Comprehensive Transcriptomic Comparison between Porcine CD8− and CD8+ Gamma Delta T Cells Revealed Distinct Immune Phenotype
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Sorting of the γδ T Cell Subsets
2.3. Performing RNA Quantification and Determining RNA Quality
2.4. Library Preparation for High-Throughput Transcriptomic Sequencing
2.5. Quality Control and Reads Mapping to the Reference Genome
2.6. Differentially Expressed Genes (DEGs) Analysis
2.7. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analyses of the DEGs
2.8. Gene Set Enrichment Analysis, Pathway Profiling, and Protein—Protein Interaction Network Analysis
3. Results
3.1. Quality Control Analysis and RNA-Seq Data
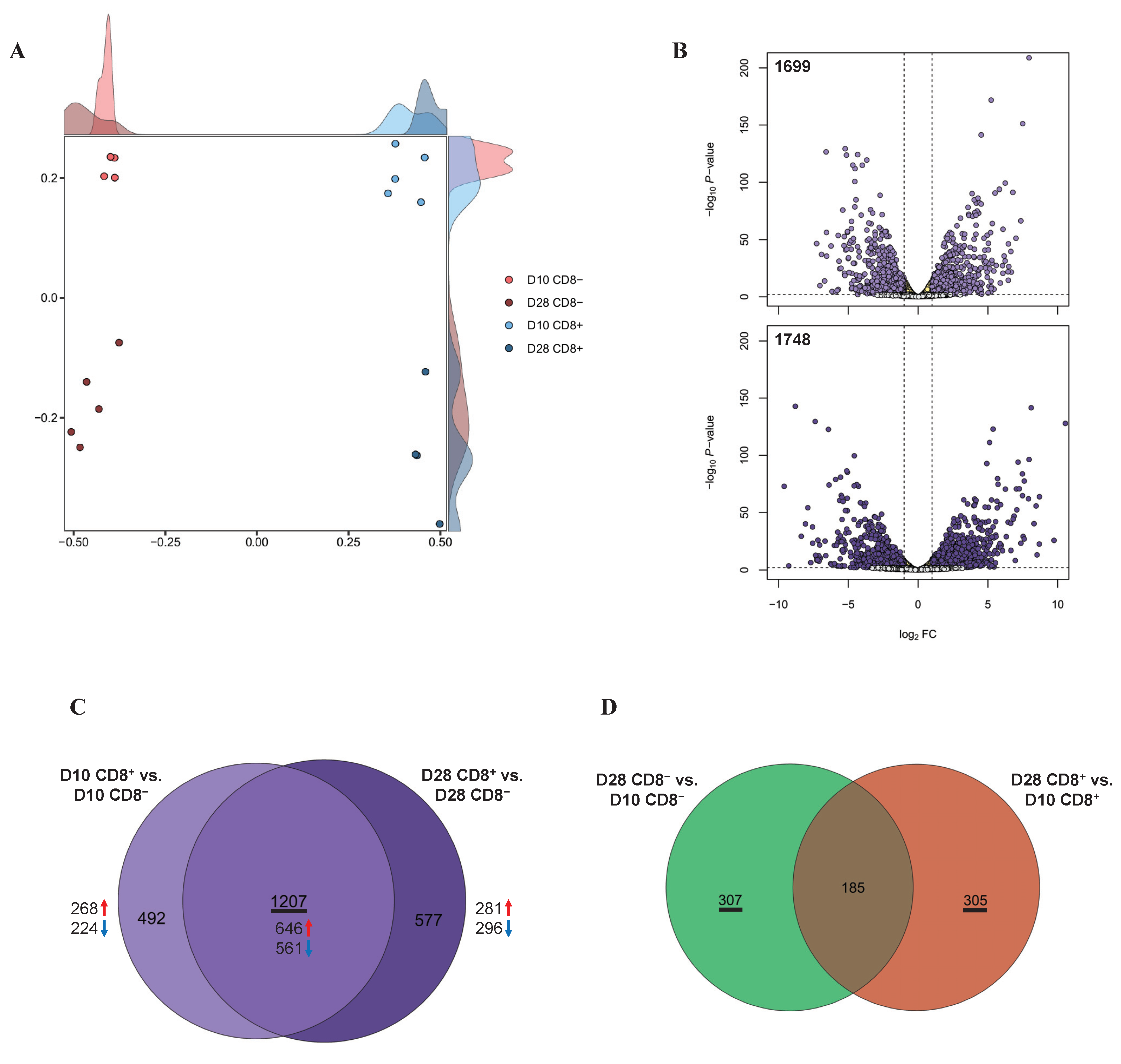
3.2. Identification of the DEGs
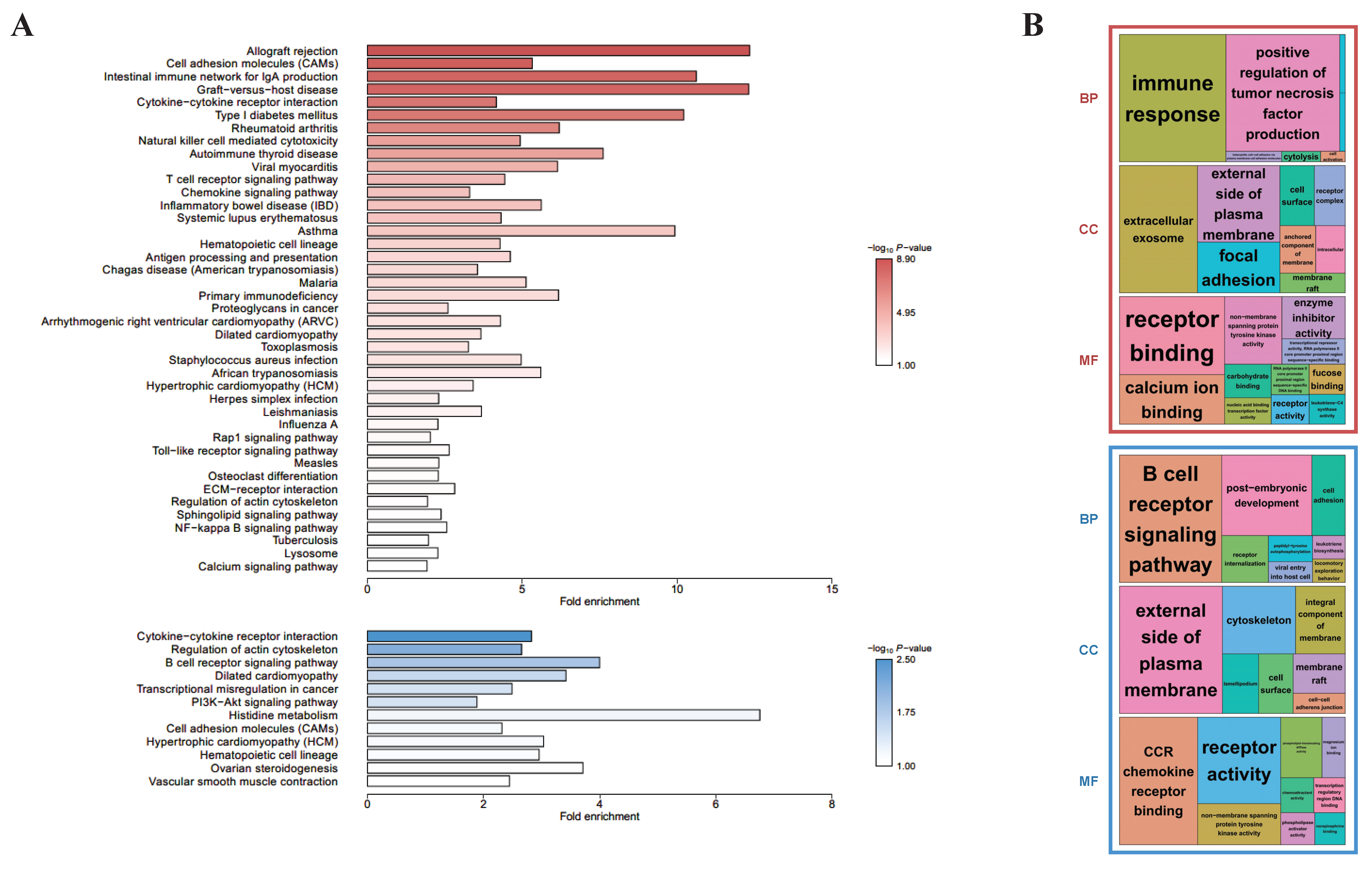
3.3. Differences in the Functional Annotations
3.4. GSEA, Pathway Profiling, and PPI Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Cromwell, G.L. Why and how antibiotics are used in swine production. Anim. Biotechnol. 2002, 13, 7–27. [Google Scholar] [CrossRef]
- Boudry, G.; Péron, V.; Le Huërou-Luron, I.; Lallès, J.P.; Sève, B. Weaning induces both transient and long-lasting modifications of absorptive, secretory, and barrier properties of piglet intestine. J. Nutr. 2004, 134, 2256–2262. [Google Scholar] [CrossRef] [PubMed]
- Gerner, W.; Käser, T.; Saalmüller, A. Porcine T lymphocytes and NK cells—An update. Dev. Comp. Immunol. 2009, 33, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Charerntantanakul, W.; Roth, J.A. Biology of porcine T lymphocytes. Anim. Health Res. Rev. 2006, 7, 81. [Google Scholar] [CrossRef] [PubMed]
- Mair, K.; Sedlak, C.; Käser, T.; Pasternak, A.; Levast, B.; Gerner, W.; Saalmüller, A.; Summerfield, A.; Gerdts, V.; Wilson, H. The porcine innate immune system: An update. Dev. Comp. Immunol. 2014, 45, 321–343. [Google Scholar] [CrossRef] [PubMed]
- Crisci, E.; Fraile, L.; Montoya, M. Cellular innate immunity against PRRSV and Swine Influenza viruses. Vet. Sci. 2019, 6, 26. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Pan, Z.; Zhang, L.; Xue, W.; Peng, M.; Hu, P.; Xu, H.; Chen, M. Increased effector γδ T cells with enhanced cytokine production are associated with inflammatory abnormalities in severe hand, foot, and mouth disease. Int. Immunopharmacol. 2019, 73, 172–180. [Google Scholar] [CrossRef]
