A., Fornasini, M., Villavicencio, A. F., Guamán, R., De Elías-Escribano, A., Pérez-Crespo, I., Artigas, P., & Mas-Coma, S. (2021). DNA Multi-Marker Genotyping and CIAS Morphometric Phenotyping of Fasciola gigantica-Sized Flukes from Ecuador, with an Analysis of the Radix Absence in the New World and the Evolutionary Lymnaeid Snail Vector Filter. Animals, 11(9), 2495. https://doi.org/10.3390/ani11092495











