Succession of Intestinal Microbial Structure of Giant Pandas (Ailuropoda melanoleuca) during Different Developmental Stages and Its Correlation with Cellulase Activity
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Object
2.2. Sample Collection and Pretreatment
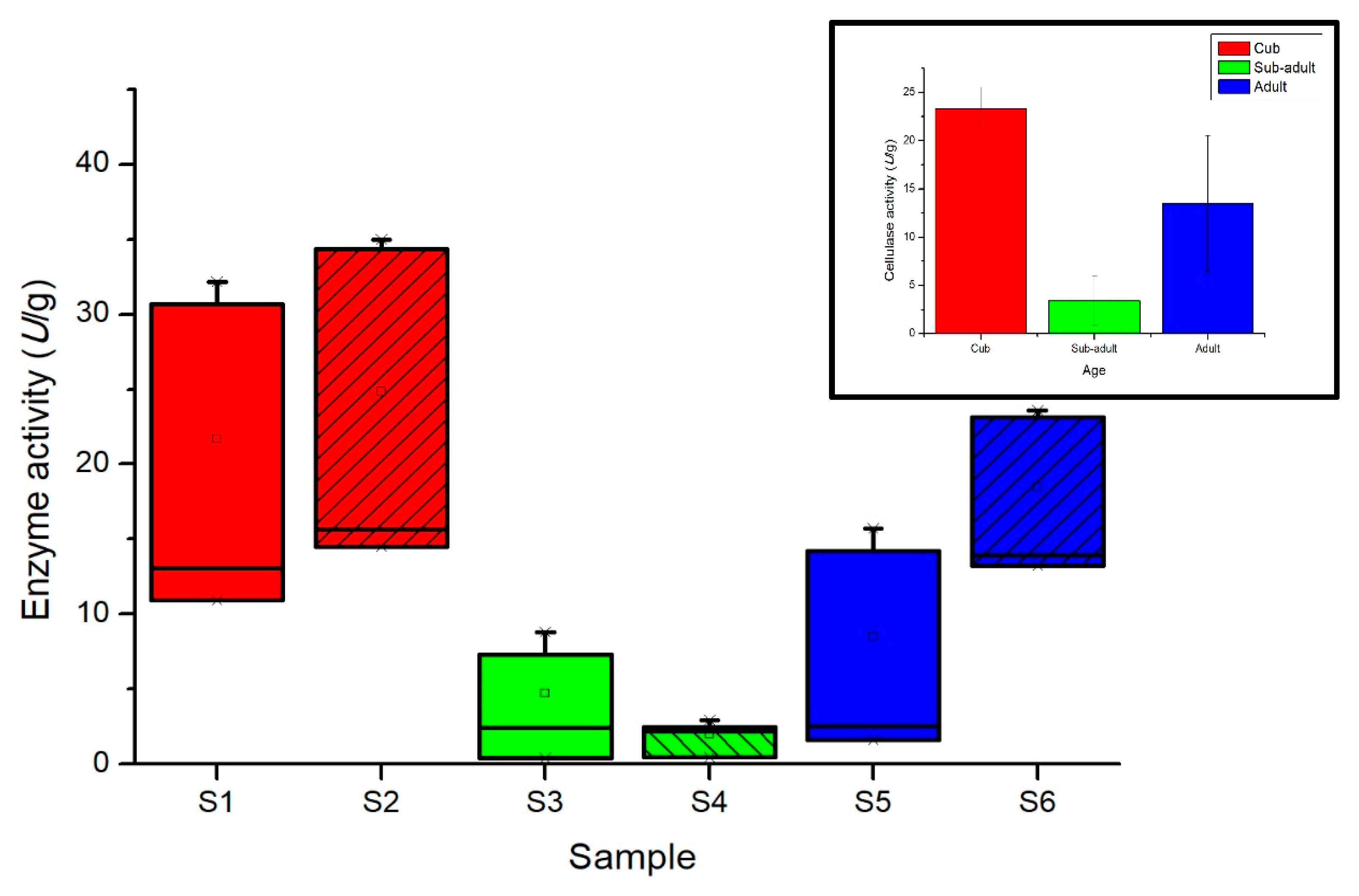
2.3. Analysis of Intestinal Cellulase Activity of the Giant Pandas
2.4. Analysis of Intestinal Microbial Diversity of Giant Pandas
2.5. Statistics Analysis
3. Results
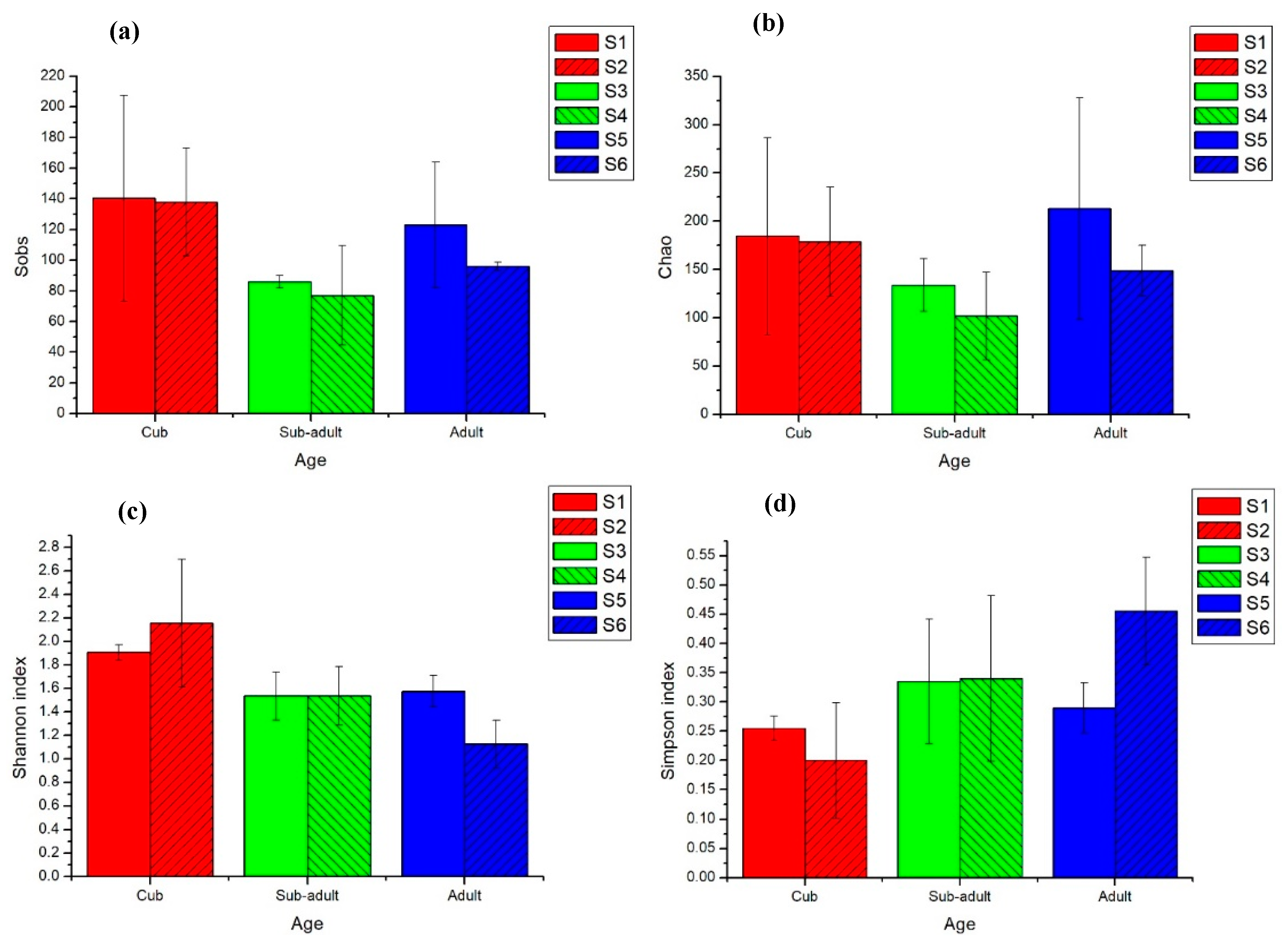
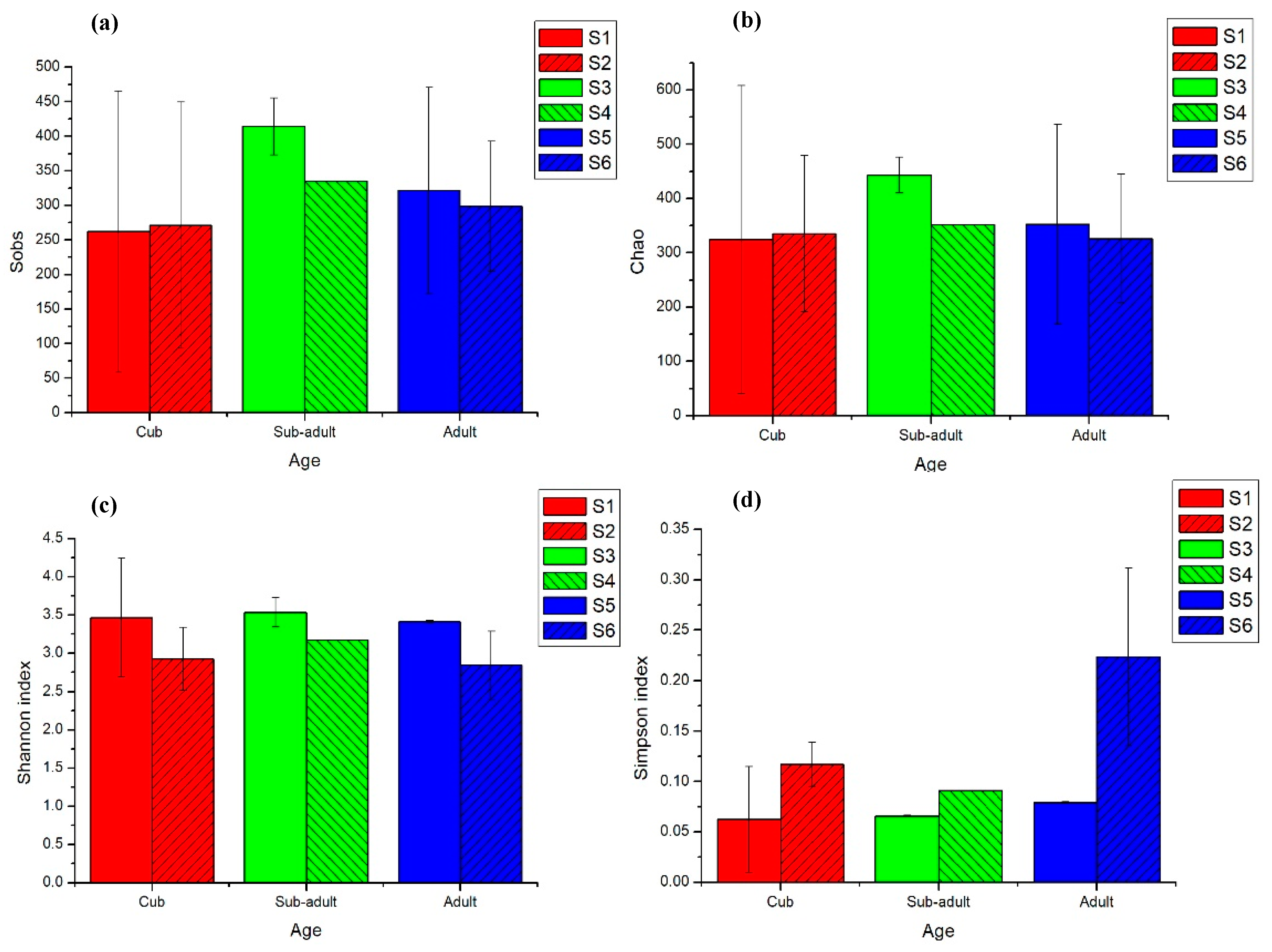
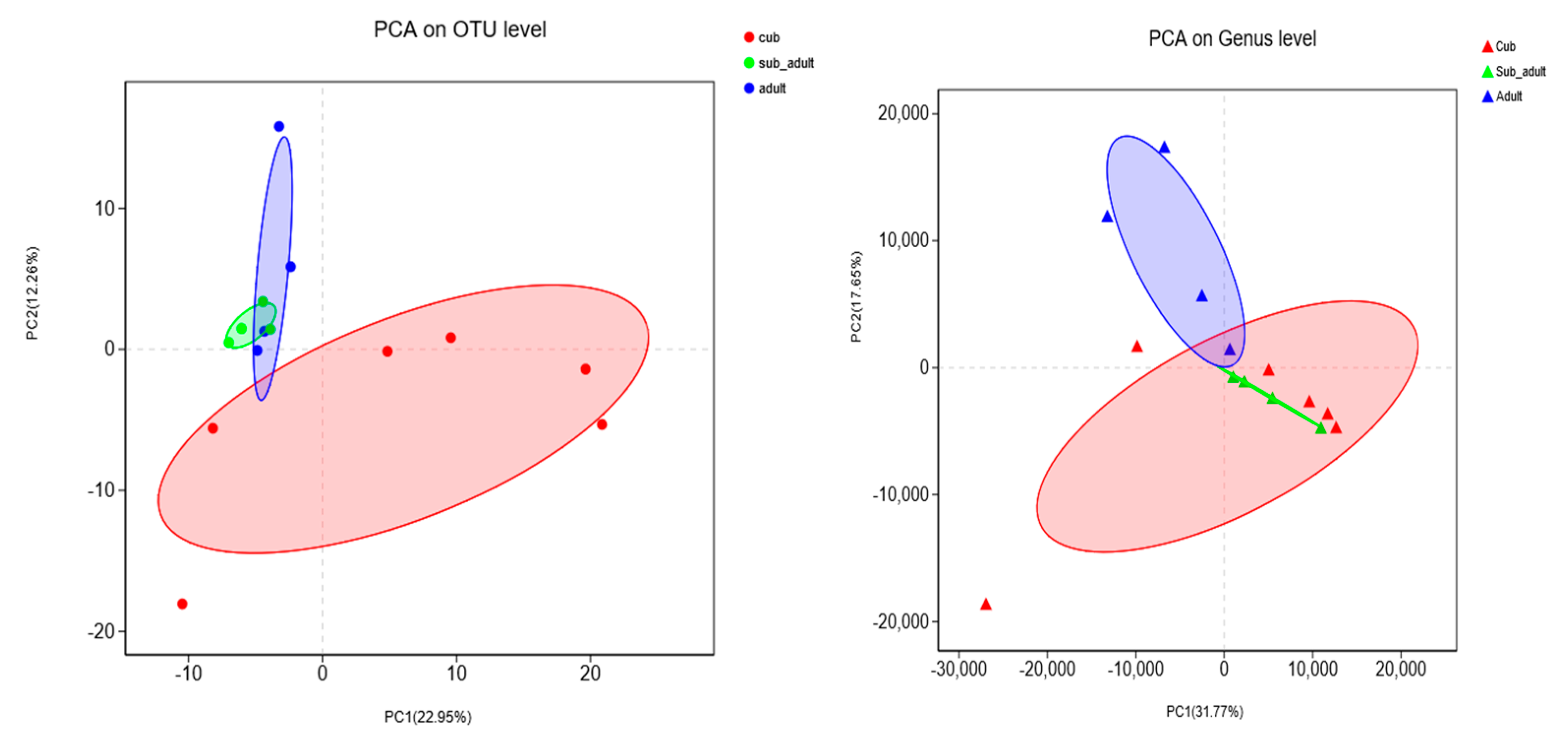
3.1. Intestinal Microbial Diversity of Giant Pandas at Different Developmental Stages
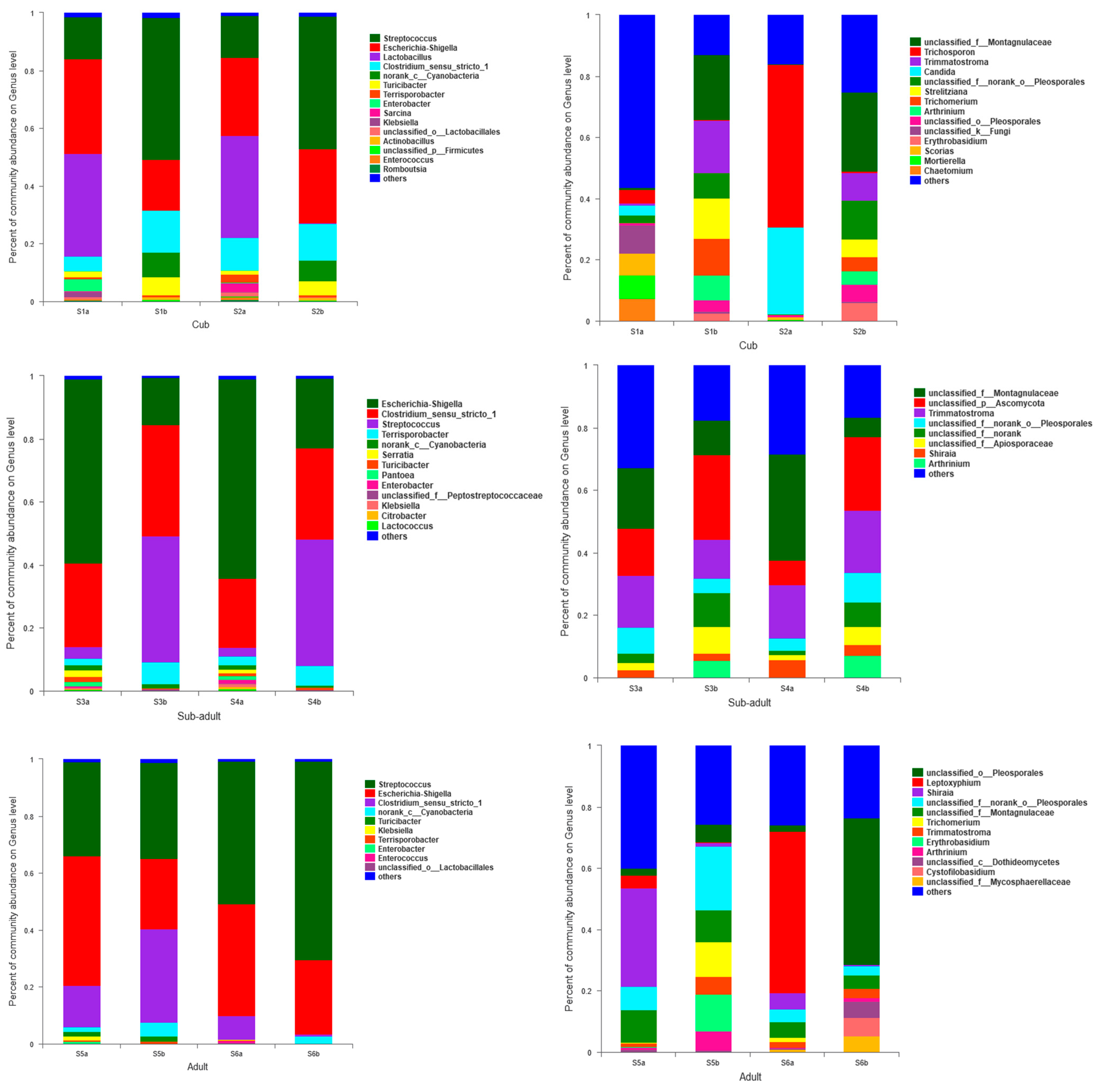
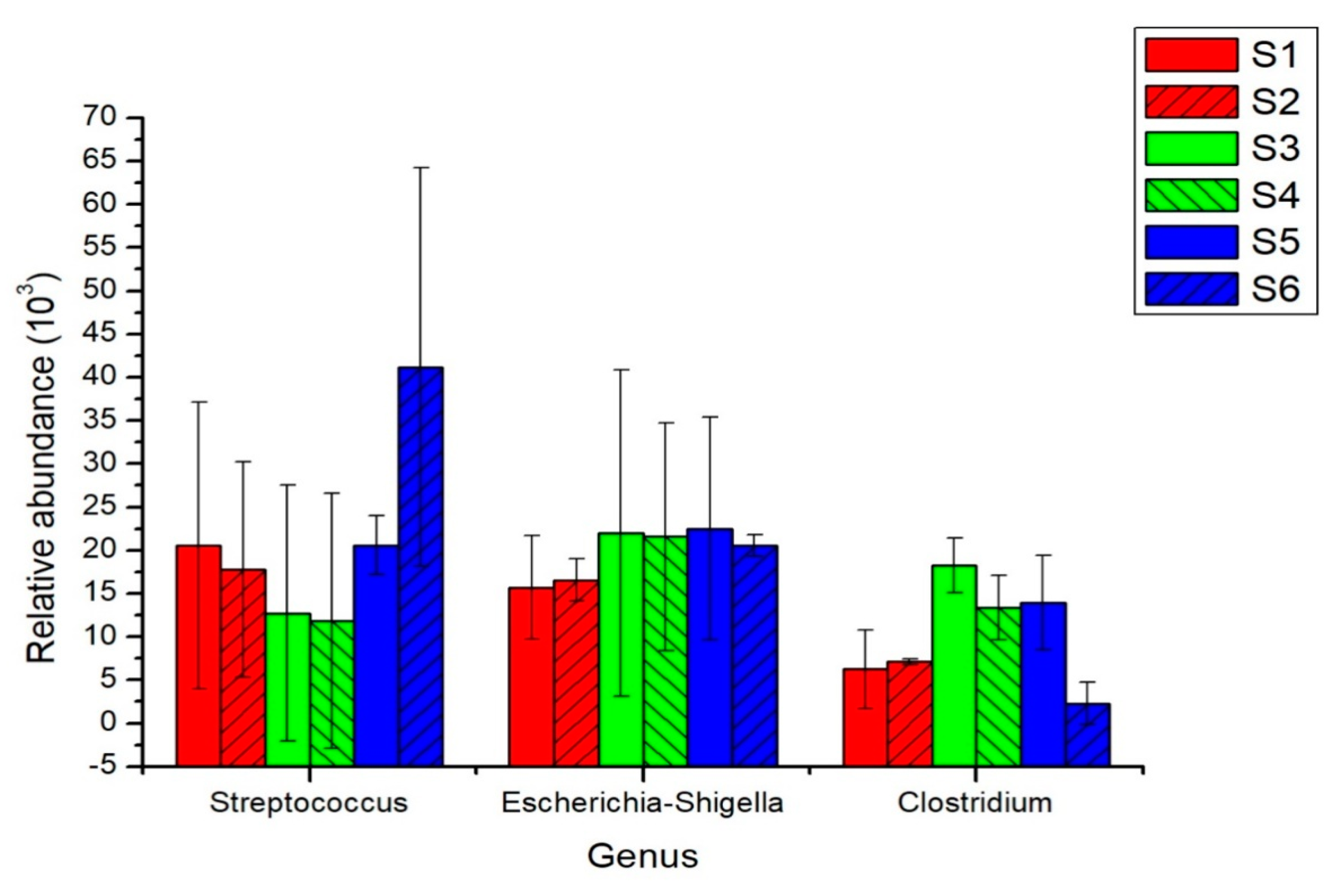
3.2. Succession of Age-Structure-Abundance of Dominant Microbes in the Intestines of Giant Pandas
3.3. Correlation between Cellulose Digestibility and Intestinal Microbial Structure in Giant Pandas
4. Discussion
4.1. Succession of Intestinal Microbial Structure and Its Potential Relationship with the Growth and Diet of Giant Pandas
4.2. Intestinal Microbial Endocellulase Activity and Cellulose Digestibility of Giant Pandas at Different Developmental Stages
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Williams, C.L.; Dill-Mcfarland, K.A. Dietary changes during weaning shape the gut microbiota of red pandas (Ailurus fulgens). Conserv. Physiol. 2018, 6, cox075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, Y.Q.; Chen, B.S. Symbiont-derived antimicrobials contribute to the control of the lepidopteran gut microbiota. Cell Chem. Biol. 2017, 24, 66–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, H.; Khosravi, A. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 2016, 352, 1116–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, A.; Debelius, J. The gut–liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef]
