Owning a Pet Is Associated with Changes in the Composition of Gut Microbiota and Could Influence the Risk of Metabolic Disorders in Humans
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Participants
2.2. Diet Assessment
2.3. Assessment of Pet Ownership
2.4. Clinical Plasma Parameters
2.5. DNA Extraction from Fecal Samples
2.6. Sequencing and Bioinformatics
2.7. Data Accesibility
2.8. Statistical Analysis
3. Results
3.1. Baseline Characteristics of the Study Participants
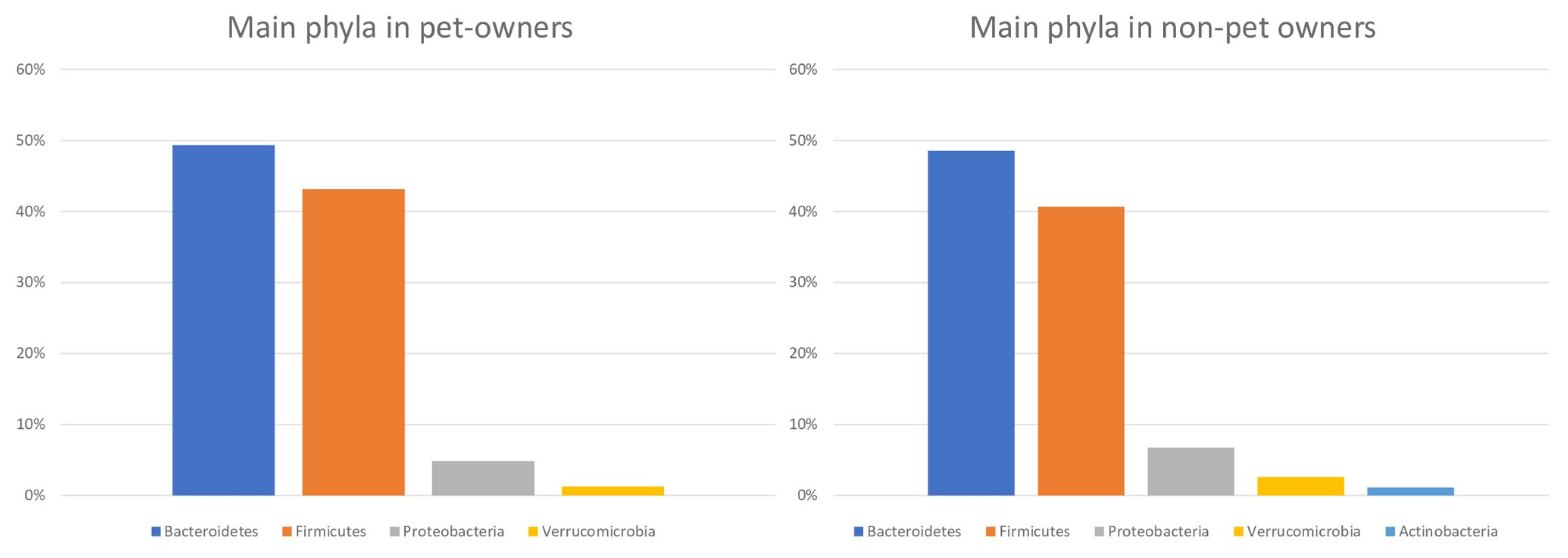
3.2. Microbiota Characteristics of the Study Participants
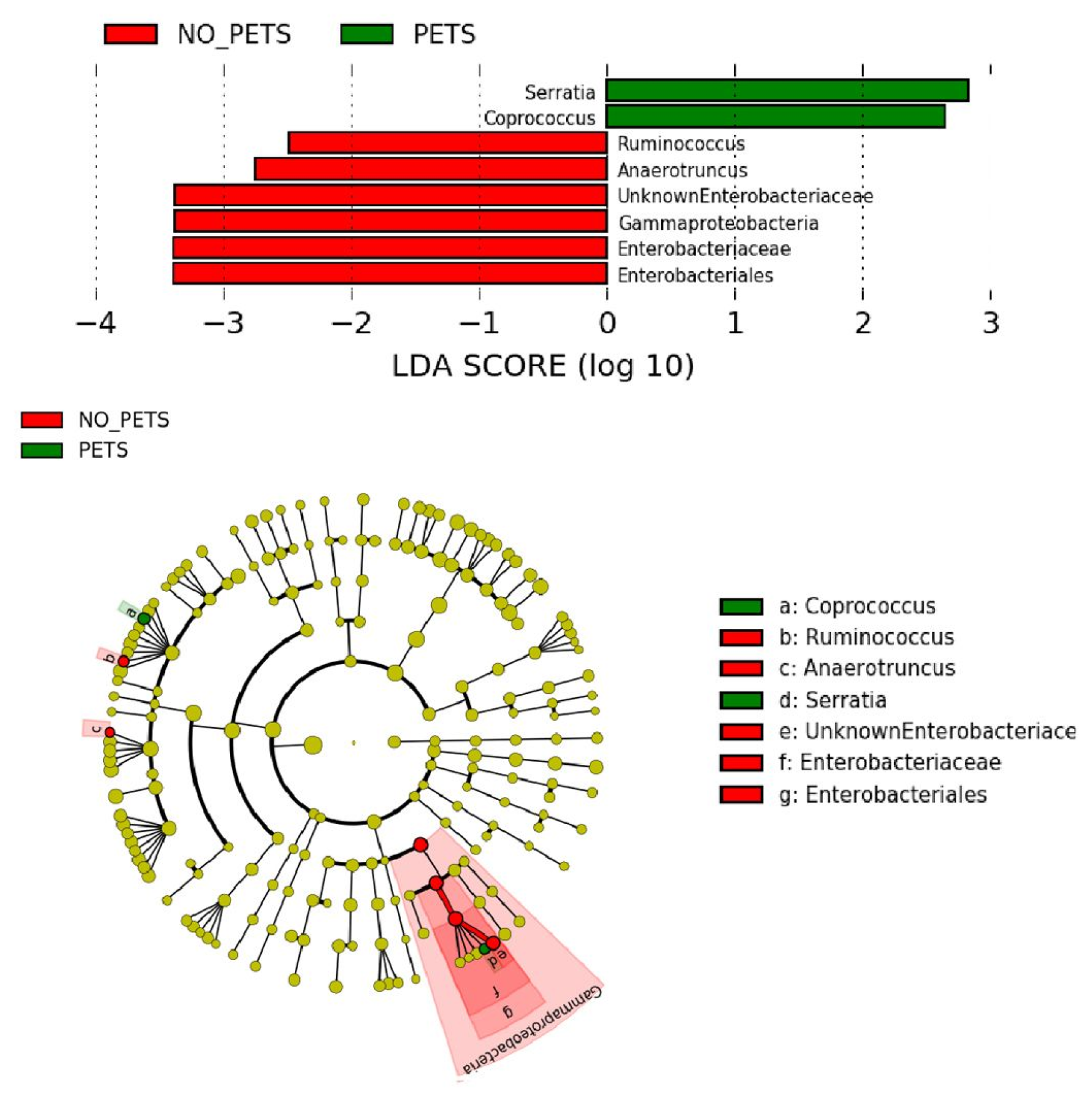
3.3. Differences in the Gut Microbiota between Pet Owners and Non-Pet Owners: LEfSe Analysis
3.4. Differences in the Gut Microbiota between Dog Owners and Non-Pet Owners: LEfSe Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bartges, J.; Kushner, R.F.; Michel, K.E.; Sallis, R.; Day, M.J. One Health Solutions to Obesity in People and Their Pets. J. Comp. Pathol. 2017, 156, 326–333. [Google Scholar] [CrossRef] [Green Version]
- Grundy, S.M.; Cleeman, J.I.; Daniels, S.R.; Donato, K.A.; Eckel, R.H.; Franklin, B.A.; Gordon, D.J.; Krauss, R.M.; Savage, P.J.; Smith, S.C., Jr.; et al. Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation 2005, 112, 2735–2752. [Google Scholar] [CrossRef] [Green Version]
- Perez-Martinez, P.; Mikhailidis, D.P.; Athyros, V.G.; Bullo, M.; Couture, P.; Covas, M.I.; de Koning, L.; Delgado-Lista, J.; Diaz-Lopez, A.; Drevon, C.A.; et al. Lifestyle recommendations for the prevention and management of metabolic syndrome: An international panel recommendation. Nutr. Rev. 2017, 75, 307–326. [Google Scholar] [CrossRef] [Green Version]
- Yeh, T.L.; Lei, W.T.; Liu, S.J.; Chien, K.L. A modest protective association between pet ownership and cardiovascular diseases: A systematic review and meta-analysis. PLoS ONE 2019, 14, e0216231. [Google Scholar] [CrossRef] [PubMed]
- Levine, G.N.; Allen, K.; Braun, L.T.; Christian, H.E.; Friedmann, E.; Taubert, K.A.; Thomas, S.A.; Wells, D.L.; Lange, R.A. Pet ownership and cardiovascular risk: A scientific statement from the American Heart Association. Circulation 2013, 127, 2353–2363. [Google Scholar] [CrossRef] [Green Version]
