Levels of Main Bacterial Phyla in the Gastrointestinal Tract of Sheep Depending on Parity and Age
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Sampling Collection
2.3. Bacterial DNA (Deoxyribonucleic Acid) Isolation
2.4. Real-Time PCR Analysis
2.5. Statistical Analysis
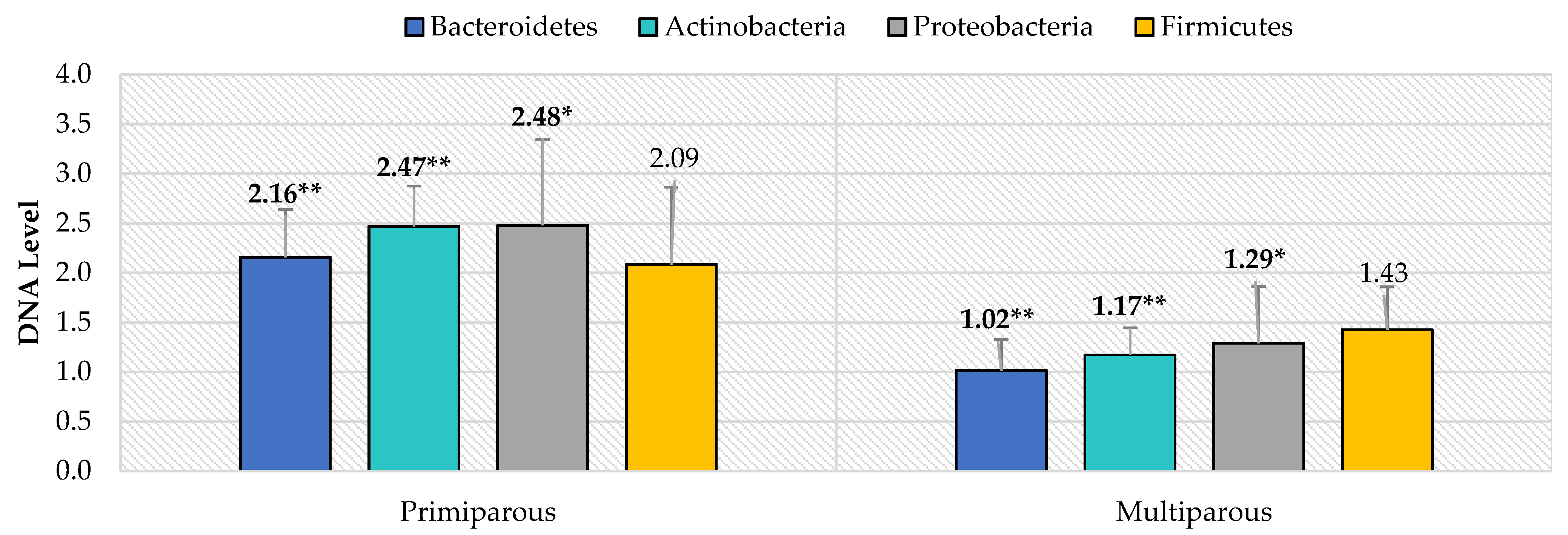
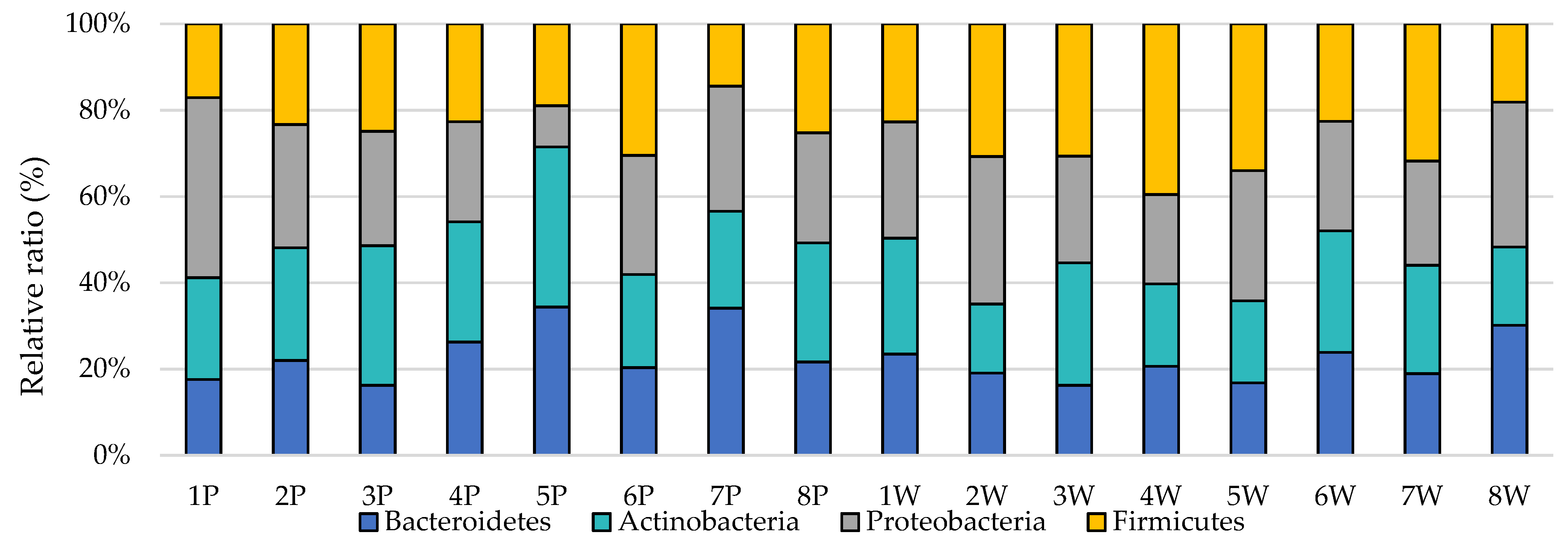
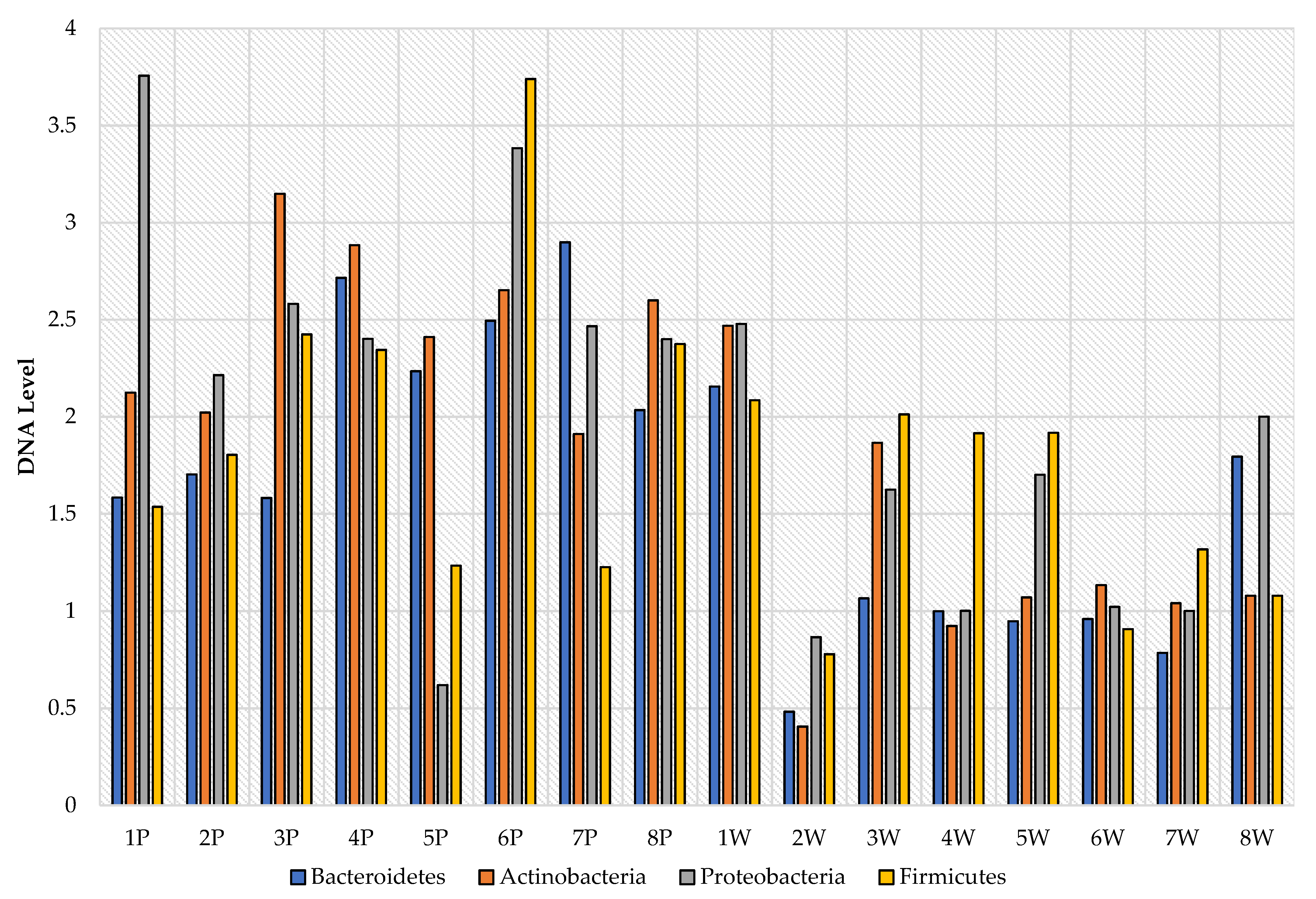
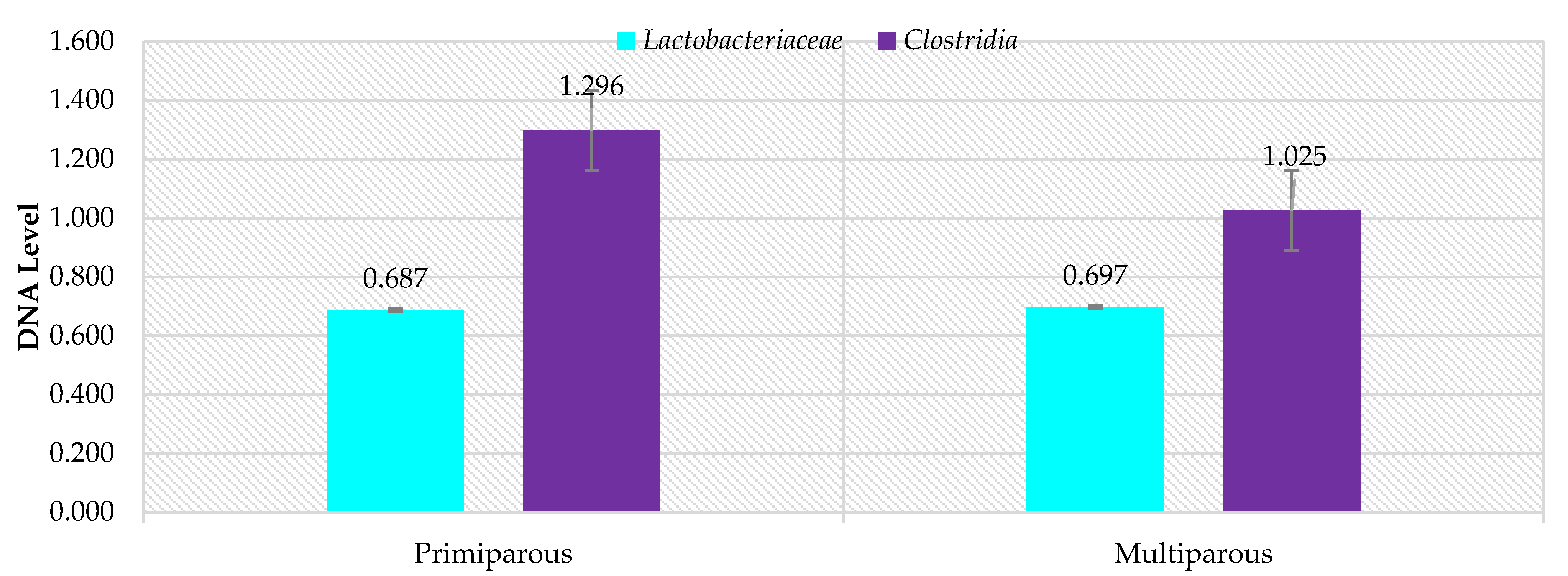
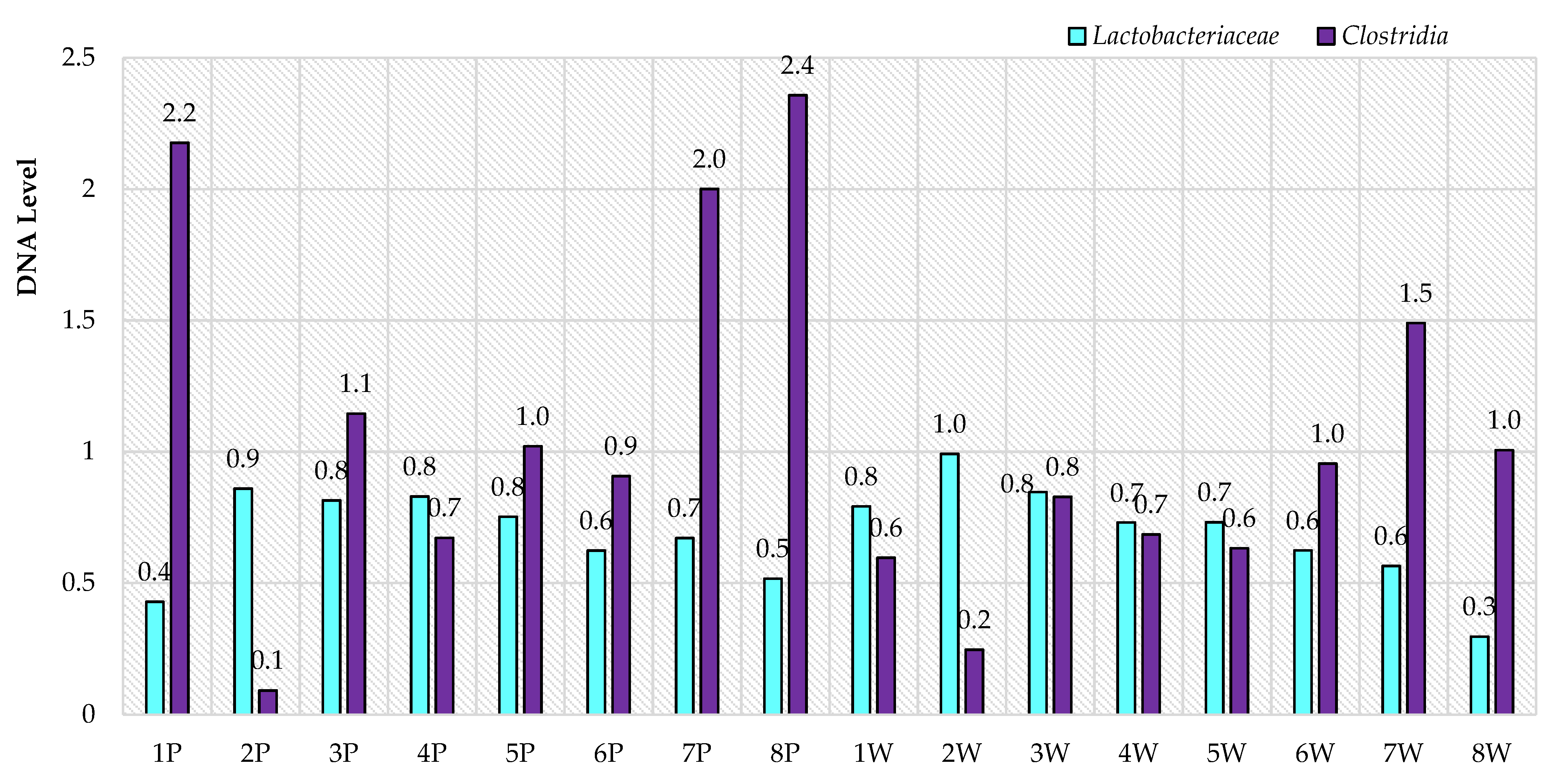
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.; Shao, M.; Huang, H.; Wang, S.; Ma, L.; Wang, H.; Zhu, R. The dynamic distribution of small-tail han sheep microbiota across different intestinal segments. Front. Microbiol. 2018, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Rey, M.; Enjalbert, F.; Combes, S.; Cauquil, L.; Bouchez, O.; Monteils, V. Establishment of ruminal bacterial community in dairy calves from birth to weaning is sequential. J. Appl. Microbiol. 2014, 116, 245–257. [Google Scholar] [CrossRef] [PubMed]
- von Engelhardt, W.; Breves, G.; Diener, M.; Gäbel, G. Physiologie der Haustiere; Georg Thieme Verlag: Stuttgart, Germany, 2015; Volume 2, pp. 40–41, 80–82. [Google Scholar]
