Isothermal Nucleic Acid Amplification Technologies for the Detection of Equine Viral Pathogens
Abstract
:Simple Summary
Abstract
1. Introduction
2. Equine Viral Diseases of Biosecurity Concern
2.1. African Horse Sickness
2.2. Equine Encephalomyelitis (Western)
2.3. Equine Infectious Anaemia
2.4. Equine Influenza
2.5. Equine Viral Arteritis
2.6. Equine Rhinopneumonitis (Caused by EHV-1)
3. Zoonotic Equine Viral Diseases of Concern
3.1. Hendra Virus
3.2. Japanese Encephalitis
3.3. Ross River Virus
3.4. West Nile Virus
4. Current Diagnostic Techniques for Equine Viral Diseases
4.1. Serological Diagnostics
4.2. Molecular Diagnostics
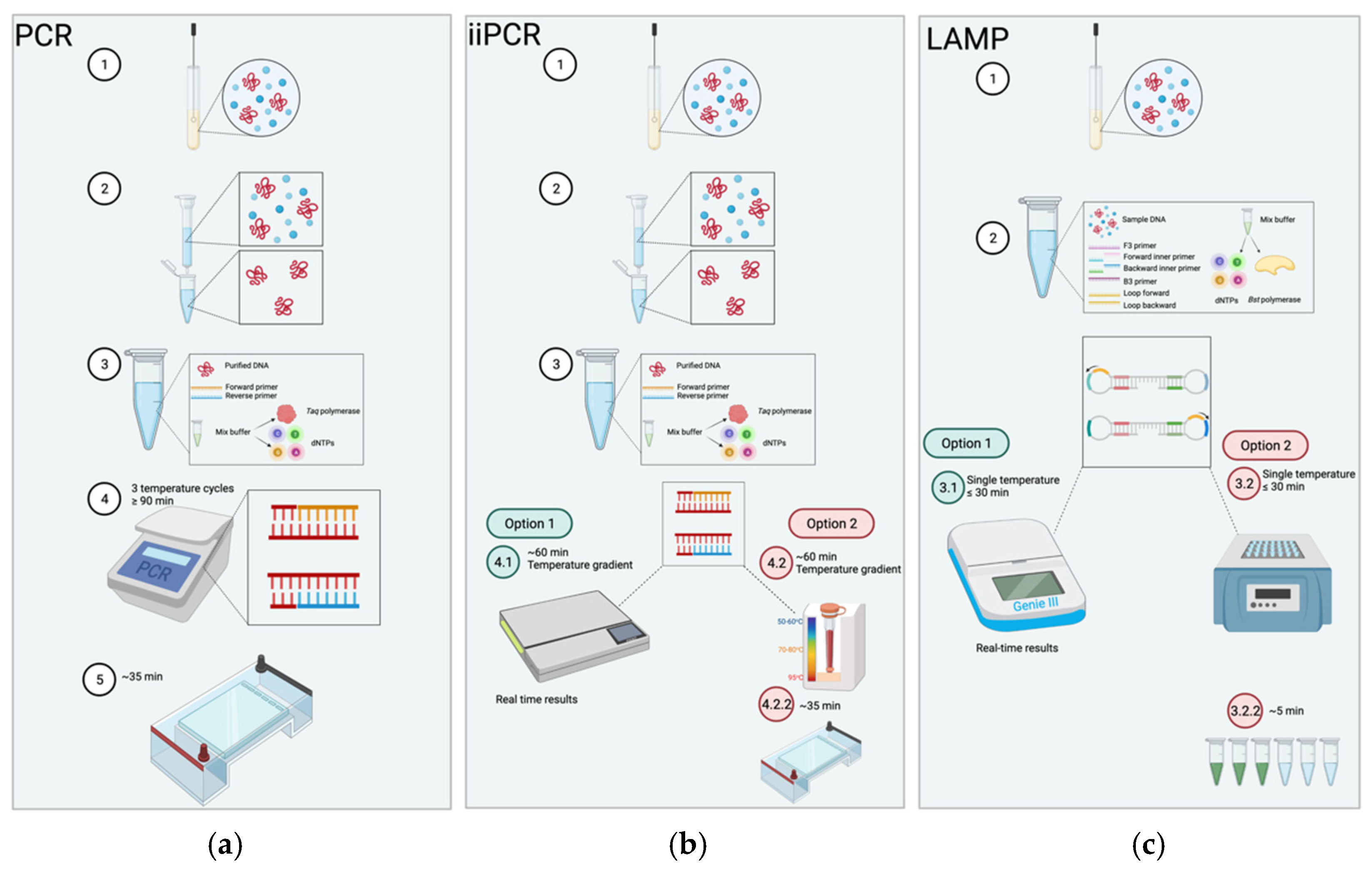
5. Isothermal Techniques
6. Application of LAMP for Equine Viral Diseases
6.1. Principles of LAMP
6.2. Application of LAMP for Equine Viral Diseases
7. Application of iiPCR for Equine Viral Diseases
7.1. Principles of iiPCR
7.2. Applications of iiPCR for Equine Viral Diseases
8. Future Applications of LAMP and iiPCR for Equine Viral Diseases
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Murray, G.; Munstermann, S.; Lam, K. Benefits and Challenges Posed by the Worldwide Expansion of Equestrian Events—New Standards for the Population of Competition Horses and Equine Disease Free Zones (EDFZ) in Countries. In Proceedings of the 81st General Session World Organisation for Animal Health, Paris, France, 26–31 May 2013. [Google Scholar]
- Littiere, T.O.; Castro, G.H.F.; Rodriguez, M.D.P.R.; Bonafé, C.M.; Magalhães, A.F.B.; Faleiros, R.R.; Vieira, J.I.G.; Santos, C.G.; Verardo, L.L. Identification and Functional Annotation of Genes Related to Horses’ Performance: From GWAS to Post-GWAS. Animals 2020, 10, 1173. [Google Scholar] [CrossRef]
- Paillot, R. Special Issue “Equine Viruses”: Old “Friends” and New Foes? Viruses 2020, 12, 153. [Google Scholar] [CrossRef] [Green Version]
- FAOSTAT. Production Statistics of the Food Agriculture Orginization of The United States. Available online: http://www.fao.org/faostat/en/#data/QA (accessed on 3 June 2021).
- Cross, P. Global Horse Statistics Internal 02 2019; HiPoint Agro Bedding Corp: Gulep, ON, Canada, 2019. [Google Scholar]
- Hatcher, F. Equine Industry Scoping Report; Regional Development Australia—Far South Coast Nowra: Nowra, NSW, Australia, 2013.
- Pritchard, J.C.; Lindberg, A.C.; Main, D.C.; Whay, H.R. Assessment of the welfare of working horses, mules and donkeys, using health and behaviour parameters. Prev. Vet. Med. 2005, 69, 265–283. [Google Scholar] [CrossRef]
- McManus, P.; Albrecht, G.; Graham, R. The Global Horseracing Industry: Social, Economic, Environmental and Ethical Perspectives. The Global Horseracing Industry: Social, Economic, Environmental and Ethical Perspectives; Routledge: London, UK, 2012; pp. 1–244. [Google Scholar] [CrossRef]
- IFHA. Annual Report 2018; International Federation of Horseracing Authorities: Boulogne, France, 2018. [Google Scholar]
- Narayan, P.K.; Smyth, R.L. The race that stops the nation: The demand for the Melbourne Cup. Econ. Rec. 2004, 80, 193–207. [Google Scholar] [CrossRef]
- Hardy, G.L.P. Measurement of Economic Impact of Australian Thoroughbred Breeding Industry 18/046; NSW: Wagga Wagga, Australia, 2019.