- Azad, M.B.; Konya, T.; Maughan, H.; Guttman, D.S.; Field, C.J.; Sears, M.R.; Becker, A.B.; Scott, J.A.; Kozyrskyj, A.L. Infant gut microbiota and the hygiene hypothesis of allergic disease: Impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin. Immunol. 2013, 9, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tun, H.M.; Konya, T.; Takaro, T.K.; Brook, J.R.; Chari, R.; Field, C.J.; Guttman, D.S.; Becker, A.B.; Mandhane, P.J.; Turvey, S.E.; et al. Exposure to household furry pets influences the gut microbiota of infant at 3–4 months following various birth scenarios. Microbiome 2017, 5, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Bajzer, M.; Seeley, R.J. Physiology: Obesity and gut flora. Nature 2006, 444, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Caesar, R.; Fåk, F.; Bäckhed, F. Effects of gut microbiota on obesity and atherosclerosis via modulation of inflammation and lipid metabolism. J. Intern. Med. 2010, 268, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Diamant, M.; Blaak, E.E.; de Vos, W.M. Do nutrient-gut-microbiota interactions play a role in human obesity, insulin resistance and type 2 diabetes? Obes. Rev. 2011, 12, 272–281. [Google Scholar] [CrossRef]
- Caugant, D.A.; Levin, B.R.; Selander, R.K. Distribution of multilocus genotypes of Escherichia coli within and between host families. J. Hyg. 1984, 92, 377–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freiman, J.A.; Chalmers, T.C.; Smith, H.; Kuebler, R.R. The importance of beta, the type II error and sample size in the design and interpretation of the randomized control trial. Survey of 71 “negative” trials. N. Engl. J. Med. 1978, 299, 690–694. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Lista, J.; Perez-Martinez, P.; Garcia-Rios, A.; Alcala-Diaz, J.F.; Perez-Caballero, A.I.; Gomez-Delgado, F.; Fuentes, F.; Quintana-Navarro, G.; Lopez-Segura, F.; Ortiz-Morales, A.M.; et al. CORonary Diet Intervention with Olive oil and cardiovascular PREVention study (the CORDIOPREV study): Rationale, methods, and baseline characteristics: A clinical trial comparing the efficacy of a Mediterranean diet rich in olive oil versus a low-fat diet on cardiovascular disease in coronary patients. Am. Heart J. 2016, 177, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Schröder, H.; Fitó, M.; Estruch, R.; Martínez-González, M.A.; Corella, D.; Salas-Salvadó, J.; Lamuela-Raventós, R.; Ros, E.; Salaverría, I.; Fiol, M.; et al. A short screener is valid for assessing Mediterranean diet adherence among older Spanish men and women. J. Nutr. 2011, 141, 1140–1145. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Ballart, J.D.; Piñol, J.L.; Zazpe, I.; Corella, D.; Carrasco, P.; Toledo, E.; Perez-Bauer, M.; Martínez-González, M.A.; Salas-Salvadó, J.; Martín-Moreno, J.M. Relative validity of a semi-quantitative food-frequency questionnaire in an elderly Mediterranean population of Spain. Br. J. Nutr. 2010, 103, 1808–1816. [Google Scholar] [CrossRef] [Green Version]