- Schulz, C.; Schütte, K. The active bacterial assemblages of the upper GI tract in individuals with and without Helicobacter infection. Gut 2016, 67, 216–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangan, K.J.; Pedicord, V.A. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science 2016, 353, 1434–1437. [Google Scholar] [CrossRef] [Green Version]
- Helena, M.H.; Stenius, F. Impact of lifestyle on the gut microbiota of healthy infants and their mothers—The ALADDIN birth cohort. FEMS Microbiol. Ecol. 2015, 90, 791–801. [Google Scholar]
- Guo, L.L.; Jason, K. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell 2014, 156, 109–122. [Google Scholar] [CrossRef] [Green Version]
- He, D.Y. Study on Bamboo Selection, Digestibility, Nutrition and Energy Countermeasures for Giant Pandas; Beijing Forestry University: Beijing, China, 2010. (In Chinese) [Google Scholar]
- Wei, F.; Wang, X. The giant panda gut microbiome. Trends Microbiol. 2015, 23, 450–452. [Google Scholar] [CrossRef]
- Zhang, W.P.; Zhang, Z.H. Macro genome development analysis of giant panda’s intestine. In Proceedings of the 13th National Symposium on Wildlife Conservation and the 6th Western China Zoological Symposium, Chengdu, China, 27–30 October 2017. (In Chinese). [Google Scholar]
- Guo, W.; Sudhanshu, M. Metagenomic study suggests that the gut microbiota of the giant panda (Ailuropoda melanoleuca) may not be specialized for fiber fermentation. Front. Microbiol. 2018, 9, 229. [Google Scholar] [CrossRef] [Green Version]