- Khatun, A.; Nazki, S.; Jeong, C.-G.; Gu, S.; Lee, S.-I.; Yang, M.-S.; Lim, B.; Kim, K.-S.; Kim, B.; Lee, K.-T. Effect of polymorphisms in porcine guanylate-binding proteins on host resistance to PRRSV infection in experimentally challenged pigs. Vet. Res. 2020, 51, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonneville, M.; O’brien, R.L.; Born, W.K. γδ T cell effector functions: A blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 2010, 10, 467–478. [Google Scholar] [CrossRef]
- Takamatsu, H.-H.; Denyer, M.; Stirling, C.; Cox, S.; Aggarwal, N.; Dash, P.; Wileman, T.; Barnett, P. Porcine γδ T cells: Possible roles on the innate and adaptive immune responses following virus infection. Vet. Immunol. Immunopathol. 2006, 112, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Šinkora, M.; Butler, J.E. The ontogeny of the porcine immune system. Dev. Comp. Immunol. 2009, 33, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.M.; Tian, D.; Heffron, C.L.; Subramaniam, S.; Opriessnig, T.; Foss, D.L.; Calvert, J.G.; Meng, X.-J. Cytotoxic T lymphocyte epitopes identified from a contemporary strain of porcine reproductive and respiratory syndrome virus enhance CD4+ CD8+ T, CD8+ T, and γδ T cell responses. Virology 2019, 538, 35–44. [Google Scholar] [CrossRef]
- Li, J.; Yu, Q.; Nie, X.; Guo, X.; Song, Q.; Li, H. Effects of porcine circovirus type 2 on expression of mRNA associated with endogenous antigen processing and presentation in pulmonary alveolar macrophages and circulating T lymphocytes in piglets. Vet. J. 2012, 193, 199–205. [Google Scholar] [CrossRef]
- Post, J.; Weesendorp, E.; Montoya, M.; Loeffen, W.L. Influence of age and dose of African swine fever virus infections on clinical outcome and blood parameters in pigs. Viral Immunol. 2017, 30, 58–69. [Google Scholar] [CrossRef]
- Butler, J.E.; Sinkora, M.; Wertz, N.; Holtmeier, W.; Lemke, C.D. Development of the neonatal B and T cell repertoire in swine: Implications for comparative and veterinary immunology. Vet. Res. 2006, 37, 417–441. [Google Scholar] [CrossRef] [Green Version]
- Johnson, W.; Roof, M.; Vaughn, E.; Christopher-Hennings, J.; Johnson, C.R.; Murtaugh, M.P. Pathogenic and humoral immune responses to porcine reproductive and respiratory syndrome virus (PRRSV) are related to viral load in acute infection. Vet. Immunol. Immunopathol. 2004, 102, 233–247. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Ginestet, C. ggplot2: Elegant graphics for data analysis. J. R. Stat. Soc. Ser. A 2011, 174, 245. [Google Scholar] [CrossRef]
- Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44. [Google Scholar]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swain, S.; Dutton, R.; Schwab, R.; Yamamoto, J. Xenogeneic human anti-mouse T cell responses are due to the activity of the same functional T cell subsets responsible for allospecific and major histocompatibility complex-restricted responses. J. Exp. Med. 1983, 157, 720–729. [Google Scholar] [CrossRef]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M. The structure of haplotype blocks in the human genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amadori, A.; Zamarchi, R.; Chieco-Bianchi, L. CD4: CD8 ratio and HIV infection: The ‘tap-and-drain’ hypothesis. Immunol. Today 1996, 17, 414–417. [Google Scholar] [CrossRef]
- Evans, D.M.; Frazer, I.H.; Martin, N.G. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res. Hum. Genet. 1999, 2, 250–257. [Google Scholar] [CrossRef]
- Shedlock, D.J.; Shen, H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science 2003, 300, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Budak, F.; Bal, S.H.; Tezcan, G.; Guvenc, F.; Akalin, E.H.; Goral, G.; Deniz, G.; Oral, H.B. MicroRNA expression patterns of CD8+ T cells in acute and chronic brucellosis. PLoS ONE 2016, 11, e0165138. [Google Scholar] [CrossRef]
- Islam, M.A.; Große-Brinkhaus, C.; Pröll, M.J.; Uddin, M.J.; Rony, S.A.; Tesfaye, D.; Tholen, E.; Hölker, M.; Schellander, K.; Neuhoff, C. Deciphering transcriptome profiles of peripheral blood mononuclear cells in response to PRRSV vaccination in pigs. BMC Genom. 2016, 17, 1–17. [Google Scholar] [CrossRef]
- Islam, M.A.; Große-Brinkhaus, C.; Pröll, M.J.; Uddin, M.J.; Aqter Rony, S.; Tesfaye, D.; Tholen, E.; Hoelker, M.; Schellander, K.; Neuhoff, C. PBMC transcriptome profiles identifies potential candidate genes and functional networks controlling the innate and the adaptive immune response to PRRSV vaccine in Pietrain pig. PLoS ONE 2017, 12, e0171828. [Google Scholar] [CrossRef] [Green Version]
- Qin, S.; Rottman, J.B.; Myers, P.; Kassam, N.; Weinblatt, M.; Loetscher, M.; Koch, A.E.; Moser, B.; Mackay, C.R. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J. Clin. Investig. 1998, 101, 746–754. [Google Scholar] [CrossRef]
- Princen, K.; Hatse, S.; Vermeire, K.; Aquaro, S.; De Clercq, E.; Gerlach, L.-O.; Rosenkilde, M.; Schwartz, T.W.; Skerlj, R.; Bridger, G. Inhibition of human immunodeficiency virus replication by a dual CCR5/CXCR4 antagonist. J. Virol. 2004, 78, 12996–13006. [Google Scholar] [CrossRef] [Green Version]