- Haro, C.; Rangel-Zúñiga, O.A.; Alcalá-Díaz, J.F.; Gómez-Delgado, F.; Pérez-Martínez, P.; Delgado-Lista, J.; Quintana-Navarro, G.M.; Landa, B.B.; Navas-Cortés, J.A.; Tena-Sempere, M.; et al. Intestinal Microbiota Is Influenced by Gender and Body Mass Index. PLoS ONE 2016, 11, e0154090. [Google Scholar] [CrossRef] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Santos-Marcos, J.A.; Haro, C.; Vega-Rojas, A.; Alcala-Diaz, J.F.; Molina-Abril, H.; Leon-Acuña, A.; Lopez-Moreno, J.; Landa, B.B.; Tena-Sempere, M.; Perez-Martinez, P.; et al. Sex Differences in the Gut Microbiota as Potential Determinants of Gender Predisposition to Disease. Mol. Nutr. Food Res. 2019, 63, e1800870. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Szumilas, M. Explaining odds ratios. J. Can. Acad. Child. Adolesc. Psychiatry 2010, 19, 227–229. [Google Scholar]
- Zuo, T.; Ng, S.C. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front. Microbiol. 2018, 9, 2247. [Google Scholar] [CrossRef] [PubMed]
- Alisi, A.; Ceccarelli, S.; Panera, N.; Nobili, V. Causative role of gut microbiota in non-alcoholic fatty liver disease pathogenesis. Front. Cell. Infect. Microbiol. 2012, 2, 132. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Festi, D.; Schiumerini, R.; Eusebi, L.H.; Marasco, G.; Taddia, M.; Colecchia, A. Gut microbiota and metabolic syndrome. World J. Gastroenterol. 2014, 20, 16079–16094. [Google Scholar] [CrossRef]
- Koren, O.; Spor, A.; Felin, J.; Fåk, F.; Stombaugh, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108, 4592–4598. [Google Scholar] [CrossRef] [Green Version]
- Oh, C.; Lee, K.; Cheong, Y.; Lee, S.-W.; Park, S.-Y.; Song, C.-S.; Choi, I.-S.; Lee, J.-B. Comparison of the Oral Microbiomes of Canines and Their Owners Using Next-Generation Sequencing. PLoS ONE 2015, 10, e0131468. [Google Scholar] [CrossRef] [Green Version]
- Zupancic, M.L.; Cantarel, B.L.; Liu, Z.; Drabek, E.F.; Ryan, K.A.; Cirimotich, S.; Jones, C.; Knight, R.; Walters, W.A.; Knights, D.; et al. Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome. PLoS ONE 2012, 7, e43052. [Google Scholar] [CrossRef]
- Kalliomäki, M.; Collado, M.C.; Salminen, S.; Isolauri, E. Early differences in fecal microbiota composition in children may predict overweight. Am. J. Clin. Nutr. 2008, 87, 534–538. [Google Scholar] [CrossRef]
