Evolutionary Comparisons of Chelonid Alphaherpesvirus 5 (ChHV5) Genomes from Fibropapillomatosis-Afflicted Green (Chelonia mydas), Olive Ridley (Lepidochelys olivacea) and Kemp’s Ridley (Lepidochelys kempii) Sea Turtles
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods and Materials
2.1. Tissue Sampling
2.2. DNA Isolation, Library Preparation and Sequencing from Tissue and eDNA Samples
2.3. Quality Control and Read Trimming
2.4. Read Alignment
2.5. Consensus Sequence Generation
2.6. Nucleotide and Gene Diversity Analysis
2.7. Phylogenetic/Phylogenomic Analysis
2.8. Patient “Yucca” Whole Genome Phylogenomics
3. Results
3.1. Sequence/Nucleotide Diversity
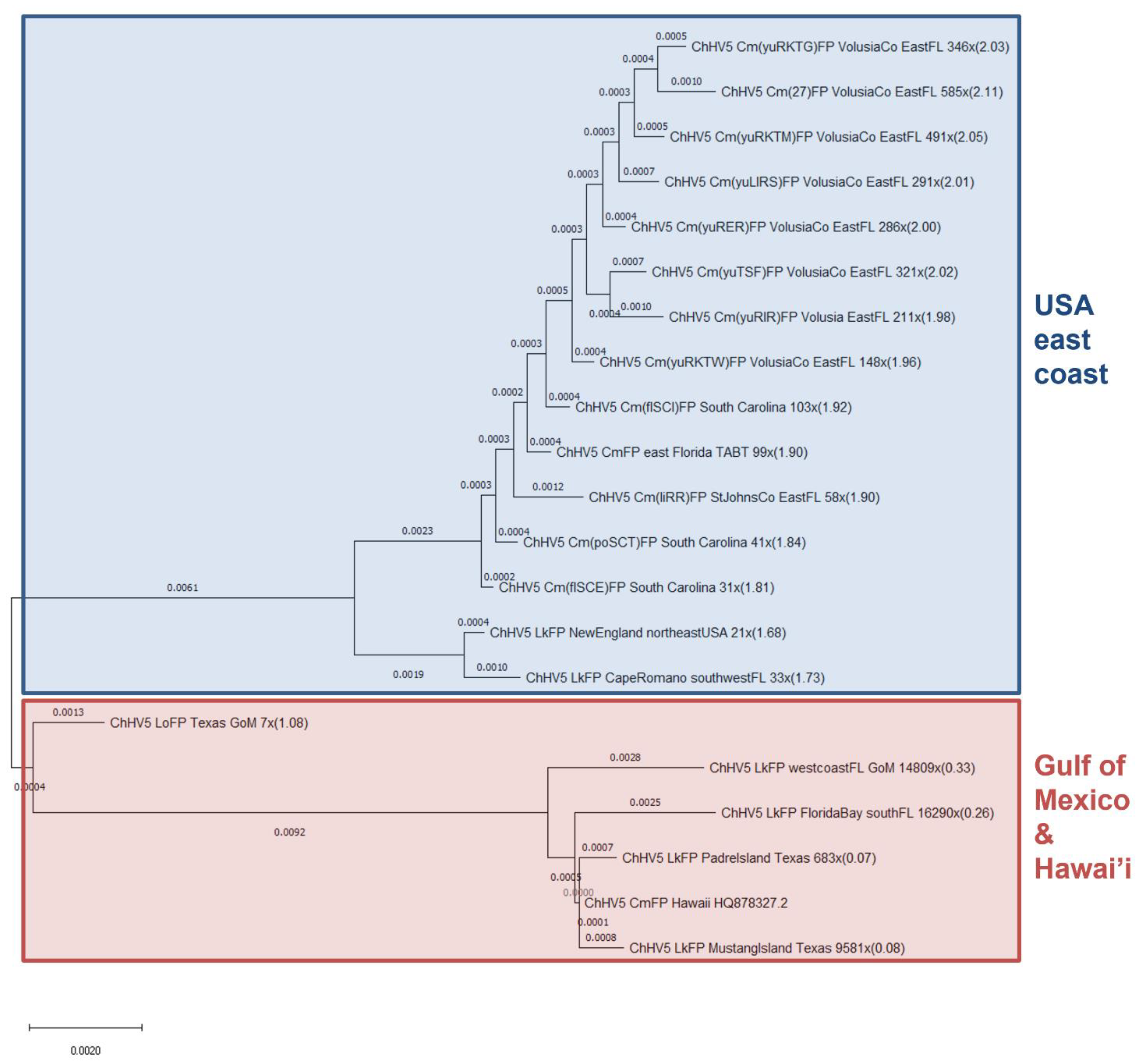
3.2. Phylogenomics Reveals Clustering of ChHV5 by Geographic Trends