- Zhan, M.Y.; Fu, X.H. Differences of the intestinal microbial structure of adult giant panda in different regions and its correlation with the digestibility of cellulose. Chin. J. Appl. Environ. Biol. 2019, 25, 736–742. (In Chinese) [Google Scholar]
- Wang, A.S.; Zhan, M.Y. Intestinal bacterial structure evolution of giant panda cubs during food transformation stage and its correlation with cellulose digestion. Chin. J. Appl. Environ. Biol. 2019, 25, 1–10. (In Chinese) [Google Scholar]
- Zhang, W.P.; Liu, W.B. Age-associated microbiome shows the giant panda lives on hemicelluloses, not on cellulose. ISME J. 2018, 12, 1319–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, R.; Xu, L. The “wildness” of the giant panda gut microbiome and its relevance to effective translocation. Glob. Ecol. Conserv. 2019, 18, e00644. [Google Scholar] [CrossRef]
- Makki, K.; Deehan, E.C.; Walter, J.; BAckhed, F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ZinÖcker, M.; Lindseth, I.A. The western diet–microbiome-host interaction and its role in metabolic disease. Nutrients 2018, 10, 365. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Chen, J. Dynamics of gut microbiome in giant panda cubs reveal transitional microbes and pathways in early life. Front. Microbiol. 2018, 9, 3138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, M.Y.; Wang, L.; Xie, C.Y.; Fu, X.H.; Zhang, S.; Wang, A.S.; Zhou, Y.M.; Xu, C.Z.; Zhang, H.M. Succession of gut Microbial structure in twin giant pandas during the dietary change stage and its role in polysaccharide metabolism. Front. Microbiol. 2020, 11, 551038. [Google Scholar] [CrossRef]