- Karlsson, C.L.; Onnerfält, J.; Xu, J.; Molin, G.; Ahrné, S.; Thorngren-Jerneck, K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity 2012, 20, 2257–2261. [Google Scholar] [CrossRef]
- Santacruz, A.; Collado, M.C.; García-Valdés, L.; Segura, M.T.; Martín-Lagos, J.A.; Anjos, T.; Martí-Romero, M.; Lopez, R.M.; Florido, J.; Campoy, C.; et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr. 2010, 104, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Fiorucci, S.; Distrutti, E. Bile Acid-Activated Receptors, Intestinal Microbiota, and the Treatment of Metabolic Disorders. Trends Mol. Med. 2015, 21, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Lippert, K.; Kedenko, L.; Antonielli, L.; Kedenko, I.; Gemeier, C.; Leitner, M.; Kautzky-Willer, A.; Paulweber, B.; Hackl, E. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef. Microbes 2017, 8, 545–556. [Google Scholar] [CrossRef]
- Org, E.; Blum, Y.; Kasela, S.; Mehrabian, M.; Kuusisto, J.; Kangas, A.J.; Soininen, P.; Wang, Z.; Ala-Korpela, M.; Hazen, S.L.; et al. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017, 18, 70. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.M.; Huang, W.C.; Weng, S.L.; Tseng, H.C.; Liang, C.; Wang, W.C.; Yang, T.; Yang, T.L.; Weng, C.T.; Chang, T.H.; et al. Systematic analysis of the association between gut flora and obesity through high-throughput sequencing and bioinformatics approaches. Biomed. Res. Int. 2014, 2014, 906168. [Google Scholar] [CrossRef]
- Ownby, D.R.; Johnson, C.C.; Peterson, E.L. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA 2002, 288, 963–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [Green Version]
- Kohl, K.D.; Amaya, J.; Passement, C.A.; Dearing, M.D.; McCue, M.D. Unique and shared responses of the gut microbiota to prolonged fasting: A comparative study across five classes of vertebrate hosts. FEMS Microbiol. Ecol. 2014, 90, 883–894. [Google Scholar] [CrossRef]
- Konikoff, T.; Gophna, U. Oscillospira: A Central, Enigmatic Component of the Human Gut Microbiota. Trends Microbiol. 2016, 24, 523–524. [Google Scholar] [CrossRef]
- Handl, S.; German, A.J.; Holden, S.L.; Dowd, S.E.; Steiner, J.M.; Heilmann, R.M.; Grant, R.W.; Swanson, K.S.; Suchodolski, J.S. Faecal microbiota in lean and obese dogs. FEMS Microbiol. Ecol. 2013, 84, 332–343. [Google Scholar] [CrossRef] [Green Version]