3.3. Phylogenetics Highlights Close Relatedness of Novel Sequences and ChHV5 Florida Variants A–C
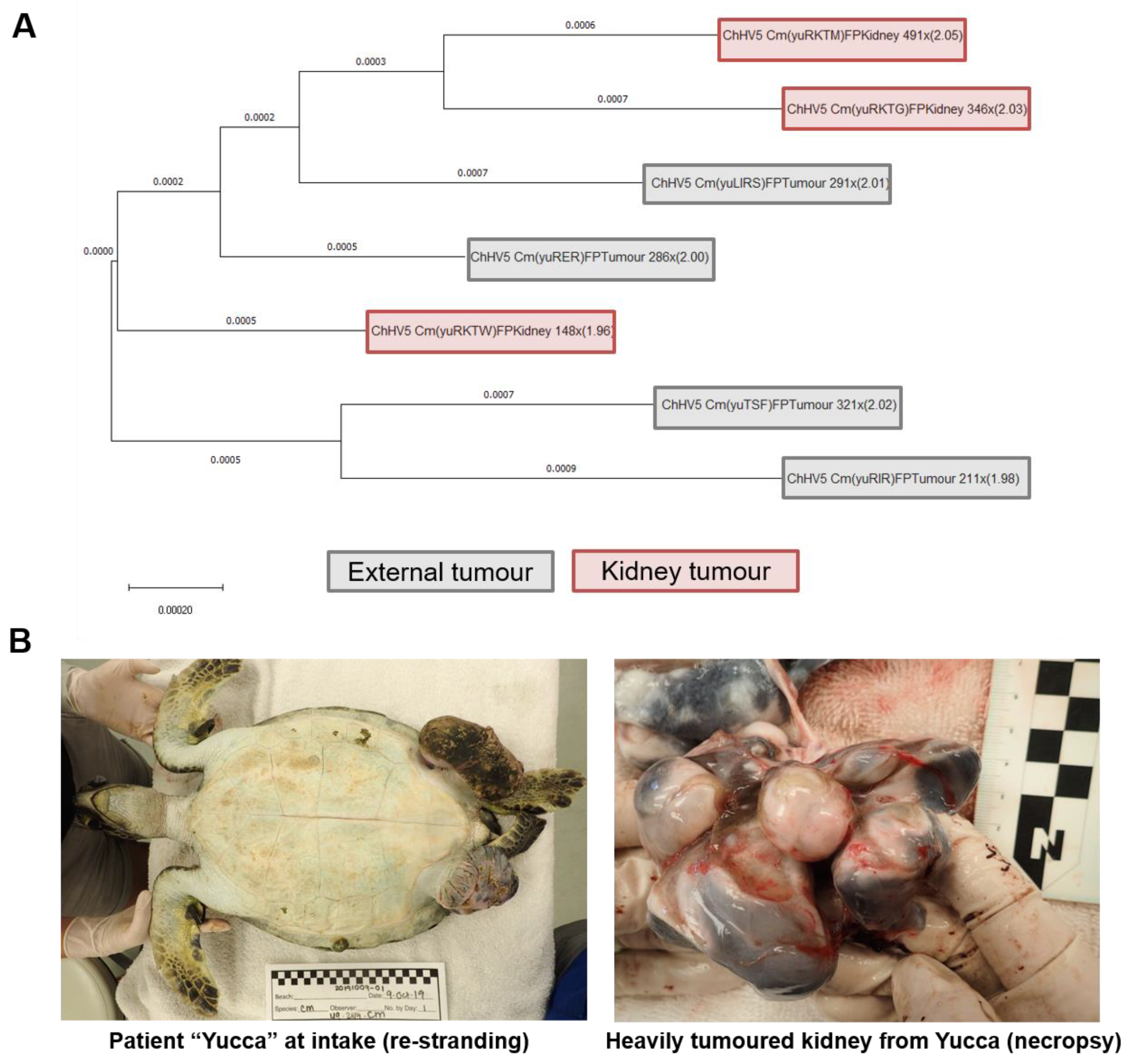
3.4. Patient “Yucca” Whole Genome Phylogenomics: Do Separate Tumours in the Same Individual Harbour Differing ChHV5 Variants?
4. Discussion
4.1. Nucleotide Diversity of ChHV5
4.2. ChHV5 Phylogenomics
4.3. ChHV5 Phylogenetics
4.4. Within-Host Viral Diversity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aguirre, A.A.; Lutz, P. Marine Turtles as Sentinels of Ecosystem Health: Is Fibropapillomatosis an Indicator? EcoHealth 2004, 1, 275–283. [Google Scholar] [CrossRef]
- IUCN. The International Union for Conservation of Nature’s (IUCN) Red List of Threatened Species. Available online: https://www.iucnredlist.org/ (accessed on 16 January 2021).