- Hudspeth, K.; Fogli, M.; Correia, D.V.; Mikulak, J.; Roberto, A.; Della Bella, S.; Silva-Santos, B.; Mavilio, D. Engagement of NKp30 on Vδ1 T cells induces the production of CCL3, CCL4, and CCL5 and suppresses HIV-1 replication. Blood 2012, 119, 4013–4016. [Google Scholar] [CrossRef] [Green Version]
- Barmania, F.; Pepper, M.S. CC chemokine receptor type five (CCR5): An emerging target for the control of HIV infection. Appl. Transl. Genom. 2013, 2, 3–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, W.; Ji, L.; Zhang, Y.; Zhen, Y.; Zhang, Q.; Xu, X.; Liu, B. Transcriptome differences in porcine alveolar macrophages from Tongcheng and Large White pigs in response to highly pathogenic porcine reproductive and respiratory syndrome virus (PRRSV) infection. Int. J. Mol. Sci. 2017, 18, 1475. [Google Scholar] [CrossRef] [Green Version]
- Lim, B.; Kim, S.; Lim, K.-S.; Jeong, C.-G.; Kim, S.-C.; Lee, S.-M.; Park, C.-K.; Te Pas, M.F.; Gho, H.; Kim, T.-H. Integrated time-serial transcriptome networks reveal common innate and tissue-specific adaptive immune responses to PRRSV infection. Vet. Res. 2020, 51, 1–18. [Google Scholar] [CrossRef]
- Robertson, M.J. Role of chemokines in the biology of natural killer cells. J. Leukoc. Biol. 2002, 71, 173–183. [Google Scholar]
- Hensbergen, P.J.; Wijnands, P.G.B.; Schreurs, M.W.; Scheper, R.J.; Willemze, R.; Tensen, C.P. The CXCR3 targeting chemokine CXCL11 has potent antitumor activity in vivo involving attraction of CD8+ T lymphocytes but not inhibition of angiogenesis. J. Immunother. 2005, 28, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chu, Y.; Wang, Y.; Guo, Q.; Xiong, S. Vaccination with IFN-inducible T cell α chemoattractant (ITAC) gene-modified tumor cell attenuates disseminated metastases of circulating tumor cells. Vaccine 2006, 24, 2966–2974. [Google Scholar] [CrossRef]
- Piqueras, B.; Connolly, J.; Freitas, H.; Palucka, A.K.; Banchereau, J. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood 2006, 107, 2613–2618. [Google Scholar] [CrossRef] [PubMed]
- Barry, M.; Bleackley, R.C. Cytotoxic T lymphocytes: All roads lead to death. Nat. Rev. Immunol. 2002, 2, 401–409. [Google Scholar] [CrossRef]
- Wen, K.; Bui, T.; Li, G.; Liu, F.; Li, Y.; Kocher, J.; Yuan, L. Characterization of immune modulating functions of γδ T cell subsets in a gnotobiotic pig model of human rotavirus infection. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 289–301. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, A.; Hühr, J.; Schwaiger, T.; Dorhoi, A.; Mettenleiter, T.C.; Blome, S.; Schröder, C.; Blohm, U. Porcine invariant natural killer T cells: Functional profiling and dynamics in steady state and viral infections. Front. Immunol. 2019, 10, 1380. [Google Scholar] [CrossRef]