- Attoui, H.; Mohd Jaafar, F. Zoonotic and emerging orbivirus infections. Rev. Sci. Tech. 2015, 34, 353–361. [Google Scholar] [CrossRef] [Green Version]
- Sack, A.; Oladunni, F.S.; Gonchigoo, B.; Chambers, T.M.; Gray, G.C. Zoonotic Diseases from Horses: A Systematic Review. Vector Borne Zoonotic. Dis. 2020, 20, 484–495. [Google Scholar] [CrossRef]
- Yuen, K.Y.; Bielefeldt-Ohmann, H. Ross River Virus Infection: A Cross-Disciplinary Review with a Veterinary Perspective. Pathogens 2021, 10, 357. [Google Scholar] [CrossRef]
- Smyth, G.B.; Dagley, K.; Tainsh, J. Insights into the economic consequences of the 2007 equine influenza outbreak in Australia. Aust. Vet. J. 2011, 89 (Suppl. 1), 151–158. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.; Brar, B.; Shah, I.; Ranjan, K.; Lambe, U.; Manimegalai, M.; Vashisht, B.; Khurana, S.; Prasad, G. Biotechnological tools for diagnosis of equine infectious diseases. J. Exp. Biol. Agric. Sci. 2016, 4, S161–S181. [Google Scholar] [CrossRef]
- Weese, J.S. Infection control and biosecurity in equine disease control. Equine Vet. J. 2014, 46, 654–660. [Google Scholar] [CrossRef]
- Slovis, N.M.; Browne, N.; Bozorgmanesh, R. Point-of-Care Diagnostics in Equine Practice. Vet. Clin. North. Am. Equine Pract 2020, 36, 161–171. [Google Scholar] [CrossRef]
- Desmettre, P. Diagnosis and prevention of equine infectious diseases: Present status, potential, and challenges for the future. Adv. Vet. Med. 1999, 41, 359–377. [Google Scholar] [CrossRef]
- Zhang, X.; Lowe, S.B.; Gooding, J.J. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP). Biosens. Bioelectron. 2014, 61, 491–499. [Google Scholar] [CrossRef]
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5. [Google Scholar] [CrossRef]
- Nagura-Ikeda, M.; Imai, K.; Tabata, S.; Miyoshi, K.; Murahara, N.; Mizuno, T.; Horiuchi, M.; Kato, K.; Imoto, Y.; Iwata, M.; et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19. J. Clin. Microbiol. 2020, 58, e01438-20. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.-F.G.; Tsai, Y.-L.; Tsai, C.-F.; Lin, C.-K.; Lee, P.-Y.; Teng, P.-H.; Su, C.; Jeng, C.-C. A thermally baffled device for highly stabilized convective PCR. Biotechnol. J. 2012, 7, 662–666. [Google Scholar] [CrossRef] [Green Version]
- Alhassan, A.; Thekisoe, O.M.; Yokoyama, N.; Inoue, N.; Motloang, M.Y.; Mbati, P.A.; Yin, H.; Katayama, Y.; Anzai, T.; Sugimoto, C.; et al. Development of loop-mediated isothermal amplification (LAMP) method for diagnosis of equine piroplasmosis. Vet. Parasitol. 2007, 143, 155–160. [Google Scholar] [CrossRef]
- Foord, A.J.; Middleton, D.; Heine, H.G. Hendra virus detection using Loop-Mediated Isothermal Amplification. J. Virol. Methods 2012, 181, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Bath, C.; Scott, M.; Sharma, P.M.; Gurung, R.B.; Phuentshok, Y.; Pefanis, S.; Colling, A.; Singanallur Balasubramanian, N.; Firestone, S.M.; Ungvanijban, S.; et al. Further development of a reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the detection of foot-and-mouth disease virus and validation in the field with use of an internal positive control. Transbound. Emerg. Dis. 2020, 67, 2494–2506. [Google Scholar] [CrossRef]
- Chua, K.H.; Lee, P.C.; Chai, H.C. Development of insulated isothermal PCR for rapid on-site malaria detection. Malar. J. 2016, 15, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, Y.L.; Wang, H.T.; Chang, H.F.; Tsai, C.F.; Lin, C.K.; Teng, P.H.; Su, C.; Jeng, C.C.; Lee, P.Y. Development of TaqMan probe-based insulated isothermal PCR (iiPCR) for sensitive and specific on-site pathogen detection. PLoS ONE 2012, 7, e45278. [Google Scholar] [CrossRef] [Green Version]
- Schemann, K.; Taylor, M.R.; Toribio, J.A.; Dhand, N.K. Horse owners’ biosecurity practices following the first equine influenza outbreak in Australia. Prev. Vet. Med. 2011, 102, 304–314. [Google Scholar] [CrossRef] [Green Version]
- OIE. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 2018; World Organisation for Animal Health (OIE): Paris, France, 2019. [Google Scholar]
- OIE. Notifable Animal Diseases. Available online: https://www.oie.int/en/what-we-do/animal-health-and-welfare/animal-diseases/ (accessed on 8 June 2021).
- Mellor, P.S.; Hamblin, C. African horse sickness. Vet. Res. 2004, 35, 445–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellor, P.S. African horse sickness: Transmission and epidemiology. Vet. Res. 1993, 24, 199–212. [Google Scholar] [PubMed]
- Sellers, R.F.; Pedgley, D.E.; Tucker, M.R. Possible spread of African horse sickness on the wind. J. Hyg. 1977, 79, 279–298. [Google Scholar] [CrossRef] [Green Version]
- Maclachlan, N.J.; Guthrie, A.J. Re-emergence of bluetongue, African horse sickness, and other orbivirus diseases. Vet. Res. 2010, 41, 35. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Bie, J.; Wang, H.; Zheng, J.; Gao, X.; Xiao, J.; Wang, H. Modelling High.-Risk Areas for African Horse Sickness Occurrence in Mainland China Along Southeast; Authorea: Hobken, NJ, USA, 2020. [Google Scholar]
- King, S.; Rajko-Nenow, P.; Ashby, M.; Frost, L.; Carpenter, S.; Batten, C. Outbreak of African horse sickness in Thailand, 2020. Transbound. Emerg. Dis. 2020, 67, 1764–1767. [Google Scholar] [CrossRef]
- Lu, G.; Pan, J.; Ou, J.; Shao, R.; Hu, X.; Wang, C.; Li, S. African horse sickness: Its emergence in Thailand and potential threat to other Asian countries. Transbound. Emerg. Dis. 2020, 67, 1751–1753. [Google Scholar] [CrossRef]
- Dennis, S.J.; Meyers, A.E.; Hitzeroth, I.I.; Rybicki, E.P. African Horse Sickness: A Review of Current Understanding and Vaccine Development. Viruses 2019, 11, 844. [Google Scholar] [CrossRef] [Green Version]
- Robin, M.; Page, P.; Archer, D.; Baylis, M. African horse sickness: The potential for an outbreak in disease-free regions and current disease control and elimination techniques. Equine. Vet. J. 2016, 48, 659–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forrester, N.L.; Kenney, J.L.; Deardorff, E.; Wang, E.; Weaver, S.C. Western Equine Encephalitis submergence: Lack of evidence for a decline in virus virulence. Virology 2008, 380, 170–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergren, N.A.; Auguste, A.J.; Forrester, N.L.; Negi, S.S.; Braun, W.A.; Weaver, S.C. Western equine encephalitis virus: Evolutionary analysis of a declining alphavirus based on complete genome sequences. J. Virol. 2014, 88, 9260–9267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robb, L.; Hartman, D.; Rice, L.; DeMaria, J.; Borland, E.; Bergren, N.; Kading, R. Continued Evidence of Decline in the Enzootic Activity of Western Equine Encephalitis Virus in Colorado. J. Med Entomol. 2018, 56, 584–588. [Google Scholar] [CrossRef]