- Quackenbush, S.L.; Work, T.M.; Balazs, G.H.; Casey, R.N.; Rovnak, J.; Chaves, A.; duToit, L.; Baines, J.D.; Parrish, C.R.; Bowser, P.R.; et al. Three Closely Related Herpesviruses Are Associated with Fibropapillomatosis in Marine Turtles. Virology 1998, 246, 392–399. [Google Scholar] [CrossRef]
- Herbst, L.; Ene, A.; Su, M.; Desalle, R.; Lenz, J. Tumor outbreaks in marine turtles are not due to recent herpesvirus mutations. Curr. Biol. 2004, 14, R697–R699. [Google Scholar] [CrossRef]
- Ariel, E.; Nainu, F.; Jones, K.; Juntunen, K.; Bell, I.; Gaston, J.; Scott, J.; Trocini, S.; Burgess, G. Phylogenetic Variation of Chelonid Alphaherpesvirus 5 (ChHV5) in Populations of Green Turtles Chelonia mydas along the Queensland Coast, Australia. J. Aquat. Anim. Health 2017, 29, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Yetsko, K.; Farrell, J.; Stammnitz, M.R.; Whitmore, L.; Whilde, J.; Eastman, C.B.; Rollinson Ramia, D.; Thomas, R.; Krstic, A.; Linser, P.; et al. Mutational, transcriptional and viral shedding dynamics of the marine turtle fibropapillomatosis tumor epizootic. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yetsko, K.; Farrell, J.A.; Blackburn, N.B.; Whitmore, L.; Stammnitz, M.R.; Whilde, J.; Eastman, C.B.; Ramia, D.R.; Thomas, R.; Krstic, A.; et al. Molecular characterization of a marine turtle tumor epizootic, profiling external, internal and postsurgical regrowth tumors. Commun. Biol. 2021, 4, 152. [Google Scholar] [CrossRef] [PubMed]
- Reséndiz, E.; Fernández-Sanz, H.; Domínguez-Contreras, J.F.; Ramos-Díaz, A.H.; Mancini, A.; Zavala-Norzagaray, A.A.; Aguirre, A.A. Molecular Characterization of Chelonid Alphaherpesvirus 5 in a Black Turtle (Chelonia mydas) Fibropapilloma from Baja California Sur, Mexico. Animals 2021, 11, 105. [Google Scholar] [CrossRef]
- Smith, G.C.; Coates, C.W. Fibro-epithelial growths of the skin in large marine turtles Chelonia mydas. Zoologica 1938, 23, 93–98. [Google Scholar]
- Williams, E.H.; Bunkley-Williams, L.; Peters, E.C.; Pinto-Rodriguez, B.; Matos-Morales, R.; Mignucci-Giannoni, A.A.; Hall, K.V.; Rueda-Almonacid, J.V.; Sybesma, J.; de Calventi, I.B.; et al. An Epizootic of Cutaneous Fibropapillomas in Green Turtles Chelonia mydas of the Caribbean: Part of a Panzootic? J. Aquat. Anim. Health 1994, 6, 70–78. [Google Scholar] [CrossRef]
- Jones, K.; Ariel, E.; Burgess, G.; Read, M. A review of fibropapillomatosis in Green turtles (Chelonia mydas). Vet. J. 2016, 212, 48–57. [Google Scholar] [CrossRef]
- Rodenbusch, C.R.; Baptistotte, C.; Werneck, M.R.; Pires, T.T.; Melo, M.T.D.; de Ataíde, M.W.; dos Reis, K.D.H.L.; Testa, P.; Alieve, M.M.; Canal, C.W. Fibropapillomatosis in green turtles Chelonia mydas in Brazil: Characteristics of tumors and virus. Dis. Aquat. Org. 2014, 111, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.; Thomas, R.; Martindale, M.Q.; Duffy, D.J. Characterisation of fibropapillomatosis tumour growth profiles in green sea turtles (Chelonia mydas). Testudo 2018, 8, 12–29. [Google Scholar]
- Ene, A.; Su, M.; Lemaire, S.; Rose, C.; Schaff, S.; Moretti, R.; Lenz, J.; Herbst, L.H. Distribution of chelonid fibropapillomatosis-associated herpesvirus variants in Florida: Molecular genetic evidence for infection of turtles following recruitment to neritic developmental habitats. J. Wildl. Dis. 2005, 41, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Teelucksingh, S.S.; Eckert, S.; Nunes, P.A.L.D. Marine turtles, ecosystem services and human welfare in the marine ecosystems of the Caribbean Sea: A discussion of key methodologies. Études Caribéennes 2010, 15. [Google Scholar] [CrossRef]
- Jones, K.; Burgess, G.; Budd, A.M.; Huerlimann, R.; Mashkour, N.; Ariel, E. Molecular evidence for horizontal transmission of chelonid alphaherpesvirus 5 at green turtle (Chelonia mydas) foraging grounds in Queensland, Australia. PLoS ONE 2020, 15, e0227268. [Google Scholar] [CrossRef] [PubMed]
- Hargrove, S.; Work, T.; Brunson, S.; Foley, A.M.; Balazs, G. Proceedings of the 2015 International Summit on Fibropapillomatosis: Global Status, Trends, and Population Impacts; NOAA Technical Memorandum NMFS-PIFSC-54; U.S. Department of Commerce, National Oceanic and Atmospheric Administration (NOAA): Washington, DC, USA, 2016; p. 85.