- Wang, J.; Li, H.X. Effects of planting patterns of double-furrow planting in dryland on the number of cellulose-decomposing bacteria and cellulase activity in soil. J. Gansu Agric. Univ. 2012, 47, 39–41. (In Chinese) [Google Scholar]
- Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [CrossRef]
- Katherine, R.A.; Carl, J.Y.; Angela, K.; Righini, N.; Carbonero, F.; Estrada, A.; Gaskins, H.R.; Stumpf, R.M.; Yildirim, S.; Torralba, M.; et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013, 7, 1344–1353. [Google Scholar]
- Thakur, V.; Kumar, V.; Kumar, S.; Singh, D. Diverse culturable bacterial communities with cellulolytic potential revealed from pristine habitat in Indian trans-Himalaya. Can. J. Microbiol. 2018, 64, 798–808. [Google Scholar] [CrossRef]
- Carlos, C.; Fan, H.; Currie, C.R. Substrate Shift Reveals Roles for Members of Bacterial Consortia in Degradation of Plant Cell Wall Polymers. Front. Microbiol. 2018, 9, 364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, A.F.A.; Neto, P.D.; Da Silva, D.F.; Pastore, G.M. Xylo-oligosaccharides from lignocellulosic materials: Chemical structure, health benefits and production by chemical and enzymatic hydrolysis. Food Res. Int. 2013, 51, 75–85. [Google Scholar] [CrossRef]
- Kumar, R.; Sinha, A.; Srivastava, S.; Mahajan, G. Evaluation of biocontrol potential of some fungal decomposers of Sesbania aculeata L. green manure against some soil-borne plant pathogens. J. Environ. Biol. 2017, 38, 37–45. [Google Scholar] [CrossRef]
- Wang, L. The Diversity of Capative Giant Panda Gut Microbiome Viewed Across Age; China West Normal University: Nanchong, China, 2019. (In Chinese) [Google Scholar]