- Wang, L.; Zhang, K.; Zhang, C.; Feng, Y.; Zhang, X.; Wang, X.; Wu, G. Dynamics and stabilization of the rumen microbiome in yearling Tibetan sheep. Scientific Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Zeng, D.; Ni, X.; Zhu, H.; Jian, P.; Zhou, Y.; Pan, K. Microbial community compositions in the gastrointestinal tract of Chinese Mongolian sheep using Illumina MiSeq sequencing revealed high microbial diversity. Amb Express. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Tanca, A.; Fraumene, C.; Manghina, V.; Palomba, A.; Abbondio, M.; Deligios, M.; Uzzau, S. Diversity and functions of the sheep faecal microbiota: A multi-omic characterization. Microb. Biotechnol. 2017, 10, 541–554. [Google Scholar] [CrossRef]
- Khafipour, E.; Li, S.; Plaizier, J.C.; Krause, D.O. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl. Environ. Microbiol. 2009, 75, 7115–7124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, S.G.; Ericsson, A.C.; Poock, S.E.; Melendez, P.; Lucy, M.C. Hot topic: 16S rRNA gene sequencing reveals the microbiome of the virgin and pregnant bovine uterus. J. Dairy Sci. 2017, 100, 4953–4960. [Google Scholar] [CrossRef] [Green Version]
- Karstrup, C.C.; Klitgaard, K.; Jensen, T.K.; Agerholm, J.S.; Pedersen, H.G. Presence of bacteria in the endometrium and placentomes of pregnant cows. Theriogenology 2017, 99, 41–47. [Google Scholar] [CrossRef]
- Sasson, G.; Ben-Shabat, S.K.; Seroussi, E.; Doron-Faigenboim, A.; Shterzer, N.; Yaacoby, S.; Mizrahi, I. Heritable bovine rumen bacteria are phylogenetically related and correlated with the cow’s capacity to harvest