- Hamann, M.; Godfrey, M.H.; Seminoff, J.A.; Arthur, K.; Barata, P.C.R.; Bjorndal, K.A.; Bolten, A.B.; Broderick, A.C.; Campbell, L.M.; Carreras, C.; et al. Global research priorities for sea turtles: Informing management and conservation in the 21st century. Endanger. Species Res. 2010, 11, 245–269. [Google Scholar] [CrossRef]
- Work, T.M.; Dagenais, J.; Weatherby, T.M.; Balazs, G.H.; Ackermann, M. In-vitro replication of Chelonid herpesvirus 5 in organotypic skin cultures from Hawaiian green turtles (Chelonia mydas). J. Virol. 2017, 91, e00404–e00417. [Google Scholar] [CrossRef] [PubMed]
- Duffy, D.J.; Schnitzler, C.; Karpinski, L.; Thomas, R.; Whilde, J.; Eastman, C.; Yang, C.; Krstic, A.; Rollinson, D.; Zirkelbach, B.; et al. Sea turtle fibropapilloma tumors share genomic drivers and therapeutic vulnerabilities with human cancers. Commun. Biol. 2018, 1, 63. [Google Scholar] [CrossRef] [PubMed]
- Page-Karjian, A.; Perrault, J.R.; Zirkelbach, B.; Pescatore, J.; Riley, R.; Stadler, M.; Zachariah, T.T.; Marks, W.; Norton, T.M. Tumor re-growth, case outcome, and tumor scoring systems in rehabilitated green turtles with fibropapillomatosis. Dis. Aquat. Org. 2019, 137, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Page-Karjian, A.; Serrano, M.E.; Cartzendafner, J.; Morgan, A.; Ritchie, B.W.; Gregory, C.R.; McNeill, J.B.; Perrault, J.R.; Christiansen, E.F.; Harms, C.A. Molecular Assessment of Chelonid Alphaherpesvirus 5 Infection in Tumor-Free Green (Chelonia mydas) and Loggerhead (Caretta caretta) Sea Turtles in North Carolina, USA, 2015–2019. Animals 2020, 10, 1964. [Google Scholar] [CrossRef]
- Page-Karjian, A.; Whitmore, L.; Stacy, B.A.; Perrault, J.R.; Farrell, J.A.; Frandsen, H.; Walker, J.S.; Shaver, D.; Rantonen, E.; Harms, C.A.; et al. Fibropapillomatosis and chelonid alphaherpesvirus 5 infection in Kemp’s ridley sea turtles (Lepidochelys kempii). Animals 2021. In review. [Google Scholar]
- Blackburn, N.B.; Leandro, A.C.; Nahvi, N.; Devlin, M.A.; Leandro, M.; Martinez Escobedo, I.; Peralta, J.M.; George, J.; Stacy, B.A.; deMaar, T.W.; et al. Transcriptomic Profiling of Fibropapillomatosis in Green Sea Turtles (Chelonia mydas) From South Texas. Front. Immunol. 2021, 12, 410. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.A.; Yetsko, K.; Whitmore, L.; Whilde, J.; Eastman, C.B.; Ramia, D.R.; Thomas, R.; Linser, P.; Creer, S.; Burkhalter, B.; et al. Environmental DNA monitoring of oncogenic viral shedding and genomic profiling of sea turtle fibropapillomatosis reveals unusual viral dynamics. Commun. Biol. 2021, 4, 565. [Google Scholar] [CrossRef] [PubMed]
- Mashkour, N.; Jones, K.; Wirth, W.; Burgess, G.; Ariel, E. The Concurrent Detection of Chelonid Alphaherpesvirus 5 and Chelonia mydas Papillomavirus 1 in Tumoured and Non-Tumoured Green Turtles. Animals 2021, 11, 697. [Google Scholar] [CrossRef] [PubMed]
- Foley, A.M.; Schroeder, B.A.; Hardy, R.; MacPherson, S.L.; Nicholas, M. Long-term behavior at foraging sites of adult female loggerhead sea turtles (Caretta caretta) from three Florida rookeries. Mar. Biol. 2014, 161, 1251–1262. [Google Scholar] [CrossRef]
- Shaver, D.J.; Walker, J.S.; Backof, T.F. Fibropapillomatosis prevalence and distribution in green turtles Chelonia mydas in Texas (USA). Dis. Aquat. Org. 2019, 136, 175–182. [Google Scholar] [CrossRef]
- da Silva-Júnior, E.S.; de Farias, D.S.D.; da Costa Bomfim, A.; da Boaviagem Freire, A.C.; Revorêdo, R.Â.; Rossi, S.; Matushima, E.R.; Grisi-Filho, J.H.H.; de Lima Silva, F.J.; Gavilan, S.A. Stranded Marine Turtles in Northeastern Brazil: Incidence and Spatial–Temporal Distribution of Fibropapillomatosis. Chelonian Conserv. Biol. 2019, 18, 249–258. [Google Scholar] [CrossRef]
- Stacy, B.A.; Foley, A.M.; Work, T.M.; Lauritsen, A.M.; Schroeder, B.A.; Hargrove, S.K.; Keene, J.L. Report of the Technical Expert Workshop: Developing Recommendations for Field Response, Captive Management, and Rehabilitation of Sea Turtles with Fibropapillomatosis; NOAA Technical Memorandum NMFS-OPR-60; National Oceanic and Atmospheric Administration (NOAA): Washington, DC, USA, 2019. [CrossRef]