- Vasemägi, A.; Visse, M.; Kisand, V. Effect of environmental factors and an emerging parasitic disease on gut microbiome of wild salmonid fish. mSphere 2017, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Hills, R.D.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan, R.C.; Wang, Z. Gut microbiome composition in the hispanic community health study/study of latinos is shaped by geographic relocation, environmental factors, and obesity. Genome Biol. 2019, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Sovran, B.; Planchais, J.; Jegou, S.; Straube, M.; Lamas, B.; Natividad, J.M.; Agus, A.; Dupraz, L.; Glodt, J.; DA Costa, G.; et al. Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome 2018, 6, 152. [Google Scholar] [CrossRef] [PubMed]
- Kapitan, M.; Niemiec, M.J. Fungi as part of the microbiota and interactions with intestinal bacteria. CT Microbiol. 2018, 422, 265–301. [Google Scholar]
- Gaurav, N.; Sivasankari, S.; Kiran, G.S.; Ninawe, A.; Selvin, J. Utilization of bioresources for sustainable biofuels: A review. Renew. Sustain. Energy Rev. 2017, 73, 205–214. [Google Scholar] [CrossRef]
- Ma, X.P.; Jiang, Y.Z. Identification, genotyping, and pathogenicity of Trichosporon, spp. Isolated from Giant pandas (Ailuropoda melanoleuca). BMC Microbiol. 2019, 19, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.M.; Bai, F.Y. Four new yeast species of the genus Sporobolomyces from plant leaves. FEMS Yeast 2004, 4, 579–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Romero, E.; Aguirre-Noyola, J.L.; Bustamante-Brito, R.; González-Román, P.; Hernández-Oaxaca, D.; Higareda-Alvear, V.; Montes-Carreto, L.M.; Martínez-Romero, J.C.; Rosenblueth, M.; Servín-Garcidueñas, L.E. We and herbivores eat endophytes. Microb. Biotechnol. 2020, 14, 1282–1299. [Google Scholar] [CrossRef]
