Transcriptional Insights into Key Genes and Pathways Underlying Muscovy Duck Subcutaneous Fat Deposition at Different Developmental Stages
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Adipose Tissue Sampling
2.3. RNA Extraction and RNA Sequencing
2.4. Bioinformatics Analysis of RNA-Seq
3. Results
3.1. Overall Assessment of RNA-Seq Data
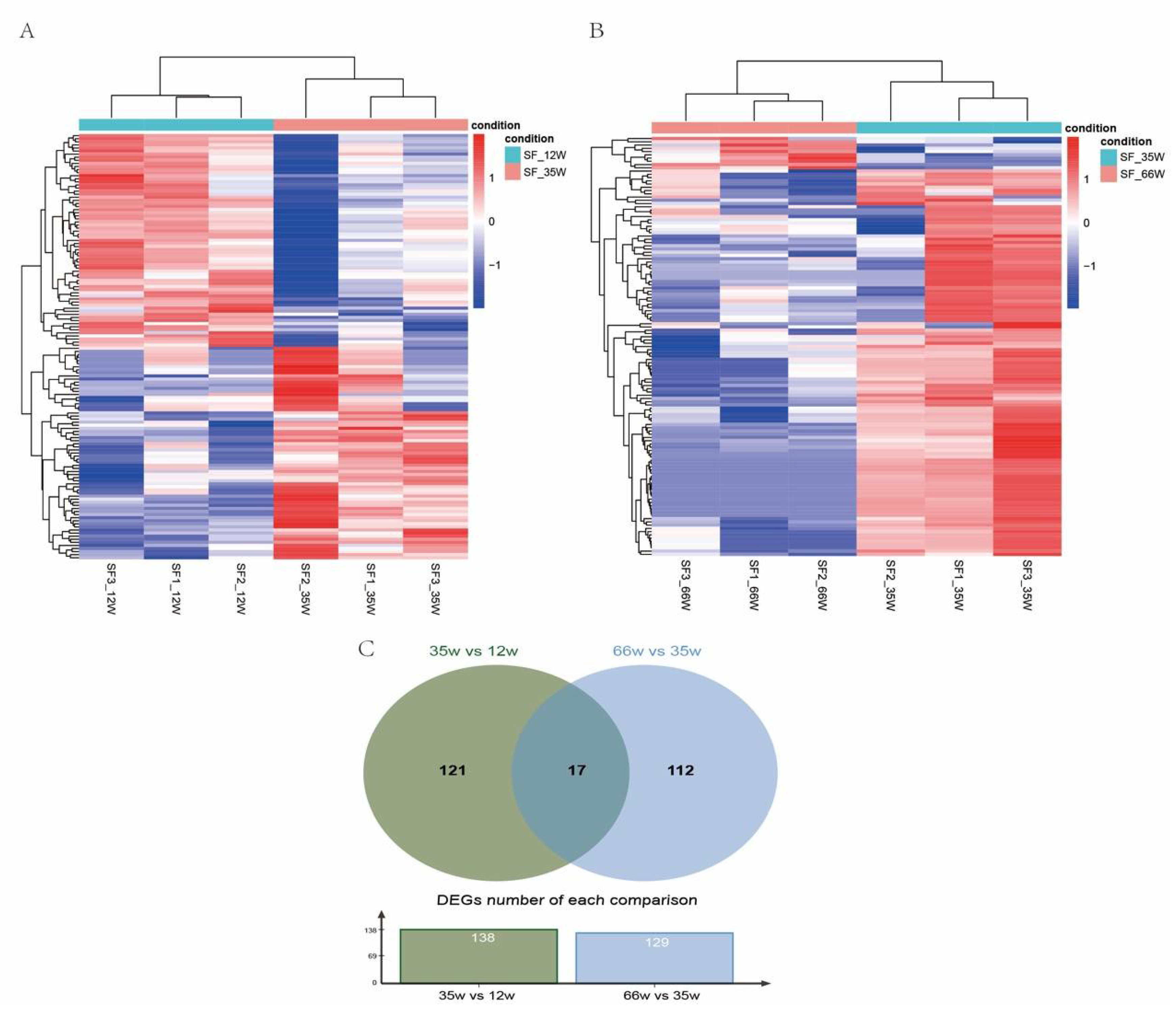
3.2. DEGs Analysis among Three Developmental Stages