- Monteiro, J.; Duarte, M.; Amadou, K.; Barbosa, C.; El Bar, N.; Madeira, F.M.; Regalla, A.; Duarte, A.; Tavares, L.; Patrício, A.R. Fibropapillomatosis and the Chelonid Alphaherpesvirus 5 in Green Turtles from West Africa. EcoHealth 2021. Advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.; Faísca, P.; Loureiro, N.S.; Rosado, R.; Gil, S.; Pereira, N.; Tavares, L. First histological and virological report of fibropapilloma associated with herpesvirus in Chelonia mydas at Príncipe Island, West Africa. Arch. Virol. 2012, 157, 1155–1159. [Google Scholar] [CrossRef]
- Reséndiz, E.; Flores-Ramírez, S.; Koch, V.; Cordero-Tapia, A. First Record of Fibropapillomatosis in a Green Turtle Chelonia mydas from the Baja California Peninsula. J. Aquat. Anim. Health 2016, 28, 252–257. [Google Scholar] [CrossRef]
- Álvarez-Varas, R.; Cárdenas, D.M.; Cucalón, R.V.; Del Río, J.; Cifuentes, F.; Ulloa, M.; Briceño, C.; Cárdenas, W.B. First report of fibropapillomatosis in an olive ridley turtle Lepidochelys olivacea from the southeastern Pacific. Dis. Aquat. Org. 2019, 135, 43–48. [Google Scholar] [CrossRef]
- Loganathan, A.L.; Palaniappan, P.; Subbiah, V.K. Evidence of Chelonid Herpesvirus 5 (ChHV5) in Green Turtles (Chelonia mydas) from Sabah, Borneo. bioRxiv 2021. [Google Scholar] [CrossRef]
- Quesada, V.; Freitas-Rodríguez, S.; Miller, J.; Pérez-Silva, J.G.; Jiang, Z.-F.; Tapia, W.; Santiago-Fernández, O.; Campos-Iglesias, D.; Kuderna, L.F.K.; Quinzin, M.; et al. Giant tortoise genomes provide insights into longevity and age-related disease. Nat. Ecol. Evol. 2019, 3, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Herbst, L.; Jacobson, E.; Moretti, R.; Brown, T.; Sundberg, J.; Klein, P. Experimental transmission of green turtle fibropapillomatosis using cell-free tumor extracts. Dis. Aquat. Org. 1995, 22, 1–12. [Google Scholar] [CrossRef]
- Herbst, L.H. Fibropapillomatosis of marine turtles. Annu. Rev. Fish. Dis. 1994, 4, 389–425. [Google Scholar] [CrossRef]
- Morrison, C.L.; Iwanowicz, L.; Work, T.M.; Fahsbender, E.; Breitbart, M.; Adams, C.; Iwanowicz, D.; Sanders, L.; Ackermann, M.; Cornman, R.S. Genomic evolution, recombination, and inter-strain diversity of chelonid alphaherpesvirus 5 from Florida and Hawaii green sea turtles with fibropapillomatosis. PeerJ 2018, 6, e4386. [Google Scholar] [CrossRef] [PubMed]
- Quackenbush, S.L.; Casey, R.N.; Murcek, R.J.; Paul, T.A.; Work, T.M.; Limpus, C.J.; Chaves, A.; duToit, L.; Perez, J.V.; Aguirre, A.A.; et al. Quantitative Analysis of Herpesvirus Sequences from Normal Tissue and Fibropapillomas of Marine Turtles with Real-Time PCR. Virology 2001, 287, 105–111. [Google Scholar] [CrossRef][Green Version]
- Page-Karjian, A.; Torres, F.; Zhang, J.; Rivera, S.; Diez, C.; Moore, P.A.; Moore, D.; Brown, C. Presence of chelonid fibropapilloma-associated herpesvirus in tumored and non-tumored green turtles, as detected by polymerase chain reaction, in endemic and non-endemic aggregations, Puerto Rico. SpringerPlus 2012, 1, 35. [Google Scholar] [CrossRef]
- Manire, C.A.; Stacy, B.A.; Kinsel, M.J.; Daniel, H.T.; Anderson, E.T.; Wellehan, J.F.X. Proliferative dermatitis in a loggerhead turtle, Caretta caretta, and a green turtle, Chelonia mydas, associated with novel papillomaviruses. Vet. Microbiol. 2008, 130, 227–237. [Google Scholar] [CrossRef]
- Herbst, L.H.; Jacobson, E.R.; Klein, P.A.; Balazs, G.H.; Moretti, R.; Brown, T.; Sundberg, J.P. Comparative Pathology and Pathogenesis of Spontaneous and Experimentally Induced Fibropapillomas of Green Turtles (Chelonia mydas). Vet. Pathol. 1999, 36, 551–564. [Google Scholar] [CrossRef]
- Espinoza, J.; Hernández, E.; Lara-Uc, M.M.; Reséndiz, E.; Alfaro-Núñez, A.; Hori-Oshima, S.; Medina-Basulto, G. Genetic Analysis of Chelonid Herpesvirus 5 in Marine Turtles from Baja California Peninsula. EcoHealth 2020, 17, 258–263. [Google Scholar] [CrossRef]
- Foley, A.M.; Schroeder, B.A.; Redlow, A.E.; Fick-Child, K.J.; Teas, W.G. Fibropapillomatosis in stranded green turtles (Chelonia mydas) from the eastern United States (1980–98): Trends and associations with environmental factors. J. Wildl. Dis. 2005, 41, 29–41. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, R.G.; Martins, A.S.; Torezani, E.; Baptistotte, C.; da Nobrega, F.J.; Horta, P.A.; Work, T.M.; Balazs, G.H. Relationship between fibropapillomatosis and environmental quality: A case study with Chelonia mydas off Brazil. Dis Aquat. Organ. 2010, 89, 87–95. [Google Scholar] [CrossRef]