- Helander, M.; Jia, R.; Huitu, O.; Sieber, T.N.; Jia, J.; Niemela, P.; Saikkonen, K. Endophytic fungi and silica content of different bamboo species in giant panda diet. Symbiosis 2013, 61, 13–22. [Google Scholar] [CrossRef]
- Carver, P.L. The battle for iron between humans and microbes. Curr. Med. Chem. 2018, 25, 85–96. [Google Scholar] [CrossRef] [PubMed]
- McKenney, E.A.; Maslanka, M.; Rodrigo, A.; Yoder, A.D. Bamboo Specialists from Two Mammalian Orders (Primates, Carnivora) Share a High Number of Low-Abundance Gut Microbes. Microb. Ecol. 2017, 76, 1–13. [Google Scholar] [CrossRef]
- Wang, W.Y.; Li, D.B. Isolation and identification of four fiber degrading bacteria from rumen and their fiber degradation characteristics. J. Anim. Husb. Vet. Med. 2016, 47, 2294–2300. (In Chinese) [Google Scholar]
- Tang, K.Y.; Wang, Z.W.; Wan, Q.H.; Fang, S.G. Metagenomics reveals seasonal functional adaptation of the gut microbiome to host feeding and fasting in the Chinese Alligator. Front. Microbiol. 2019, 10, 2409. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.M.; Mai, G.Q. Metagenomic insights into a cellulose-rich niche reveal microbial cooperation in cellulose degradation. Front. Microbiol. 2019, 10, 618. [Google Scholar] [CrossRef]
- Robertson, R.C.; Manges, A.R. The human microbiome and child growth-first 1000 days and beyond. Trends Microbiol. 2019, 27, 131–147. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.C.; Zhang, Z.Z. Giant panda foraging and movement patterns in response to bamboo shoot growth. Environ. Sci. Pollut. Res. 2018, 25, 8638–8643. [Google Scholar] [CrossRef] [Green Version]