energy from its feed. MBio 2017, 8, 00703–00717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cholewińska, P.; Górniak, W.; Wojnarowski, K. Impact of selected environmental factors on microbiome of the digestive tract of ruminants. BMC Vet. Res. 2021, 17, 25. [Google Scholar] [CrossRef]
- Kawęcka, A.; Krupiński, J.; Sikora, J. Polish Pogórza sheep—Genetic resources conservation programme. Wiad. Zoot. 2014, 4, 11–17. [Google Scholar]
- Dowd, S.E.; Callaway, T.R.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Hagevoort, R.G.; Edrington, T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (BTEFAP). BMC Microbiol. 2008, 8, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Gregoris, T.B.; Aldred, N.; Clare, A.S.; Burgess, J.G. Improvement of phylum-And class-specific primers for real-time PCR quantification of bacterial taxa. J. Microbiol. Methods 2011, 86, 351–356. [Google Scholar] [CrossRef]
- Mitsumori, M.; Ajisaka, N.; Tajima, K.; Kajikawa, H.; Kurihara, M. Detection of Proteobacteria from the rumen by PCR using methanotroph-specific primers. Lett. Appl. Microbiol. 2002, 35, 251–255. [Google Scholar] [CrossRef]
- Blackwood, C.B.; Oaks, A.; Buyer, J.S. Phylum-And class-specific PCR primers for general microbial community analysis. Appl. Environ. Microbiol. 2005, 71, 6193–6198. [Google Scholar] [CrossRef] [Green Version]