- Van Houtan, K.S.; Hargrove, S.K.; Balazs, G.H. Land Use, Macroalgae, and a Tumor-Forming Disease in Marine Turtles. PLoS ONE 2010, 5, e12900. [Google Scholar] [CrossRef]
- Patrício, A.R.; Herbst, L.H.; Duarte, A.; Vélez-Zuazo, X.; Santos Loureiro, N.; Pereira, N.; Tavares, L.; Toranzos, G.A. Global phylogeography and evolution of chelonid fibropapilloma-associated herpesvirus. J. Gen. Virol. 2012, 93, 1035–1045. [Google Scholar] [CrossRef]
- Rodenbusch, C.R.; Almeida, L.L.; Marks, F.S.; Ataíde, M.W.; Alievi, M.M.; Tavares, M.; Pereira, R.A.; Canal, C.W. Detection and characterization of fibropapilloma associated herpesvirus of marine turtles in Rio Grande do Sul, Brazil. Pesqui. Vet. Bras. 2012, 32, 1179–1183. [Google Scholar] [CrossRef]
- Monezi, T.A.; Mehnert, D.U.; de Moura, E.M.M.; Müller, N.M.G.; Garrafa, P.; Matushima, E.R.; Werneck, M.R.; Borella, M.I. Chelonid herpesvirus 5 in secretions and tumor tissues from green turtles (Chelonia mydas) from Southeastern Brazil: A ten-year study. Vet. Microbiol. 2016, 186, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Li, T.-H.; Hsu, W.-L.; Lan, Y.-C.; Balazs, G.-H.; Work, T.M.; Tseng, C.-T.; Chang, C.-C. Identification of Chelonid herpesvirus 5 (ChHV5) in endangered green turtles (Chelonia mydas) with fibropapillomatosis in Asia. Bull. Mar. Sci. 2017, 93, 1011–1022. [Google Scholar] [CrossRef]
- Greenblatt, R.J.; Quackenbush, S.L.; Casey, R.N.; Rovnak, J.; Balazs, G.H.; Work, T.M.; Casey, J.W.; Sutton, C.A. Genomic Variation of the Fibropapilloma-Associated Marine Turtle Herpesvirus across Seven Geographic Areas and Three Host Species. J. Virol. 2005, 79, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Koriabine, M.; Hartmann-Fritsch, F.; de Jong, P.J.; Lewis, T.D.; Schetle, N.; Work, T.M.; Dagenais, J.; Balazs, G.H.; Leong, J.-A.C. The Genome of Chelonid Herpesvirus 5 Harbors Atypical Genes. PLoS ONE 2012, 7, e46623. [Google Scholar] [CrossRef] [PubMed]
- Origgi, F.C.; Tecilla, M.; Pilo, P.; Aloisio, F.; Otten, P.; Aguilar-Bultet, L.; Sattler, U.; Roccabianca, P.; Romero, C.H.; Bloom, D.C.; et al. A Genomic Approach to Unravel Host-Pathogen Interaction in Chelonians: The Example of Testudinid Herpesvirus 3. PLoS ONE 2015, 10, e0134897. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, H.R.; Wilson, H.M.; Walker, S.; Purvin, C.M.; Dutton, P.; Lacasella, E.L.; Stacy, B.A.; Whitmore, L.; Farrell, J.A.; Duffy, D.J.; et al. First olive ridley sea turtle (Lepidochelys olivacea) stranding in Texas, USA and identification of chelonid alphaherpesvirus 5 (ChHV5) variant present in tumor tissue. Herpetol. Rev. 2021. In press. [Google Scholar]
- Břinda, K.; Boeva, V.; Kucherov, G. Ococo: An online variant and consensus caller. arXiv 2017, arXiv:1712.01146. [Google Scholar]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Biswas, S.; Akey, J.M. Genomic insights into positive selection. Trends Genet. 2006, 22, 437–446. [Google Scholar] [CrossRef]
- Smiley, J.R. Herpes Simplex Virus Virion Host Shutoff Protein: Immune Evasion Mediated by a Viral RNase? J. Virol. 2004, 78, 1063–1068. [Google Scholar] [CrossRef]
- Yao, X.-D.; Rosenthal, K.L. Herpes simplex virus type 2 virion host shutoff protein suppresses innate dsRNA antiviral pathways in human vaginal epithelial cells. J. Gen. Virol. 2011, 92, 1981–1993. [Google Scholar] [CrossRef]
- Yang, L.; Wang, M.; Cheng, A.; Yang, Q.; Wu, Y.; Jia, R.; Liu, M.; Zhu, D.; Chen, S.; Zhang, S.; et al. Innate Immune Evasion of Alphaherpesvirus Tegument Proteins. Front. Immunol. 2019, 10, 2196. [Google Scholar] [CrossRef] [PubMed]
- Daszak, P.; Cunningham, A.A.; Hyatt, A.D. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop. 2001, 78, 103–116. [Google Scholar] [CrossRef]
- Whilde, J.; Martindale, M.Q.; Duffy, D.J. Precision wildlife medicine: Applications of the human-centred precision medicine revolution to species conservation. Glob. Chang. Biol. 2017, 23, 1792–1805. [Google Scholar] [CrossRef] [PubMed]
- Schmeller, D.S.; Courchamp, F.; Killeen, G. Biodiversity loss, emerging pathogens and human health risks. Biodivers. Conserv. 2020, 29, 3095–3102. [Google Scholar] [CrossRef] [PubMed]