- Scoma, A.; Khor, W.C.; Coma, M.; Heyer, R.; Props, R.; Schoelynck, J.; Bouts, T.; Benndorf, D.; Li, D.H.; Zhang, H.M.; et al. Substrate-Dependent Fermentation of Bamboo in Giant Panda Gut Microbiomes: Leaf Primarily to Ethanol and Pith to Lactate. Front. Microbiol. 2020, 11, 530. [Google Scholar] [CrossRef]
- Wei, F.W. Giant panda evolutionary conservation biology. Sci. Sin. Vitae 2018, 48, 1048–1053. [Google Scholar] [CrossRef]
- Matt, S.; Marcus, C. Dietary evolution: The panda paradox. Curr. Biol. 2019, 29, R417–R419. [Google Scholar]
- Luo, C.B.; Li, Y.Q. B24amboo lignocellulose degradation by gut symbiotic microbiota of the bamboo snout beetle Cyrtotrachelus buqueti. Biotechnol. Biofuels 2019, 12, 70. [Google Scholar] [CrossRef] [PubMed]
| Name | Lineage | Date of Birth | Sex | Weight (m/kg) | Location | Health Condition | SampleNumber |
|---|---|---|---|---|---|---|---|
| Yueyue | 1052 | 4 October 2016 | Male | 33.6 | Shanghai wild animal park | Health | S1 |
| Banban | 1053 | 4 October 2016 | Female | 32.4 | Shanghai wild animal park | Health | S2 |
| Xinger | 900 | 23 August 2013 | Male | 112.3 | Shanghai zoo | Health | S3 |
| Yaer | 904 | 27 August 2013 | Male | 116.8 | Shanghai zoo | Health | S4 |
| Yaao | 583 | 13 August 2004 | Male | 136.0 | Shanghai wild animal park | Health | S5 |
| Youyou | 474 | 14 July 1998 | Female | 108.0 | Shanghai wild animal park | Health | S6 |
| Sample | Aug (2017) | Feb (2018) | Average |
|---|---|---|---|
| S1 | 12.02 ± 1.518 | 31.43 ± 1.037 | 21.72 ± 13.722 |
| S2 | 15.08 ± 0.815 | 34.67 ± 0.444 | 24.89 ± 13.851 |
| S3 | 2.32 ± 0.130 | 8.07 ± 1.037 | 5.19 ± 4.064 |
| S4 | 0.45 ± 0.037 | 2.72 ± 0.296 | 1.58 ± 1.612 |
| S5 | 2.07 ± 0.667 | 14.98 ±1.037 | 8.52 ± 9.129 |