- Abramson, A.L.; Isenberg, H.D.; McDermott, L.M. Microbiology of the canine nasal cavities. Rhinology 1980, 18, 143–150. [Google Scholar] [CrossRef]
- Schmidt, M.; Unterer, S.; Suchodolski, J.S.; Honneffer, J.B.; Guard, B.C.; Lidbury, J.A.; Steiner, J.M.; Fritz, J.; Kölle, P. The fecal microbiome and metabolome differs between dogs fed Bones and Raw Food (BARF) diets and dogs fed commercial diets. PLoS ONE 2018, 13, e0201279. [Google Scholar] [CrossRef] [Green Version]
- Santos-Marcos, J.A.; Perez-Jimenez, F.; Camargo, A. The role of diet and intestinal microbiota in the development of metabolic syndrome. J. Nutr. Biochem. 2019, 70, 1–27. [Google Scholar] [CrossRef]

| - | Patients | Pets | No Pets | p-Value |
|---|---|---|---|---|
| n | 162 | 83 | 79 | n/a |
| Men/Women (n) | 133/29 | 72/11 | 61/18 | 0.114 |
| Diet (LF vs. MED) | 68/94 | 35/48 (LF vs. MED) | 33/46 (LF vs. MED) | 0.959 |
| T2DM (No vs. T2DM) | 49/113 | 27/56 (No vs. T2DM) | 22/57 (No vs. T2DM) | 0.517 |
| Metabolic syndrome (No vs. MetS) | 101/61 | 59/24 (No vs. MetS) | 42/37 (No vs. MetS) | 0.019 |
| Obesity (No vs. Obesity) | 77/85 | 46/37 (No vs. Obesity) | 31/48 (No vs. Obesity) | 0.039 |
| Arterial hypertension (No vs. AHT) | 53/109 | 33/50 (No vs. AHT) | 20/59 (No vs. AHT) | 0.050 |
| Age (years) | 63.32 ± 8.45 | 60.86 ± 8.21 | 65.92 ± 7.96 | <0.001 |
| Weight (Kg) | 82.75 ± 13.35 | 83.15 ± 14.2 | 82.35 ± 12.5 | 0.708 |
| BMI (Kg/m2) | 30.36 ± 3.94 | 29.88 ± 3.88 | 30.85 ± 3.96 | 0.123 |
| Serum triacylglycerols (mg/dL) | 129.32 ± 88.49 | 115.71 ± 46.41 | 143.44 ± 115.89 | 0.237 |
| Total cholesterol (mg/dL) | 160.47 ± 34.24 | 156.87 ± 31.73 | 164.20 ± 36.5 | 0.191 |
| HDL-cholesterol (mg/dL) | 40.98 ± 9.64 | 41.29 ± 9.83 | 40.66 ± 9.49 | 0.745 |
| LDL-cholesterol (mg/dL) | 93.63 ± 27.92 | 92.02 ± 26.54 | 95.37 ± 29.42 | 0.456 |
| CRP (mg/dL) | 2.77 ± 3.81 | 2.51 ± 2.94 | 3.03 ± 4.54 | 0.106 |
| ISI | 4.07 ± 2.62 | 4.29 ± 2.86 | 3.80 ± 2.29 | 0.356 |
| Systolic BP | 136.55 ± 19.02 | 134.00 ± 17.58 | 139.34 ± 20.23 | 0.084 |
| Diastolic BP | 76.86 ± 11.22 | 77.11 ± 9.89 | 76.59 ± 12.59 | 0.776 |
| - | Dogs | No Pets | p-Value |
|---|---|---|---|
| n | 28 | 79 | n/a |
| Men/Women (n) | 24/4 | 61/18 | 0.339 |
| Diet (LF vs. MED) | 17/11 (LF vs. MED) | 33/46 (LF vs. MED) | 0.084 |
| T2DM (No vs. T2DM) | 8/20 (No vs. T2DM) | 22/57 (No vs. T2DM) | 0.942 |
| Metabolic syndrome (No vs. MetS) | 21/7 (No vs. MetS) | 42/37 (No vs. MetS) | 0.044 |
| Obesity (No vs. Obesity) | 17/11 (No vs. Obesity) | 31/48 (No vs. Obesity) | 0.049 |