- Bergren, N.A.; Haller, S.; Rossi, S.L.; Seymour, R.L.; Huang, J.; Miller, A.L.; Bowen, R.A.; Hartman, D.A.; Brault, A.C.; Weaver, S.C. “Submergence” of Western equine encephalitis virus: Evidence of positive selection argues against genetic drift and fitness reductions. PLoS Pathog 2020, 16, e1008102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calisher, C.H.; Monath, T.P.; Mitchell, C.J.; Sabattini, M.S.; Cropp, C.B.; Kerschner, J.; Hunt, A.R.; Lazuick, J.S. Arbovirus investigations in Argentina, 1977-1980. III. Identification and characterization of viruses isolated, including new subtypes of western and Venezuelan equine encephalitis viruses and four new bunyaviruses (Las Maloyas, Resistencia, Barranqueras, and Antequera). Am. J. Trop Med. Hyg. 1985, 34, 956–965. [Google Scholar]
- Avilés, G.; Sabattini, M.S.; Mitchell, C.J. Transmission of western equine encephalomyelitis virus by Argentine Aedes albifasciatus (Diptera: Culicidae). J. Med. Entomol. 1992, 29, 850–853. [Google Scholar] [CrossRef]
- Stromberg, Z.R.; Fischer, W.; Bradfute, S.B.; Kubicek-Sutherland, J.Z.; Hraber, P. Vaccine Advances against Venezuelan, Eastern, and Western Equine Encephalitis Viruses. Vaccines 2020, 8, 273. [Google Scholar] [CrossRef]
- Anderson, B.A. Focal neurologic signs in western equine encephalitis. Can. Med. Assoc. J. 1984, 130, 1019–1021. [Google Scholar] [PubMed]
- Reed, D.S.; Larsen, T.; Sullivan, L.J.; Lind, C.M.; Lackemeyer, M.G.; Pratt, W.D.; Parker, M.D. Aerosol exposure to western equine encephalitis virus causes fever and encephalitis in cynomolgus macaques. J. Infect. Dis. 2005, 192, 1173–1182. [Google Scholar] [CrossRef]
- Pellegrini-Masini, A.; Livesey, L.C. Meningitis and encephalomyelitis in horses. Vet. Clin. North. Am. Equine. Pract. 2006, 22, 553–589. [Google Scholar] [CrossRef]
- CDC. Arboviral disease—United States, 1994. MMWR Morb Mortal Wkly. Rep. 1995, 44, 641–644. [Google Scholar]
- Bleck, T. Arboviruses Affecting the Central Nervous System. Goldman’s Cecil Medicine, 24th ed.; Elsevier Saunders: Philadelphia, PA, USA, 2011; Volume 2, pp. 2161–2168. [Google Scholar] [CrossRef]
- Lambert, A.J.; Martin, D.A.; Lanciotti, R.S. Detection of North American eastern and western equine encephalitis viruses by nucleic acid amplification assays. J. Clin. Microbiol. 2003, 41, 379–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sellon, D.C. Equine infectious anemia. Vet. Clin. North. Am. Equine. Pract. 1993, 9, 321–336. [Google Scholar] [CrossRef]
- Craigo, J.K.; Montelaro, R.C. Lessons in AIDS vaccine development learned from studies of equine infectious, anemia virus infection and immunity. Viruses 2013, 5, 2963–2976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cursino, A.E.; Vilela, A.P.P.; Franco-Luiz, A.P.M.; de Oliveira, J.G.; Nogueira, M.F.; Júnior, J.P.A.; de Aguiar, D.M.; Kroon, E.G. Equine infectious anemia virus in naturally infected horses from the Brazilian Pantanal. Arch. Virol. 2018, 163, 2385–2394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lupulovic, D.; Savić, S.; Gaudaire, D.; Berthet, N.; Grgić, Ž.; Matović, K.; Deshiere, A.; Hans, A. Identification and genetic characterization of equine infectious anemia virus in Western Balkans. BMC Vet. Res. 2021, 17, 168. [Google Scholar] [CrossRef]
- Cook, R.F.; Leroux, C.; Issel, C.J. Equine infectious anemia and equine infectious anemia virus in 2013: A review. Vet. Microbiol. 2013, 167, 181–204. [Google Scholar] [CrossRef]
- Cruz, F.; Fores, P.; Ireland, J.; Moreno, M.A.; Newton, R. Freedom from equine infectious anaemia virus infection in Spanish Purebred horses. Vet. Rec. Open 2015, 2, e000074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.-N.; Rao, D.; Fu, X.-Q.; Hu, M.-M.; Dong, J.-G. Equine infectious anemia virus in China. Oncotarget 2017, 9, 1356–1364. [Google Scholar] [CrossRef] [Green Version]
- Issel, C.J.; Coggins, L. Equine infectious anemia: Current knowledge. J. Am. Vet. Med. Assoc. 1979, 174, 727–733. [Google Scholar]
- Bolfa, P.; Barbuceanu, F.; Leau, S.E.; Leroux, C. Equine infectious anaemia in Europe: Time to re-examine the efficacy of monitoring and control protocols? Equine Vet. J. 2016, 48, 140–142. [Google Scholar] [CrossRef] [Green Version]
- Espasandin, A.G.; Cipolini, M.F.; Forletti, A.; Díaz, S.; Soto, J.; Martínez, D.E.; Storani, C.A.; Monzón, N.M.; Beltrame, J.I.; Lucchesi, E.; et al. Comparison of serological techniques for the diagnosis of equine infectious Anemia in an endemic area of Argentina. J. Virol. Methods 2021, 291, 114101. [Google Scholar] [CrossRef]
- McConnico, R.S.; Issel, C.J.; Cook, S.J.; Cook, R.F.; Floyd, C.; Bisson, H. Predictive methods to define infection with equine infectious anemia virus in foals out of reactor mares. J. Equine Vet. Sci. 2000, 20, 387–392. [Google Scholar] [CrossRef]
- Sack, A.; Cullinane, A.; Daramragchaa, U.; Chuluunbaatar, M.; Gonchigoo, B.; Gray, G.C. Equine Influenza Virus—A Neglected, Reemergent Disease Threat. Emerg. Infect. Dis. 2019, 25, 1185–1191. [Google Scholar] [CrossRef] [Green Version]
- van Maanen, C.; Cullinane, A. Equine influenza virus infections: An update. Vet. Q 2002, 24, 79–94. [Google Scholar] [CrossRef]
- Taylor, M.R.; Agho, K.E.; Stevens, G.J.; Raphael, B. Factors influencing psychological distress during a disease epidemic: Data from Australia’s first outbreak of equine influenza. BMC Public Health 2008, 8, 347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowled, B.; Ward, M.P.; Hamilton, S.; Garner, G. The equine influenza epidemic in Australia: Spatial and temporal descriptive analyses of a large propagating epidemic. Prev. Vet. Med. 2009, 92, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Elton, D.; Bryant, N. Facing the threat of equine influenza. Equine Vet. J. 2011, 43, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Mena, J.; Brito, B.; Moreira, R.; Tadich, T.; González, I.; Cruces, J.; Ortega, R.; van Bakel, H.; Rathnasinghe, R.; Pizarro-Lucero, J.; et al. Reemergence of H3N8 Equine Influenza A virus in Chile, 2018. Transbound. Emerg. Dis. 2018, 65, 1408–1415. [Google Scholar] [CrossRef]
- Cullinane, A.; Elton, D.; Mumford, J. Equine influenza—Surveillance and control. Influenza Other Respir. Viruses 2010, 4, 339–344. [Google Scholar] [CrossRef] [Green Version]
- Balasuriya, U.B.; Carossino, M. Reproductive effects of arteriviruses: Equine arteritis virus and porcine reproductive and respiratory syndrome virus infections. Curr. Opin. Virol. 2017, 27, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Del Piero, F. Equine viral arteritis. Vet. Pathol. 2000, 37, 287–296. [Google Scholar] [CrossRef]