| S6 | 13.61 ± 0.500 | 23.36 ± 0.296 | 18.48 ± 6.898 |
| Variable (xi) | Simple Correlation Coefficient with y (riy) | Direct Path Coefficient (Piy) | Indirect Path Coefficient (xj) | Total ∑(rij × Pjy) | |||
|---|---|---|---|---|---|---|---|
| X1 (ri1 × P1y) | X2 (ri2 × P2y) | X3 (ri3 × P3y) | X4 (ri4 × P4y) | ||||
| X1 | −0.954 | −1.112 | - | 0.563 | −0.753 | 0.348 | 0.158 |
| X2 | 0.857 | −0.655 | 0.955 | - | 0.889 | −0.333 | 1.511 |
| X3 | −0.852 | −1.056 | −0.792 | 0.552 | - | 0.444 | 0.203 |
| X4 | −0.835 | 0.484 | −0.798 | 0.450 | −0.969 | - | −1.317 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, A.; Zhan, M.; Pei, E. Succession of Intestinal Microbial Structure of Giant Pandas (Ailuropoda melanoleuca) during Different Developmental Stages and Its Correlation with Cellulase Activity. Animals 2021, 11, 2358. https://doi.org/10.3390/ani11082358
Wang A, Zhan M, Pei E. Succession of Intestinal Microbial Structure of Giant Pandas (Ailuropoda melanoleuca) during Different Developmental Stages and Its Correlation with Cellulase Activity. Animals. 2021; 11(8):2358. https://doi.org/10.3390/ani11082358
Chicago/Turabian StyleWang, Aishan, Mingye Zhan, and Enle Pei. 2021. "Succession of Intestinal Microbial Structure of Giant Pandas (Ailuropoda melanoleuca) during Different Developmental Stages and Its Correlation with Cellulase Activity" Animals 11, no. 8: 2358. https://doi.org/10.3390/ani11082358
APA StyleWang, A., Zhan, M., & Pei, E. (2021). Succession of Intestinal Microbial Structure of Giant Pandas (Ailuropoda melanoleuca) during Different Developmental Stages and Its Correlation with Cellulase Activity. Animals, 11(8), 2358. https://doi.org/10.3390/ani11082358