| Arterial hypertension (No vs. AHT) | 10/18 (No vs. AHT) | 20/59 (No vs. AHT) | 0.293 |
| Age (years) | 60.17 ± 7.70 | 65.92 ± 7.96 | <0.001 |
| Weight (Kg) | 81.85 ± 12.16 | 82.35 ± 12.50 | 0.860 |
| BMI (Kg/m2) | 29.40 ± 3.15 | 30.85 ± 3.96 | 0.093 |
| Serum triacylglycerols (mg/dL) | 115.32 ± 49.89 | 143.44 ± 115.89 | 0.218 |
| Total cholesterol (mg/dL) | 155.89 ± 30.53 | 164.20 ± 36.50 | 0.284 |
| HDL-cholesterol (mg/dL) | 40.50 ± 10.97 | 40.66 ± 9.49 | 0.777 |
| LDL-cholesterol (mg/dL) | 91.96 ± 24.56 | 95.37 ± 29.42 | 0.586 |
| CRP (mg/dL) | 2.57 ± 2.40 | 3.03 ± 4.54 | 0.608 |
| ISI | 4.03 ± 2.13 | 3.80 ± 2.29 | 0.715 |
| Systolic BP | 134.11 ± 17.48 | 139.34 ± 20.24 | 0.232 |
| Diastolic BP | 76.46 ± 11.60 | 76.59 ± 12.59 | 0.962 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arenas-Montes, J.; Perez-Martinez, P.; Vals-Delgado, C.; Romero-Cabrera, J.L.; Cardelo, M.P.; Leon-Acuña, A.; Quintana-Navarro, G.M.; Alcala-Diaz, J.F.; Lopez-Miranda, J.; Camargo, A.; et al. Owning a Pet Is Associated with Changes in the Composition of Gut Microbiota and Could Influence the Risk of Metabolic Disorders in Humans. Animals 2021, 11, 2347. https://doi.org/10.3390/ani11082347
Arenas-Montes J, Perez-Martinez P, Vals-Delgado C, Romero-Cabrera JL, Cardelo MP, Leon-Acuña A, Quintana-Navarro GM, Alcala-Diaz JF, Lopez-Miranda J, Camargo A, et al. Owning a Pet Is Associated with Changes in the Composition of Gut Microbiota and Could Influence the Risk of Metabolic Disorders in Humans. Animals. 2021; 11(8):2347. https://doi.org/10.3390/ani11082347
Chicago/Turabian StyleArenas-Montes, Javier, Pablo Perez-Martinez, Cristina Vals-Delgado, Juan Luis Romero-Cabrera, Magdalena P. Cardelo, Ana Leon-Acuña, Gracia M. Quintana-Navarro, Juan F. Alcala-Diaz, Jose Lopez-Miranda, Antonio Camargo, and et al. 2021. "Owning a Pet Is Associated with Changes in the Composition of Gut Microbiota and Could Influence the Risk of Metabolic Disorders in Humans" Animals 11, no. 8: 2347. https://doi.org/10.3390/ani11082347
APA StyleArenas-Montes, J., Perez-Martinez, P., Vals-Delgado, C., Romero-Cabrera, J. L., Cardelo, M. P., Leon-Acuña, A., Quintana-Navarro, G. M., Alcala-Diaz, J. F., Lopez-Miranda, J., Camargo, A., & Perez-Jimenez, F. (2021). Owning a Pet Is Associated with Changes in the Composition of Gut Microbiota and Could Influence the Risk of Metabolic Disorders in Humans. Animals, 11(8), 2347. https://doi.org/10.3390/ani11082347