- Timoney, P.J.; McCollum, W.H. Equine viral arteritis—Epidemiology and control. J. Equine Vet. Sci. 1988, 8, 54–59. [Google Scholar] [CrossRef]
- Timoney, P.J.; McCollum, W.H. Equine viral arteritis. Vet. Clin. North. Am. Equine Pract. 1993, 9, 295–309. [Google Scholar] [CrossRef]
- Balasuriya, U.B.R.; Carossino, M.; Timoney, P.J. Equine viral arteritis: A respiratory and reproductive disease of significant economic importance to the equine industry. Equine Vet. Educ. 2018, 30, 497–512. [Google Scholar] [CrossRef]
- Ruiz-Saenz, J. Equine Viral Arteritis: Epidemiological and intervention perspectives. Revista Colombiana Ciencias Pecuarias 2010, 23, 501. [Google Scholar]
- Pronost, S.; Pitel, P.H.; Miszczak, F.; Legrand, L.; Marcillaud-Pitel, C.; Hamon, M.; Tapprest, J.; Balasuriya, U.B.; Freymuth, F.; Fortier, G. Description of the first recorded major occurrence of equine viral arteritis in France. Equine Vet. J. 2010, 42, 713–720. [Google Scholar] [CrossRef]
- Crabb, B.S.; Studdert, M.J. Equine herpesviruses 4 (equine rhinopneumonitis virus) and 1 (equine abortion virus). Adv. Virus Res. 1995, 45, 153–190. [Google Scholar] [CrossRef] [PubMed]
- Dunowska, M. A review of equid herpesvirus 1 for the veterinary practitioner. Part B: Pathogenesis and epidemiology. N. Z. Vet. J. 2014, 62, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Balasuriya, U.B.; Crossley, B.M.; Timoney, P.J. A review of traditional and contemporary assays for direct and indirect detection of Equid herpesvirus 1 in clinical samples. J. Vet. Diagn Investig. 2015, 27, 673–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzpatrick, D.R.; Studdert, M.J. Immunologic relationships between equine herpesvirus type 1 (equine abortion virus) and type 4 (equine rhinopneumonitis virus). Am. J. Vet. Res. 1984, 45, 1947–1952. [Google Scholar]
- Patel, J.R.; Heldens, J. Equine herpesviruses 1 (EHV-1) and 4 (EHV-4)—Epidemiology, disease and immunoprophylaxis: A brief review. Vet. J. 2005, 170, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Lunn, D.P.; Davis-Poynter, N.; Flaminio, M.J.; Horohov, D.W.; Osterrieder, K.; Pusterla, N.; Townsend, H.G. Equine herpesvirus-1 consensus statement. J. Vet. Intern. Med. 2009, 23, 450–461. [Google Scholar] [CrossRef]
- Ma, G.; Azab, W.; Osterrieder, N. Equine herpesviruses type 1 (EHV-1) and 4 (EHV-4)—Masters of co-evolution and a constant threat to equids and beyond. Vet. Microbiol. 2013, 167, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Kydd, J.H.; Townsend, H.G.; Hannant, D. The equine immune response to equine herpesvirus-1: The virus and its vaccines. Vet. Immunol. Immunopathol. 2006, 111, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.G. Viral respiratory disease of the horse. Vet. Clin. N. Am. Equine Pract 1991, 7, 27–52. [Google Scholar] [CrossRef]
- Reed, S.M.; Toribio, R.E. Equine herpesvirus 1 and 4. Vet. Clin. N. Am. Equine Pract. 2004, 20, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Oladunni, F.S.; Horohov, D.W.; Chambers, T.M. EHV-1: A Constant Threat to the Horse Industry. Front. Microbiol. 2019, 10, 140–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Field, H.; de Jong, C.; Melville, D.; Smith, C.; Smith, I.; Broos, A.; Kung, Y.H.N.; McLaughlin, A.; Zeddeman, A. Hendra virus infection dynamics in Australian fruit bats. PLoS ONE 2011, 6, e28678. [Google Scholar] [CrossRef]
- Khusro, A.; Aarti, C.; Pliego, A.B.; Cipriano-Salazar, M. Hendra Virus Infection in Horses: A Review on Emerging Mystery Paramyxovirus. J. Equine Vet. Sci. 2020, 91, 103149. [Google Scholar] [CrossRef]
- Mahalingam, S.; Herrero, L.J.; Playford, E.G.; Spann, K.; Herring, B.; Rolph, M.S.; Middleton, D.; McCall, B.; Field, H.; Wang, L.F. Hendra virus: An emerging paramyxovirus in Australia. Lancet Infect. Dis. 2012, 12, 799–807. [Google Scholar] [CrossRef]
- Playford, G.; McCall, B.; Smith, G.; Slinko, V.; Allen, G.; Smith, I.; Moore, F.; Taylor, C.; Kung, N.; Field, H. Human Hendra Virus Encephalitis Associated with Equine Outbreak, Australia, 2008. Emerg Infect. Dis. 2010, 16, 219–223. [Google Scholar] [CrossRef]
- Mendez, D.H.; Judd, J.; Speare, R. Unexpected result of Hendra virus outbreaks for veterinarians, Queensland, Australia. Emerg Infect. Dis. 2012, 18, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Hii, C.; Dhand, N.K.; Toribio, J.-A.L.M.L.; Taylor, M.R.; Wiethoelter, A.; Schembri, N.; Sawford, K.; Kung, N.; Moloney, B.; Wright, T.; et al. Information delivery and the veterinarian-horse owner relationship in the context of Hendra virus in Australia. Prev. Vet. Med. 2020, 179, 104988. [Google Scholar] [CrossRef] [PubMed]
- Whitley, R.J.; Gnann, J.W. Viral encephalitis: Familiar infections and emerging pathogens. Lancet 2002, 359, 507–513. [Google Scholar] [CrossRef]
- Mackenzie, J.S.; Johansen, C.A.; Ritchie, S.A.; van den Hurk, A.F.; Hall, R.A. Japanese encephalitis as an emerging virus: The emergence and spread of Japanese encephalitis virus in Australasia. Curr. Top. Microbiol. Immunol. 2002, 267, 49–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, K.; Ellis, T.; Williams, D.; Lunt, R.; Daniels, P.; Watkins, K.; Riggs, C. Japanese encephalitis in a racing Thoroughbred gelding in Hong Kong. Vet. Rec. 2005, 157, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, K.L.; Hernández-Triana, L.M.; Banyard, A.C.; Fooks, A.R.; Johnson, N. Japanese encephalitis virus infection, diagnosis and control in domestic animals. Vet. Microbiol. 2017, 201, 85–92. [Google Scholar] [CrossRef]
- van den Hurk, A.F.; Ritchie, S.A.; Mackenzie, J.S. Ecology and geographical expansion of Japanese encephalitis virus. Annu. Rev. Entomol. 2009, 54, 17–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, R.C. Ross River virus: Ecology and distribution. Annu. Rev. Entomol. 2002, 47, 1–31. [Google Scholar] [CrossRef]
- Johnson, B.J.; Robbins, A.; Gyawali, N.; Ong, O.; Loader, J.; Murphy, A.K.; Hanger, J.; Devine, G.J. The environmental and ecological determinants of elevated Ross River Virus exposure in koalas residing in urban coastal landscapes. Sci. Rep. 2021, 11, 4419. [Google Scholar] [CrossRef]
- Liu, X.; Mutso, M.; Cherkashchenko, L.; Zusinaite, E.; Herrero, L.J.; Doggett, S.L.; Haniotis, J.; Merits, A.; Herring, B.L.; Taylor, A.; et al. Identification of Natural Molecular Determinants of Ross River Virus Type I Interferon Modulation. J. Virol. 2020, 94, e01788-01719. [Google Scholar] [CrossRef] [PubMed]