- Price, G.E.; Huang, L.; Ou, R.; Zhang, M.; Moskophidis, D. Perforin and Fas cytolytic pathways coordinately shape the selection and diversity of CD8+-T-cell escape variants of influenza virus. J. Virol. 2005, 79, 8545–8559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoenborn, J.R.; Wilson, C.B. Regulation of interferon-γ during innate and adaptive immune responses. Adv. Immunol. 2007, 96, 41–101. [Google Scholar]
- Lederman, S.; Yellin, M.; Krichevsky, A.; Belko, J.; Lee, J.; Chess, L. Identification of a novel surface protein on activated CD4+ T cells that induces contact-dependent B cell differentiation (help). J. Exp. Med. 1992, 175, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Okutani, M.; Tsukahara, T.; Kato, Y.; Fukuta, K.; Inoue, R. Gene expression profiles of CD 4/CD 8 double-positive T cells in porcine peripheral blood. Anim. Sci. J. 2018, 89, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.R.; Rosenthal, K.S.; Pfaller, M.A. Medical Microbiology E-Book; Elsevier Health Sciences: Amsterdam, The Netherlands, 2020. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Lim, B.; Mattoo, S.-u.-S.; Oh, E.-Y.; Jeong, C.-G.; Kim, W.-I.; Lee, K.-T.; Lee, S.-M.; Kim, J.-M. Comprehensive Transcriptomic Comparison between Porcine CD8− and CD8+ Gamma Delta T Cells Revealed Distinct Immune Phenotype. Animals 2021, 11, 2165. https://doi.org/10.3390/ani11082165
Kim S, Lim B, Mattoo S-u-S, Oh E-Y, Jeong C-G, Kim W-I, Lee K-T, Lee S-M, Kim J-M. Comprehensive Transcriptomic Comparison between Porcine CD8− and CD8+ Gamma Delta T Cells Revealed Distinct Immune Phenotype. Animals. 2021; 11(8):2165. https://doi.org/10.3390/ani11082165
Chicago/Turabian StyleKim, Sangwook, Byeonghwi Lim, Sameer-ul-Salam Mattoo, Eun-Young Oh, Chang-Gi Jeong, Won-Il Kim, Kyung-Tai Lee, Sang-Myeong Lee, and Jun-Mo Kim. 2021. "Comprehensive Transcriptomic Comparison between Porcine CD8− and CD8+ Gamma Delta T Cells Revealed Distinct Immune Phenotype" Animals 11, no. 8: 2165. https://doi.org/10.3390/ani11082165
APA StyleKim, S., Lim, B., Mattoo, S.-u.-S., Oh, E.-Y., Jeong, C.-G., Kim, W.-I., Lee, K.-T., Lee, S.-M., & Kim, J.-M. (2021). Comprehensive Transcriptomic Comparison between Porcine CD8− and CD8+ Gamma Delta T Cells Revealed Distinct Immune Phenotype. Animals, 11(8), 2165. https://doi.org/10.3390/ani11082165