- Walter, J.; Hertel, C.; Tannock, G.W.; Lis, C.M.; Munro, K.; Hammes, W.P. Detection of lactobacillus, pediococcus, leuconostoc, and weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2001, 67, 2578–2585. [Google Scholar] [CrossRef] [Green Version]
- Amit-Romach, E.; Sklan, D.; Uni, Z. Microflora ecology of the chicken intestine using 16S ribosomal DNA primers. Poult. Sci. 2004, 83, 1093–1098. [Google Scholar] [CrossRef]
- Szeligowska, N.; Cholewińska, P.; Czyż, K.; Wojnarowski, K.; Janczak, M. Inter and intraspecies comparison of the level of selected bacterial phyla in in cattle and sheep based on feces. BMC Vet. Res. 2021, 17, 1–9. [Google Scholar] [CrossRef]
- Cholewińska, P.; Czyż, K.; Nowakowski, P.; Wyrostek, A. The microbiome of the digestive system of ruminants—A review. Anim. Health Res. Rev. 2020, 21, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Mamun, M.A.A.; Sandeman, M.; Rayment, P.; Brook-Carter, P.; Scholes, E.; Kasinadhuni, N.; Greenhill, A.R. The composition and stability of the faecal microbiota of Merino sheep. J. Appl. Microbiol. 2020, 128, 280–291. [Google Scholar] [CrossRef] [Green Version]
- Hallmaier-Wacker, L.K.; Lueert, S.; Roos, C.; Knauf, S. The influence of sex on the urogenital microbiome of rhesus monkeys. bioRxiv 2019, 555771. [Google Scholar]