- Hall, N.L.; Barnes, S.; Canuto, C.; Nona, F.; Redmond, A.M. Climate change and infectious diseases in Australia’s Torres Strait Islands. Aust. N. Z. J. Public Health 2021, 45, 122–128. [Google Scholar] [CrossRef]
- Azuolas, J.K. Ross River Virus Disease of Horses. Aust. Equine Vet. 1988, 16, 3. [Google Scholar]
- El-Hage, C.M.; Bamford, N.J.; Gilkerson, J.R.; Lynch, S.E. Ross River Virus Infection of Horses: Appraisal of Ecological and Clinical Consequences. J. Equine Vet. Sci. 2020, 93, 103143. [Google Scholar] [CrossRef]
- Chapman, G.E.; Baylis, M.; Archer, D.; Daly, J.M. The challenges posed by equine arboviruses. Equine Vet. J. 2018, 50, 436–445. [Google Scholar] [CrossRef]
- Harley, D.; Sleigh, A.; Ritchie, S. Ross River virus transmission, infection, and disease: A cross-disciplinary review. Clin. Microbiol. Rev. 2001, 14, 909–932. [Google Scholar] [CrossRef] [Green Version]
- Campbell, G.L.; Marfin, A.A.; Lanciotti, R.S.; Gubler, D.J. West Nile virus. Lancet Infect. Dis. 2002, 2, 519–529. [Google Scholar] [CrossRef]
- Petersen, L.R.; Brault, A.C.; Nasci, R.S. West Nile virus: Review of the literature. JAMA 2013, 310, 308–315. [Google Scholar] [CrossRef]
- Hayes, E.B.; Komar, N.; Nasci, R.S.; Montgomery, S.P.; O’Leary, D.R.; Campbell, G.L. Epidemiology and transmission dynamics of West Nile virus disease. Emerg Infect. Dis. 2005, 11, 1167–1173. [Google Scholar] [CrossRef]
- Sharifi, Z.; Mahmoodian Shooshtari, M.; Talebian, A. A study of West Nile virus infection in Iranian blood donors. Arch. Iran. Med. 2010, 13, 1–4. [Google Scholar]
- Zeller, H.G.; Schuffenecker, I. West Nile virus: An overview of its spread in Europe and the Mediterranean basin in contrast to its spread in the Americas. Eur. J. Clin. Microbiol. Infect. Dis. 2004, 23, 147–156. [Google Scholar] [CrossRef]
- Cantile, C.; Di Guardo, G.; Eleni, C.; Arispici, M. Clinical and neuropathological features of West Nile virus equine encephalomyelitis in Italy. Equine Vet. J. 2000, 32, 31–35. [Google Scholar] [CrossRef]
- Bertram, F.M.; Thompson, P.N.; Venter, M. Epidemiology and Clinical Presentation of West Nile Virus Infection in Horses in South Africa, 2016–2017. Pathogens 2020, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Salazar, P.; Morley, P.; Wilmot, D.; Steffen, D.; Cunningham, W.; Salman, M. Outcome of equids with clinical signs of West Nile virus infection and factors associated with death. J. Am. Vet. Med Assoc. 2004, 225, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Bosco-Lauth, A.M.; Bowen, R.A. West Nile Virus: Veterinary Health and Vaccine Development. J. Med. Entomol. 2019, 56, 1463–1466. [Google Scholar] [CrossRef]
- Ulbert, S. West Nile virus vaccines—Current situation and future directions. Hum. Vaccin Immunother. 2019, 15, 2337–2342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gubler, D.J.; Campbell, G.L.; Nasci, R.; Komar, N.; Petersen, L.; Roehrig, J.T. West Nile virus in the United States: Guidelines for detection, prevention, and control. Viral. Immunol. 2000, 13, 469–475. [Google Scholar] [CrossRef]
- Kramer, L.D.; Li, J.; Shi, P.-Y. West Nile virus. Lancet Neurol. 2007, 6, 171–181. [Google Scholar] [CrossRef]
- Sambri, V.; Capobianchi, M.R.; Cavrini, F.; Charrel, R.; Donoso-Mantke, O.; Escadafal, C.; Franco, L.; Gaibani, P.; Gould, E.A.; Niedrig, M.; et al. Diagnosis of west nile virus human infections: Overview and proposal of diagnostic protocols considering the results of external quality assessment studies. Viruses 2013, 5, 2329–2348. [Google Scholar] [CrossRef] [Green Version]
- Rizzoli, A.; Jimenez-Clavero, M.A.; Barzon, L.; Cordioli, P.; Figuerola, J.; Koraka, P.; Martina, B.; Moreno, A.; Nowotny, N.; Pardigon, N.; et al. The challenge of West Nile virus in Europe: Knowledge gaps and research priorities. Eurosurveillance 2015, 20. [Google Scholar] [CrossRef] [Green Version]
- Eidson, M.; Komar, N.; Sorhage, F.; Nelson, R.; Talbot, T.; Mostashari, F.; McLean, R.; West Nile Virus Avian Mortality Surveillance, G. Crow deaths as a sentinel surveillance system for West Nile virus in the northeastern United States, 1999. Emerg Infect. Dis. 2001, 7, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.D.; Meece, J.K.; Henkel, J.S.; Shukla, S.K. Birds, migration and emerging zoonoses: West nile virus, lyme disease, influenza A and enteropathogens. Clin. Med. Res. 2003, 1, 5–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvey, L.A.; Donnelly, J.A.; Lindsay, M.D.; PottumarthyBoddu, S.; D’Abrera, V.C.; Smith, D.W. Ross River virus infection surveillance in the Greater Perth Metropolitan area--has there been an increase in cases in the winter months? Commun. Dis. Intell. Q. Rep. 2014, 38, E114–E122. [Google Scholar] [PubMed]
- Kinsley, R.; Scott, S.D.; Daly, J.M. Controlling equine influenza: Traditional to next generation serological assays. Vet. Microbiol. 2016, 187, 15–20. [Google Scholar] [CrossRef]
- Zimmerman, K.L.; Crisman, M.V. Diagnostic equine serology. Vet. Clin. N. Am. Equine Pract. 2008, 24, 311–334. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, K.; Prasad, M.; Prasad, G. Application of Molecular and Serological Diagnostics in Veterinary Parasitology. J. Adv. Parasitol. 2016, 2, 80–99. [Google Scholar] [CrossRef]
- Cappelli, K.; Capomaccio, S.; Cook, F.R.; Felicetti, M.; Marenzoni, M.L.; Coppola, G.; Verini-Supplizi, A.; Coletti, M.; Passamonti, F. Molecular detection, epidemiology, and genetic characterization of novel European field isolates of equine infectious anemia virus. J. Clin. Microbiol. 2011, 49, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Jaffer, O.; Abdishakur, F.; Hakimuddin, F.; Riya, A.; Wernery, U.; Schuster, R.K. A comparative study of serological tests and PCR for the diagnosis of equine piroplasmosis. Parasitol. Res. 2010, 106, 709–713. [Google Scholar] [CrossRef]
- Mott, J.; Rikihisa, Y.; Zhang, Y.; Reed, S.M.; Yu, C.Y. Comparison of PCR and culture to the indirect fluorescent-antibody test for diagnosis of Potomac horse fever. J. Clin. Microbiol. 1997, 35, 2215–2219. [Google Scholar] [CrossRef] [Green Version]
- Varrasso, A.; Dynon, K.; Ficorilli, N.; Hartley, C.A.; Studdert, M.J.; Drummer, H.E. Identification of equine herpesviruses 1 and 4 by polymerase chain reaction. Aust. Vet. J. 2001, 79, 563–569. [Google Scholar] [CrossRef]
- Sachse, K. Specificity and performance of PCR detection assays for microbial pathogens. Mol. Biotechnol. 2004, 26, 61–80. [Google Scholar] [CrossRef]