- Lorentzen, H.F.; Benfield, T.; Stisen, S.; Rahbek, C. COVID-19 is possibly a consequence of the anthropogenic biodiversity crisis and climate changes. Dan. Med. J. 2020, 67, A205025. [Google Scholar]
- Work, T.M.; Dagenais, J.; Balazs, G.H.; Schettle, N.; Ackermann, M. Dynamics of Virus Shedding and In Situ Confirmation of Chelonid Herpesvirus 5 in Hawaiian Green Turtles with Fibropapillomatosis. Vet. Pathol. 2015, 52, 1195–1201. [Google Scholar] [CrossRef]
- Domiciano, I.G.; Broadhurst, M.K.; Domit, C.; Flaiban, K.K.M.C.; Goldberg, D.W.; Fritzen, J.T.T.; Bracarense, A.P.F.R.L. Chelonid Alphaherpesvirus 5 DNA in Fibropapillomatosis-Affected Chelonia mydas. EcoHealth 2019, 16, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Forster, P.; Forster, L.; Renfrew, C.; Forster, M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 9241–9243. [Google Scholar] [CrossRef]
- Stein, L.D. The case for cloud computing in genome informatics. Genome Biol. 2010, 11, 207. [Google Scholar] [CrossRef] [PubMed]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Smart, A.S.; Weeks, A.R.; van Rooyen, A.R.; Moore, A.; McCarthy, M.A.; Tingley, R. Assessing the cost-efficiency of environmental DNA sampling. Methods Ecol. Evol. 2016, 7, 1291–1298. [Google Scholar] [CrossRef]
- Duffy, D.J.; Martindale, M.Q. Perspectives on the expansion of human precision oncology and genomic approaches to sea turtle fibropapillomatosis. Commun. Biol. 2019, 2, 54. [Google Scholar] [CrossRef] [PubMed]
- Roizman, B.; Campadelli-Fiume, G. Chapter 6. Alphaherpes viral genes and their functions. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Li, C.; Wang, M.; Cheng, A.; Jia, R.; Yang, Q.; Wu, Y.; Zhu, D.; Zhao, X.; Chen, S.; Liu, M.; et al. The Roles of Envelope Glycoprotein M in the Life Cycle of Some Alphaherpesviruses. Front. Microbiol. 2021, 12, 226. [Google Scholar] [CrossRef]
- Lawrance, M.F.; Mansfield, K.L.; Sutton, E.; Savage, A.E. Molecular evolution of fibropapilloma-associated herpesviruses infecting juvenile green and loggerhead sea turtles. Virology 2018, 521, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.J. Molecular Evolution of Alphaherpesviruses: Varicella Zoster Virus; Cambridge University Press: Cambridge, UK, 2000; pp. 25–50. [Google Scholar]
- Lythgoe, K.A.; Hall, M.; Ferretti, L.; de Cesare, M.; MacIntyre-Cockett, G.; Trebes, A.; Andersson, M.; Otecko, N.; Wise, E.L.; Moore, N.; et al. SARS-CoV-2 within-host diversity and transmission. Science 2021, 372, eabg0821. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Zu, W.; Liu, Y.; Li, J.; Li, Z.; Wen, Y.; Wang, H.; Yuan, J.; Cheng, L.; Zhang, S.; et al. Quasispecies of SARS-CoV-2 revealed by single nucleotide polymorphisms (SNPs) analysis. Virulence 2021, 12, 1209–1226. [Google Scholar] [CrossRef] [PubMed]


| Sample | Stranding Location | Species | Tissue Type | Sequencing Strategy | Total Reads | % ChHV5 Alignment | Total ChHV5 Aligning Reads | ChHV5 RPTM | Genome Coverage |
|---|---|---|---|---|---|---|---|---|---|
| 27L1Fdna | Ormond Beach, east Florida | Cm | Lung tumour | No enrichment (host and viral DNA) | 6.88 × 108 | 0.038 | 257,692.5 | 3843 | 584.6× |
| fISCEYFdna | South Carolina | Cm | External eye tumour | No enrichment (host and viral DNA) | 1.31 × 108 | 0.011 | 13,627 | 1064 | 30.9× |
| fISCINFdna | External tumour | No enrichment (host and viral DNA) | 1.52 × 108 | 0.03 | 45,272 | 3031 | 102.7× | ||
| poSCTFdna | South Carolina | Cm | External tumour | No enrichment (host and viral DNA) | 1.52 × 108 | 0.012 | 18,133 | 1224 | 41.1× |
| yuLIRSFdna | Halifax Harbour, east Florida | Cm | External tumour | No enrichment (host and viral DNA) | 6.21 × 108 | 0.021 | 128,132 | 2119 | 290.7× |
| yuRERFdna | External tumour | No enrichment (host and viral DNA) | 6.53 × 108 | 0.02 | 125,910 | 1976 | 285.7× | ||
| yuRIRSFdna | External tumour | No enrichment (host and viral DNA) | 7.93 × 108 | 0.012 | 93,086 | 1198 | 211.2× | ||
| yuRKTGFdna | Kidney tumour | No enrichment (host and viral DNA) | 7.25 × 108 | 0.021 | 152,495 | 2143 | 346.0× | ||
| yuRKTMFdna | Kidney tumour | No enrichment (host and viral DNA) | 7.07 × 108 | 0.031 | 216,336 | 3127 | 490.8× | ||
| yuRKTW1Fdna | Kidney tumour | No enrichment (host and viral DNA) | 6.30 × 108 | 0.011 | 65,453 | 1063 | 148.5× | ||