- Markle, J.G.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podsiadła-Urban, G.; Kiernicka, M.; Wysokińska-Miszczuk, J. The Influence of Estrogen and Progesterone on the State of Female Periodontium in Each Stage of Women’s Life—Review of the Literature. Dent. Med. Probl. 2010, 47, 89–96. [Google Scholar]
- Edwards, S.M.; Cunningham, S.A.; Dunlop, A.L.; Corwin, E.J. The maternal gut microbiome during pregnancy. MCN. Am. J. Matern. Child Nurs. 2017, 42, 310. [Google Scholar] [CrossRef] [PubMed]
- Menon, R.; Watson, S.E.; Thomas, L.N.; Allred, C.D.; Dabney, A.; Azcarate-Peril, M.A.; Sturino, J.M. Diet complexity and estrogen receptor β status affect the composition of the murine intestinal microbiota. Appl. Environ. Microbiol. 2013, 79, 5763–5773. [Google Scholar] [CrossRef] [Green Version]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.; Laitinen, K.; Bäckhed, H.K.; Bäckhed, F. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012, 150, 470–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forsum, E.; Sadurskis, A.; Wager, J. Estimation of body fat in healthy Swedish women during pregnancy and lactation. Am. J. Clin. Nutr. 1989, 50, 465–473. [Google Scholar] [CrossRef]
- Knecht, D.; Cholewińska, P.; Jankowska-Mąkosa, A.; Czyż, K. Development of Swine’s Digestive Tract Microbiota and Its Relation to Production Indices—A Review. Animals 2020, 10, 527. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.; Ma, T.; Tang, W.; Li, D.; Mishra, S.K.; Xu, Z.; Wang, Q.; Jie, H. Gut Microbiome of Chinese Forest Musk Deer Examined across Gender and Age. BioMed Res. Internat. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Yáñez-Ruiz, D.R.; Abecia, L.; Newbold, C.J. Manipulating rumen microbiome and fermentation through interventions during early life: A review. Front. Microbiol. 2015, 6, 1133. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Zhou, M.; Adamowicz, E.; Basarab, J.A. Characterization of bovine ruminal epithelial bacterial communities using 16S rRNA sequencing, PCR-DGGE, and qRT-PCR analysis. Vet. Microbiol. 2012, 155, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, K.; Li, C.; Wang, X.; Chen, Y.; Yang, Y. Characterization and comparison of microbiota in the gastrointestinal tracts of the goat (capra hircus) during preweaning development. Front. Microbiol. 2019, 10, 2125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buford, T.W. (Dis) Trust your gut: The gut microbiome in age-related inflammation, health, and disease. Microbiome 2017, 5, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Name | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Universal Eubacterial Genes [13] | 530F (5′-GTC CCA GCM GCN GCG G) | 1100R (5′-GGG TTN CGN TCG TTG) |
| Firmicutes [14] | 928F-Firm (5′-TGA AAC TYA AAG GAA TTG ACG) | 1040FirmR (5′-ACC ATG CAC CAC CTG TC) |
| Bacteroidetes [14] | 798cfbF (5′-CRA ACA GGA TTA GAT ACC CT) | cfb967R (5′-GGT AAG GGT TCC TCG CGT AT) |