- Wilson, I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997, 63, 3741–3751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, N.D.; Martin, L.J.; Simpson, R.B.; Wallis, D.E.; Neibergs, H.L. Comparison of polymerase chain reaction and microbiological culture for detection of salmonellae in equine feces and environmental samples. Am. J. Vet. Res. 1996, 57, 780–786. [Google Scholar] [PubMed]
- Furr, M.; MacKay, R.; Granstrom, D.; Schott, H., 2nd; Andrews, F. Clinical diagnosis of equine protozoal myeloencephalitis (EPM). J. Vet. Intern. Med. 2002, 16, 618–621. [Google Scholar] [CrossRef]
- Paxson, J. Evaluating polymerase chain reaction-based tests for infectious pathogens. Compend. Equine 2008, 3, 308–320. [Google Scholar]
- Pusterla, N.; Madigan, J.E.; Leutenegger, C.M. Real-time polymerase chain reaction: A novel molecular diagnostic tool for equine infectious diseases. J. Vet. Intern. Med. 2006, 20, 3–12. [Google Scholar] [CrossRef]
- Diallo, I.S.; Hewitson, G.; Wright, L.; Rodwell, B.J.; Corney, B.G. Detection of equine herpesvirus type 1 using a real-time polymerase chain reaction. J. Virol. Methods 2006, 131, 92–98. [Google Scholar] [CrossRef]
- Kim, C.M.; Blanco, L.B.; Alhassan, A.; Iseki, H.; Yokoyama, N.; Xuan, X.; Igarashi, I. Diagnostic real-time PCR assay for the quantitative detection of Theileria equi from equine blood samples. Vet. Parasitol. 2008, 151, 158–163. [Google Scholar] [CrossRef]
- Fernández-Pinero, J.; Fernández-Pacheco, P.; Rodríguez, B.; Sotelo, E.; Robles, A.; Arias, M.; Sánchez-Vizcaíno, J.M. Rapid and sensitive detection of African horse sickness virus by real-time PCR. Res. Vet. Sci. 2009, 86, 353–358. [Google Scholar] [CrossRef]
- Aeschbacher, S.; Santschi, E.; Gerber, V.; Stalder, H.P.; Zanoni, R.G. Development of a real-time RT-PCR for detection of equine influenza virus. Schweiz Arch. Tierheilkd 2015, 157, 191–201. [Google Scholar] [CrossRef]
- Zanoli, L.M.; Spoto, G. Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors 2012, 3, 18–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bodulev, O.L.; Sakharov, I.Y. Isothermal Nucleic Acid Amplification Techniques and Their Use in Bioanalysis. Biochemistry 2020, 85, 147–166. [Google Scholar] [CrossRef] [PubMed]
- Obande, G.A.; Banga Singh, K.K. Current and Future Perspectives on Isothermal Nucleic Acid Amplification Technologies for Diagnosing Infections. Infect. Drug Resist. 2020, 13, 455–483. [Google Scholar] [CrossRef] [Green Version]
- Vincent, M.; Xu, Y.; Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004, 5, 795–800. [Google Scholar] [CrossRef]
- Balasuriya, U.B.R.; Lee, P.-Y.A.; Tiwari, A.; Skillman, A.; Nam, B.; Chambers, T.M.; Tsai, Y.-L.; Ma, L.-J.; Yang, P.-C.; Chang, H.-F.G.; et al. Rapid detection of equine influenza virus H3N8 subtype by insulated isothermal RT-PCR (iiRT-PCR) assay using the POCKIT™ Nucleic Acid Analyzer. J. Virol. Methods 2014, 207, 66–72. [Google Scholar] [CrossRef]
- Parida, M.; Posadas, G.; Inoue, S.; Hasebe, F.; Morita, K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 2004, 42, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Dean, F.B.; Hosono, S.; Fang, L.; Wu, X.; Faruqi, A.F.; Bray-Ward, P.; Sun, Z.; Zong, Q.; Du, Y.; Du, J.; et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA 2002, 99, 5261–5266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Compton, J. Nucleic acid sequence-based amplification. Nature 1991, 350, 91–92. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.Q. Rolling replication of short DNA circles. Proc. Natl. Acad. Sci. USA 1995, 92, 4641–4645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.T.; Fraiser, M.S.; Schram, J.L.; Little, M.C.; Nadeau, J.G.; Malinowski, D.P. Strand displacement amplification--an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992, 20, 1691–1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carossino, M.; Lee, P.Y.; Nam, B.; Skillman, A.; Shuck, K.M.; Timoney, P.J.; Tsai, Y.L.; Ma, L.J.; Chang, H.F.; Wang, H.T.; et al. Development and evaluation of a reverse transcription-insulated isothermal polymerase chain reaction (RT-iiPCR) assay for detection of equine arteritis virus in equine semen and tissue samples using the POCKIT™ system. J. Virol. Methods 2016, 234, 7–15. [Google Scholar] [CrossRef]
- Nagamine, K.; Watanabe, K.; Ohtsuka, K.; Hase, T.; Notomi, T. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin. Chem. 2001, 47, 1742–1743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, Y.; Notomi, T. Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009, 15, 62–69. [Google Scholar] [CrossRef]
- Parida, M.; Sannarangaiah, S.; Dash, P.K.; Rao, P.V.; Morita, K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008, 18, 407–421. [Google Scholar] [CrossRef]
- Wheeler, S.S.; Ball, C.S.; Langevin, S.A.; Fang, Y.; Coffey, L.L.; Meagher, R.J. Surveillance for Western Equine Encephalitis, St. Louis Encephalitis, and West Nile Viruses Using Reverse Transcription Loop-Mediated Isothermal Amplification. PLoS ONE 2016, 11, e0147962. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Pardee, K.; Pena, L. Loop-Mediated Isothermal Amplification (LAMP) for the Diagnosis of Zika Virus: A Review. Viruses 2019, 12, 19. [Google Scholar] [CrossRef] [Green Version]
- Dukes, J.P.; King, D.P.; Alexandersen, S. Novel reverse transcription loop-mediated isothermal amplification for rapid detection of foot-and-mouth disease virus. Arch. Virol. 2006, 151, 1093–1106. [Google Scholar] [CrossRef]
- Han, Q.; Zhang, S.; Liu, D.; Yan, F.; Wang, H.; Huang, P.; Bi, J.; Jin, H.; Feng, N.; Cao, Z.; et al. Development of a Visible Reverse Transcription-Loop-Mediated Isothermal Amplification Assay for the Detection of Rift Valley Fever Virus. Front. Microbiol. 2020, 11, 590732. [Google Scholar] [CrossRef] [PubMed]
- Fowler, V.L.; Howson, E.L.A.; Flannery, J.; Romito, M.; Lubisi, A.; Agüero, M.; Mertens, P.; Batten, C.A.; Warren, H.R.; Castillo-Olivares, J. Development of a Novel Reverse Transcription Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of African Horse Sickness Virus. Transbound. Emerg. Dis. 2017, 64, 1579–1588. [Google Scholar] [CrossRef] [Green Version]
- Nemoto, M.; Tsujimura, K.; Yamanaka, T.; Kondo, T.; Matsumura, T. Loop-mediated isothermal amplification assays for detection of Equid herpesvirus 1 and 4 and differentiating a gene-deleted candidate vaccine strain from wild-type Equid herpesvirus 1 strains. J. Vet. Diagn Investig. 2010, 22, 30–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Zhang, H.; Tang, Q.; Liu, T.C.; Xu, J.; Li, L.R.; Yu, G.; Wang, Y.; Yoa, X.P.; Yang, Z.X. Preliminary study of LAMP method for the detection of equine infectious anemia virus. In Proceedings of the 2017 2nd International Conference on Biomedical and Biological Engineering (BBE2017), Guilin, China, 26–28 May 2017; pp. 492–499. [Google Scholar]