| yuTSFdna | External tumour | No enrichment (host and viral DNA) | 8.59× 108 | 0.017 | 141,446 | 1682 | 320.9× | ||
| TABT-Cm | Anchorage Marina, east Florida | Cm | Bladder tumour | No enrichment (host and viral DNA) | 1.69 × 108 | 0.03 | 43,563 | 2573 | 98.8× |
| liRRF4dna | Marineland Beach, east Florida | Cm | External tumour | No enrichment (host and viral DNA) | 4.27 × 108 | 0.01 | 38,503 | 391 | 58.2× |
| LoTXFdna | Texas | Lo | External tumour (deceased) | No enrichment (host and viral DNA) | 1.59 × 108 | 0.002 | 3018 | 192 | 6.9× |
| LkNEFdna | New England, Cape Cod, Massachusetts | Lk | External tumour | No enrichment (host and viral DNA) | 1.12 × 108 | 0.01 | 9062 | 811 | 20.6× |
| LkFLMCKFdna | Cape Romano, SW Florida | Lk | External tumour | No enrichment (host and viral DNA) | 1.50 × 108 | 0.01 | 14,929 | 992 | 33.9× |
| 20170226AFA | Mustang Island, Texas | Lk | External tumour | Viral enrichment | 3.45 × 107 | 14.61 | 4,222,944 | 1,461,284 | 9580.7× |
| LLE-419 | Padre Island, Texas | Lk | External tumour | Viral enrichment | 1.46 × 107 | 2.2 | 301,155 | 219,649 | 683.2× |
| MCK2015011701 | West Coast, Florida | Lk | External tumour | Viral enrichment | 5.89 × 107 | 13.1 | 6,527,585 | 1,309,565 | 14,809.3× |
| NMFS14_313 | Florida Bay, Florida | Lk | External tumour | Viral enrichment | 3.69 × 107 | 23.2 | 7,180,057 | 2,320,447 | 16,289.6× |
| Gene | ChHV5 Genome Position | Tajima’s D Statistic | Predicted Feature |
|---|---|---|---|
| F-UL37 | 98894–102139 | 1.31 | Tegument protein |
| F-UL36 | 91944–98897 | 0.84 | VP1/2 tegument protein |
| F-UL25 | 72184–73857 | 0.81 | 99% ID with gb|AAU93323.1 minor capsid protein |
| HP17 | 57297–57764 | 0.79 | Hypothetical protein (HP) |
| F-US8 | 12220–13842 | 0.77 | Glycoprotein e (gE) |
| F-UL8 | 43118–45361 | 0.77 | Herpesvirus DNA helicase/primase complex associated protein |
| HP16 | 50429–51067 | 0.66 | HP; predicted bipartite NLS |
| F-UL15B | 55718–56788 | 0.61 | Probable DNA packing protein, C-terminus |
| F-UL41 | 104719–105897 | 0.56 | Close similarity to gb|AER28066.1. Tegument host shutoff protein |
| F-US4 | 14752–15549 | 0.51 | Similar to glycoprotein D (gD) |
| HP34 | 126003–126476 | −1.64 | HP |
| F-UL35 | 91561–91926 | −1.65 | VP26 basic phosphorylated capsid protein |
| F-UL10 | 47779–49044 | −1.94 | Glycoprotein M (gM) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Whitmore, L.; Yetsko, K.; Farrell, J.A.; Page-Karjian, A.; Daniel, W.; Shaver, D.J.; Frandsen, H.R.; Walker, J.S.; Crowder, W.; Bovery, C.; et al. Evolutionary Comparisons of Chelonid Alphaherpesvirus 5 (ChHV5) Genomes from Fibropapillomatosis-Afflicted Green (Chelonia mydas), Olive Ridley (Lepidochelys olivacea) and Kemp’s Ridley (Lepidochelys kempii) Sea Turtles. Animals 2021, 11, 2489. https://doi.org/10.3390/ani11092489
Whitmore L, Yetsko K, Farrell JA, Page-Karjian A, Daniel W, Shaver DJ, Frandsen HR, Walker JS, Crowder W, Bovery C, et al. Evolutionary Comparisons of Chelonid Alphaherpesvirus 5 (ChHV5) Genomes from Fibropapillomatosis-Afflicted Green (Chelonia mydas), Olive Ridley (Lepidochelys olivacea) and Kemp’s Ridley (Lepidochelys kempii) Sea Turtles. Animals. 2021; 11(9):2489. https://doi.org/10.3390/ani11092489
Chicago/Turabian StyleWhitmore, Liam, Kelsey Yetsko, Jessica A. Farrell, Annie Page-Karjian, Whitney Daniel, Donna J. Shaver, Hilary R. Frandsen, Jennifer Shelby Walker, Whitney Crowder, Caitlin Bovery, and et al. 2021. "Evolutionary Comparisons of Chelonid Alphaherpesvirus 5 (ChHV5) Genomes from Fibropapillomatosis-Afflicted Green (Chelonia mydas), Olive Ridley (Lepidochelys olivacea) and Kemp’s Ridley (Lepidochelys kempii) Sea Turtles" Animals 11, no. 9: 2489. https://doi.org/10.3390/ani11092489
APA StyleWhitmore, L., Yetsko, K., Farrell, J. A., Page-Karjian, A., Daniel, W., Shaver, D. J., Frandsen, H. R., Walker, J. S., Crowder, W., Bovery, C., Rollinson Ramia, D., Burkhalter, B., Ryan, E., & Duffy, D. J. (2021). Evolutionary Comparisons of Chelonid Alphaherpesvirus 5 (ChHV5) Genomes from Fibropapillomatosis-Afflicted Green (Chelonia mydas), Olive Ridley (Lepidochelys olivacea) and Kemp’s Ridley (Lepidochelys kempii) Sea Turtles. Animals, 11(9), 2489. https://doi.org/10.3390/ani11092489