| Proteobacteria [15] | 27F (5′-GAGTTTGATCMTGGCTCAG-3′) | 1529R(5′-CAKAAAGGAGGTGATCC-3′) |
| Actinobacteria [16] | Act1159R TCCGAGTTRACCCCGGC | Eub338F ACGGGCGGTGTGTACA |
| Lactobacillaceae [17] | lac1 forward (5′-AGC AGT AGG GAA TCT TCC A) | Lac2Seq (5′-ATTTCACCGCTACACATG) |
| Clostridia [18] | Clos58-fAAAGGAAGATTAATACCGCATAA | Clos780-r ATCTTGCGACCGTACTCCCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smoliński, J.; Szeligowska, N.; Cholewińska, P.; Czyż, K.; Janczak, M. Levels of Main Bacterial Phyla in the Gastrointestinal Tract of Sheep Depending on Parity and Age. Animals 2021, 11, 2203. https://doi.org/10.3390/ani11082203
Smoliński J, Szeligowska N, Cholewińska P, Czyż K, Janczak M. Levels of Main Bacterial Phyla in the Gastrointestinal Tract of Sheep Depending on Parity and Age. Animals. 2021; 11(8):2203. https://doi.org/10.3390/ani11082203
Chicago/Turabian StyleSmoliński, Jakub, Natalia Szeligowska, Paulina Cholewińska, Katarzyna Czyż, and Marzena Janczak. 2021. "Levels of Main Bacterial Phyla in the Gastrointestinal Tract of Sheep Depending on Parity and Age" Animals 11, no. 8: 2203. https://doi.org/10.3390/ani11082203
APA StyleSmoliński, J., Szeligowska, N., Cholewińska, P., Czyż, K., & Janczak, M. (2021). Levels of Main Bacterial Phyla in the Gastrointestinal Tract of Sheep Depending on Parity and Age. Animals, 11(8), 2203. https://doi.org/10.3390/ani11082203