- Nemoto, M.; Yamanaka, T.; Bannai, H.; Tsujimura, K.; Kondo, T.; Matsumura, T. Development and evaluation of a reverse transcription loop-mediated isothermal amplification assay for H3N8 equine influenza virus. J. Virol. Methods 2011, 178, 239–242. [Google Scholar] [CrossRef]
- Nemoto, M.; Yamanaka, T.; Bannai, H.; Tsujimura, K.; Kondo, T.; Matsumura, T. Development of a reverse transcription loop-mediated isothermal amplification assay for H7N7 equine influenza virus. J. Vet. Med. Sci. 2012, 74, 929–931. [Google Scholar] [CrossRef] [Green Version]
- Nemoto, M.; Morita, Y.; Niwa, H.; Bannai, H.; Tsujimura, K.; Yamanaka, T.; Kondo, T. Rapid detection of equine coronavirus by reverse transcription loop-mediated isothermal amplification. J. Virol. Methods 2015, 215-216, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Vissani, M.A.; Becerra, M.L.; Olguín Perglione, C.; Tordoya, M.S.; Miño, S.; Barrandeguy, M. Neuropathogenic and non-neuropathogenic genotypes of Equid Herpesvirus type 1 in Argentina. Vet. Microbiol. 2009, 139, 361–364. [Google Scholar] [CrossRef]
- Waters, R.A.; Fowler, V.L.; Armson, B.; Nelson, N.; Gloster, J.; Paton, D.J.; King, D.P. Preliminary validation of direct detection of foot-and-mouth disease virus within clinical samples using reverse transcription loop-mediated isothermal amplification coupled with a simple lateral flow device for detection. PLoS ONE 2014, 9, e105630. [Google Scholar] [CrossRef] [PubMed]
- Howson, E.L.A.; Armson, B.; Madi, M.; Kasanga, C.J.; Kandusi, S.; Sallu, R.; Chepkwony, E.; Siddle, A.; Martin, P.; Wood, J.; et al. Evaluation of Two Lyophilized Molecular Assays to Rapidly Detect Foot-and-Mouth Disease Virus Directly from Clinical Samples in Field Settings. Transbound. Emerg. Dis. 2017, 64, 861–871. [Google Scholar] [CrossRef]
- Brault, A.C.; Fang, Y.; Reisen, W.K. Multiplex qRT-PCR for the Detection of Western Equine Encephalomyelitis, St. Louis Encephalitis, and West Nile Viral RNA in Mosquito Pools (Diptera: Culicidae). J. Med. Entomol. 2015, 52, 491–499. [Google Scholar] [CrossRef]
- Tsai, Y.L.; Lin, Y.C.; Chou, P.H.; Teng, P.H.; Lee, P.Y. Detection of white spot syndrome virus by polymerase chain reaction performed under insulated isothermal conditions. J. Virol. Methods 2012, 181, 134–137. [Google Scholar] [CrossRef]
- Wilkes, R.P.; Anis, E.; Dunbar, D.; Lee, P.A.; Tsai, Y.L.; Lee, F.C.; Chang, H.G.; Wang, H.T.; Graham, E.M. Rapid and sensitive insulated isothermal PCR for point-of-need feline leukaemia virus detection. J. Feline Med. Surg. 2018, 20, 362–369. [Google Scholar] [CrossRef] [Green Version]
- Vissani, M.A.; Tordoya, M.S.; Tsai, Y.L.; Lee, P.Y.A.; Shen, Y.H.; Lee, F.C.; Wang, H.T.T.; Parreño, V.; Barrandeguy, M. On-site detection of equid alphaherpesvirus 3 in perineal and genital swabs of mares and stallions. J. Virol. Methods 2018, 257, 29–32. [Google Scholar] [CrossRef]
- Balasuriya, U.B.; Lee, P.A.; Tsai, Y.L.; Tsai, C.F.; Shen, Y.H.; Chang, H.G.; Skillman, A.; Wang, H.T.; Pronost, S.; Zhang, Y. Translation of a laboratory-validated equine herpesvirus-1 specific real-time PCR assay into an insulated isothermal polymerase chain reaction (iiPCR) assay for point-of-need diagnosis using POCKIT™ nucleic acid analyzer. J. Virol. Methods 2017, 241, 58–63. [Google Scholar] [CrossRef]
- Cook, R.F.; Barrandeguy, M.; Lee, P.A.; Tsai, C.F.; Shen, Y.H.; Tsai, Y.L.; Chang, H.G.; Wang, H.T.; Balasuriya, U.B.R. Rapid detection of equine infectious anaemia virus nucleic acid by insulated isothermal RT-PCR assay to aid diagnosis under field conditions. Equine Vet. J. 2019, 51, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Balasuriya, U.B.R.; Leutenegger, C.M.; Topol, J.B.; McCollum, W.H.; Timoney, P.J.; MacLachlan, N.J. Detection of equine arteritis virus by real-time TaqMan® reverse transcription-PCR assay. J. Virol. Methods 2002, 101, 21–28. [Google Scholar] [CrossRef]
- Tomlinson, J. In-field diagnostics using loop-mediated isothermal amplification. Methods Mol. Biol. 2013, 938, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Schar, D.; Padungtod, P.; Tung, N.; O’Leary, M.; Kalpravidh, W.; Claes, F. New frontiers in applied veterinary point-of-capture diagnostics: Toward early detection and control of zoonotic influenza. Influenza Other Respir. Viruses 2019, 13, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Moehling, T.J.; Choi, G.; Dugan, L.C.; Salit, M.; Meagher, R.J. LAMP Diagnostics at the Point-of-Care: Emerging Trends and Perspectives for the Developer Community. Expert Rev. Mol. Diagn 2021, 21, 43–61. [Google Scholar] [CrossRef]
- Minka, N.S.; Ayo, J.O. Effects of loading behaviour and road transport stress on traumatic injuries in cattle transported by road during the hot-dry season. Livestock Sci. 2007, 107, 91–95. [Google Scholar] [CrossRef]
- Nayak, S.; Blumenfeld, N.R.; Laksanasopin, T.; Sia, S.K. Point-of-Care Diagnostics: Recent Developments in a Connected Age. Anal. Chem. 2017, 89, 102–123. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, N.; Singh, N.P.; Mishra, A.; Rajneesh, R.; Jamwal, S. A detailed review of transportation stress in livestock and its mitigation techniques. Int. J. Livest. Res. 2021, 1, 30–41. [Google Scholar] [CrossRef]
- Vashist, S.K.; Luppa, P.B.; Yeo, L.Y.; Ozcan, A.; Luong, J.H.T. Emerging Technologies for Next-Generation Point-of-Care Testing. Trends Biotechnol. 2015, 33, 692–705. [Google Scholar] [CrossRef]
- St John, A.; Price, C.P. Existing and Emerging Technologies for Point-of-Care Testing. Clin. Biochem. Rev. 2014, 35, 155–167. [Google Scholar]
- Kemleu, S.; Guelig, D.; Eboumbou Moukoko, C.; Essangui, E.; Diesburg, S.; Mouliom, A.; Melingui, B.; Manga, J.; Donkeu, C.; Epote, A.; et al. A Field-Tailored Reverse Transcription Loop-Mediated Isothermal Assay for High Sensitivity Detection of Plasmodium falciparum Infections. PLoS ONE 2016, 11, e0165506. [Google Scholar] [CrossRef]
- Sathish Kumar, T.; Radhika, K.; Joseph Sahaya Rajan, J.; Makesh, M.; Alavandi, S.V.; Vijayan, K.K. Closed-tube field-deployable loop-mediated isothermal amplification (LAMP) assay based on spore wall protein (SWP) for the visual detection of Enterocytozoon hepatopenaei (EHP). J. Invertebr. Pathol. 2021, 183, 107624. [Google Scholar] [CrossRef]
- Ambagala, A.; Fisher, M.; Goolia, M.; Nfon, C.; Furukawa-Stoffer, T.; Ortega Polo, R.; Lung, O. Field-Deployable Reverse Transcription-Insulated Isothermal PCR (RT-iiPCR) Assay for Rapid and Sensitive Detection of Foot-and-Mouth Disease Virus. Transbound. Emerg. Dis. 2017, 64, 1610–1623. [Google Scholar] [CrossRef]
- Dudley, D.M.; Newman, C.M.; Weiler, A.M.; Ramuta, M.D.; Shortreed, C.G.; Heffron, A.S.; Accola, M.A.; Rehrauer, W.M.; Friedrich, T.C.; O’Connor, D.H. Optimizing direct RT-LAMP to detect transmissible SARS-CoV-2 from primary nasopharyngeal swab samples. PLoS ONE 2020, 15, e0244882. [Google Scholar] [CrossRef]
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Ambagala, A.; Pahari, S.; Fisher, M.; Lee, P.A.; Pasick, J.; Ostlund, E.N.; Johnson, D.J.; Lung, O. A Rapid Field-Deployable Reverse Transcription-Insulated Isothermal Polymerase Chain Reaction Assay for Sensitive and Specific Detection of Bluetongue Virus. Transbound. Emerg. Dis. 2017, 64, 476–486. [Google Scholar] [CrossRef]
- Carossino, M.; Li, Y.; Lee, P.-Y.A.; Tsai, C.-F.; Chou, P.-H.; Williams, D.; Skillman, A.; Frank Cook, R.; Brown, G.; Chang, H.-F.G.; et al. Evaluation of a field-deployable reverse transcription-insulated isothermal PCR for rapid and sensitive on-site detection of Zika virus. BMC Infect. Dis. 2017, 17, 778. [Google Scholar] [CrossRef]
| Disease | Prescribed Diagnostic Test/s [31] |
|---|---|
| OIE listed notifiable equine viral diseases [32] | |
| African horse sickness | RT-PCR 1 Virus isolation |
| Equine encephalomyelitis (Western) | RT-PCR Virus isolation |
| Equine infectious anaemia | AGID 2 |
| Equine influenza | ELISA 3 RT-PCR |
| Equine viral arteritis | CF 4 PCR VN 5 Virus isolation |
| Equine viral rhinopneumonitis (EHV-1) | PCR VN Virus isolation |
| Zoonotic equine viral diseases of concern | |
| Hendra virus | RT-PCR Virus isolation |
| Japanese encephalitis | RT-PCR Virus isolation |
| Ross River virus | RT-PCR [126] Virus isolation [126] |
| West Nile virus | RT-PCR |
| Technique | Template | Temperature 1 | Enzyme | Reference |
|---|---|---|---|---|
| Helicase-dependent amplification (HDA) | DNA | 65 °C | Helicase | [147] |
| Insulated isothermal PCR (iiPCR) | DNA | 95 °C | Taq DNA polymerase | [24] |
| Insulated isothermal reverse-transcription PCR (iiRT-PCR) | RNA | 95 °C | Taq DNA polymerase M-MLV reverse transcription | [148] |
| Loop-mediated isothermal amplification (LAMP) | DNA | 65 °C | Bst DNA polymerase | [23] |
| Reverse transcription loop-mediated isothermal amplification (RT-LAMP) | RNA | 65 °C | Bst DNA polymerase AMV reverse transcription | [149] |
| Multiple displacement amplification (MDA) | DNA | 30 °C | Φ29 DNA polymerase | [150] |
| Nucleic acid sequence-based amplification (NASBA) | RNA | 50 °C | T7 RNA polymerase RNase H AMV reverse transcription | [151] |
| Rolling circular amplification (RCA) | DNA | 30 °C | Phi29 Bst DNA polymerase Vent exo-DNA polymerase T7 RNA polymerase | [152] |
| Recombinase polymerase amplification (RPA) | DNA RNA | 37 °C | DNA polymerase | [153] |
| Strand displacement amplification (SDA) | DNA | 60 °C | DNA polymerase | [154] |
| Disease | Type | Vector-Borne | Target Gene | Sample | Detection Limit | In-Field | Ref |
|---|---|---|---|---|---|---|---|
| African horse sickness | dsRNA | Yes—Midges, Mosquito | Vp7 | Horse—Blood | n/a | Yes | [163] |
| Equine herpesvirus 1 | dsDNA | No | Glycoprotein C | Horse—Nasal swab 1 | 1 pfu/rxn | No 1 | [164] |
| Glycoprotein E | Horse—Nasal swab 1 | 1 pfu/rxn | |||||
| Equine herpesvirus 4 | dsDNA | No | Glycoprotein C | Horse—Nasal swab 1 | 1 pfu/rxn | No 1 | [164] |
| Equine infectious anaemia | ssRNA | Yes—Horse and deer flies | Gag nsP | Recombinant plasmid | 0.1 pfu/rxn | No | [165] |
| Equine influenza (H3N8) | ssRNA | No | HA | Horse—Nasal swab | 10−5 copies/rxn | Yes | [166] |
| Equine influenza (H7N7) | ssRNA | No | HA | Horse—Nasal swab | 10−4 copies/rxn | Yes | [167] |
| Equine coronavirus | ssRNA | No | Nucleocapsid | Horse—Nasal swab, fecal samples | 101.8 copies/rxn | Yes | [168] |
| Hendra virus | ssRNA | No | P | Horse—Nasal swab 1 | 10−5 copies/rxn | No 1 | [26] |
| St Louis encephalitis | ssRNA | Yes—mosquito | UTR | Mosquito | <0.1 pfu/rxn | Yes | [159] |
| Western equine encephalitis | ssRNA | Yes—mosquito | nsP4 | Mosquito | 100 pfu/ml | Yes | [159] |
| West Nile virus | ssRNA | Yes—mosquito | E | Mosquito | 0.1 pfu/ml | Yes 2 | [149] |
| Disease | Type | Vector-Borne | Target Gene | Sample | Detection Limit | In-Field | Ref |
|---|---|---|---|---|---|---|---|
| Equine viral arteritis | ssRNA | No | ORF7 | Horse—Tissue, semen | 10 copies/rxn | Yes | [155] |
| Equine herpesvirus 3 | dsDNA | No | gG | Horse—Perineal and genital swabs | 6 copies/rxn | Yes | [175] |
| Equine herpesvirus myeloencephalopathy (EHM) caused by EHV-1 | dsDNA | No | ORF3 | Horse—Tissue | 13 copies/rxn | Yes | [176] |
| Equine infectious anaemia | ssRNA | Yes—Horse and deer flies | 5′ UTR Exon 1 of tat gene | Horse—Tissue | 8 copies/rxn | Yes | [177] |
| Equine influenza (H3N8) | ssRNA | No | HA | Horse—Nasal swab | 11 copies/rxn | Yes | [148] |
| Properties | PCR | iiPCR | LAMP |
|---|---|---|---|
| Temperature | Cycles through 3 temperatures 55–95 °C | Constant temperature drives temperature gradient 15–30 °C | Constant temperature 60–65 °C |
| Equipment | Thermocycler | Specialized reaction tube Fluorescence-based detector | Heat source |
| Field-deployable | No | Yes | Yes |
| Reaction time | At least 90 min | ≤60 min | <30 min |
| Sensitivity | Starts at nanograms | Starts at nanograms | Starts at femtograms |
| Specificity | Requires specific primer design Prone to errors | Requires specific primer design Prone to errors | Tolerates combination of primer designs |
| Visualization | Only through gel electrophoresis | Real-time available | Real-time available |
| Template prep | Requires purification | Requires purification | Tolerates impurities |
| Cost | $$$ | $$ | $ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knox, A.; Beddoe, T. Isothermal Nucleic Acid Amplification Technologies for the Detection of Equine Viral Pathogens. Animals 2021, 11, 2150. https://doi.org/10.3390/ani11072150
Knox A, Beddoe T. Isothermal Nucleic Acid Amplification Technologies for the Detection of Equine Viral Pathogens. Animals. 2021; 11(7):2150. https://doi.org/10.3390/ani11072150
Chicago/Turabian StyleKnox, Alexandra, and Travis Beddoe. 2021. "Isothermal Nucleic Acid Amplification Technologies for the Detection of Equine Viral Pathogens" Animals 11, no. 7: 2150. https://doi.org/10.3390/ani11072150
APA StyleKnox, A., & Beddoe, T. (2021). Isothermal Nucleic Acid Amplification Technologies for the Detection of Equine Viral Pathogens. Animals, 11(7), 2150. https://doi.org/10.3390/ani11072150