3.3. Functional Enrichment of the DEGs between 35 wk and 12 wk
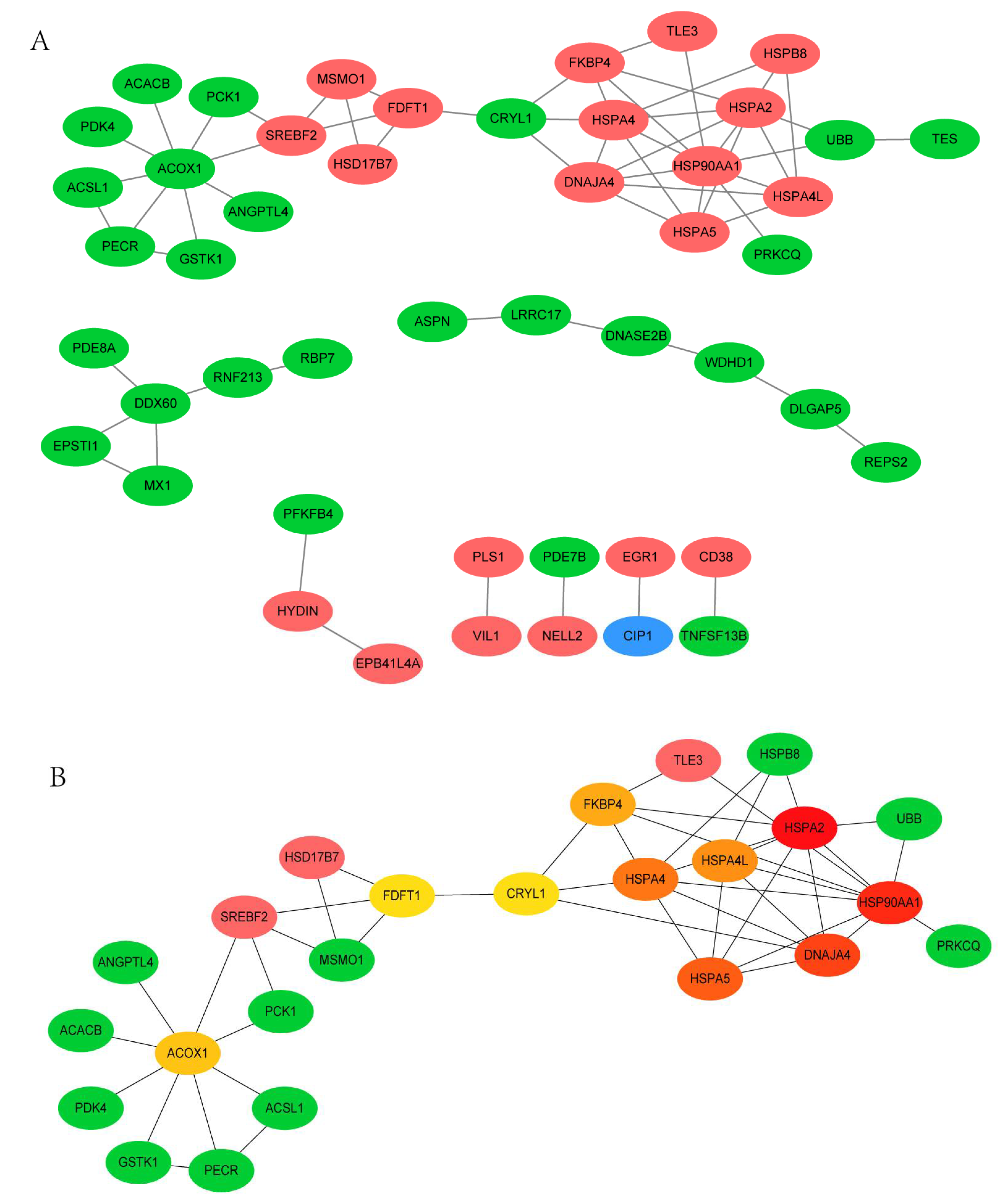
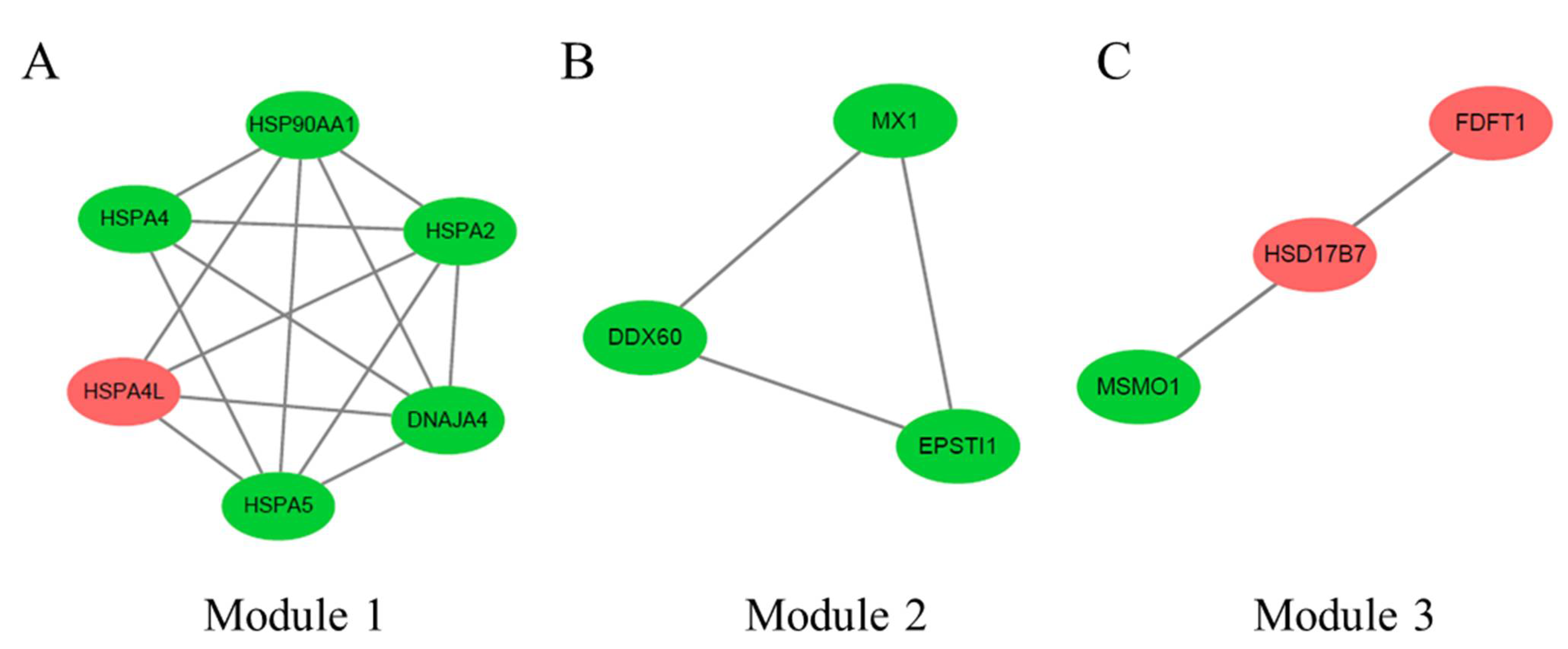
3.4. PPI Network Analysis of DEGs between 35 wk and 12 wk
3.5. Functional Enrichment Analysis of the DEGs between 66 wk and 35 wk
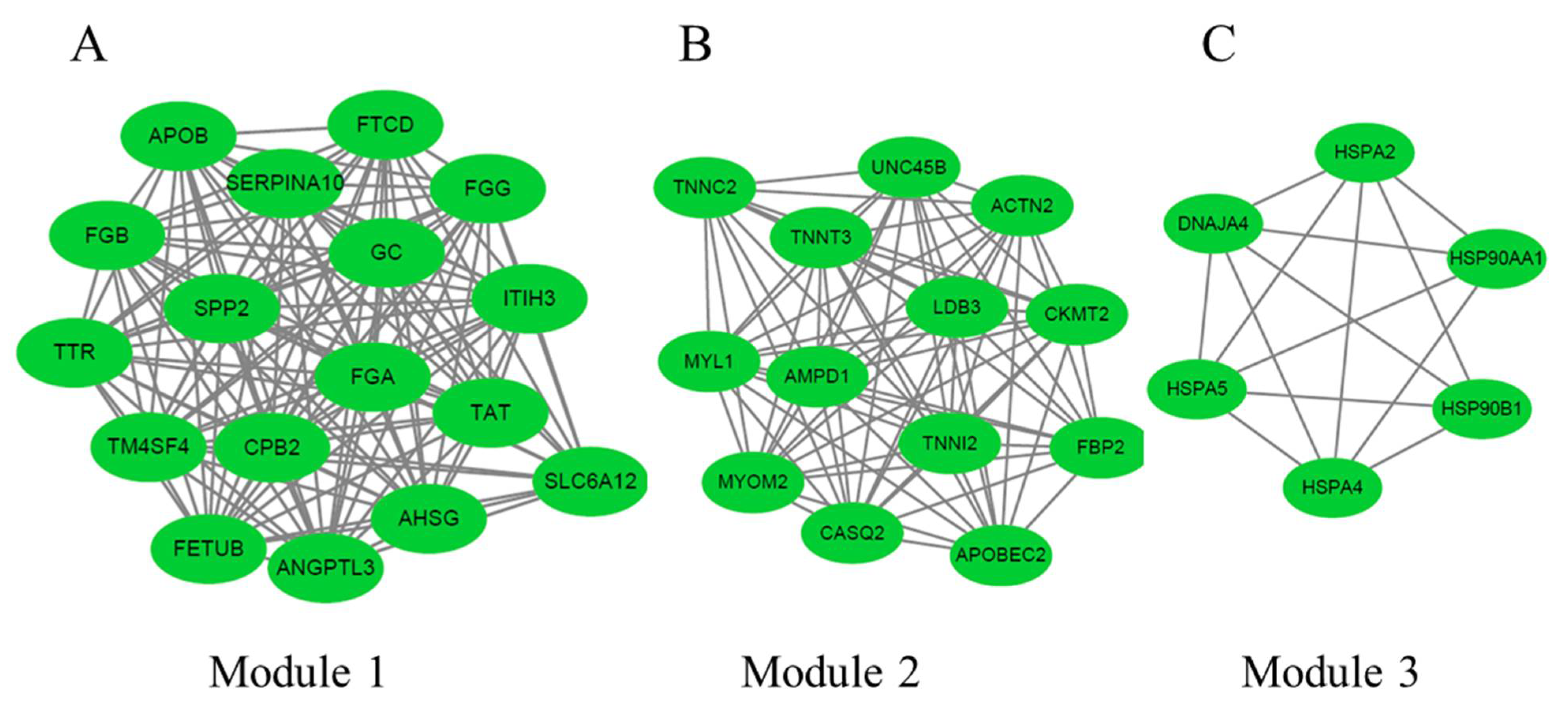
3.6. PPI Network Analysis of DEGs between 66 wk and 35 wk
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Tamsil, H. Genetic Resource of Muscovy Duck (Cairina moschata): Profile and Potential Production as Meat Producer. Indones. Bull. Anim. Vet. Sci. 2018, 28, 129–138. [Google Scholar] [CrossRef]
- Baeza, E. Nutritional requirements and feed management of meat type ducks. Worlds Poult. Sci. J. 2016, 72, 5–19. [Google Scholar] [CrossRef]
- Shin, D.M.; Kim, D.; Yune, J.H.; Kwon, H.C.; Kim, H.J.; Seo, H.G.; Han, S.G. Oxidative Stability and Quality Characteristics of Duck, Chicken, Swine and Bovine Skin Fats Extracted by Pressurized Hot Water Extraction. Food Sci. Anim. Resour. 2019, 39, 446–458. [Google Scholar] [CrossRef]
- Raj, S.; Skiba, G.; Weremko, D.; Fandrejewski, H.; Migdal, W.; Borowiec, F.; Polawska, E. The relationship between the chemical composition of the carcass and the fatty acid composition of intramuscular fat and backfat of several pig breeds slaughtered at different weights. Meat Sci. 2010, 86, 324–330. [Google Scholar] [CrossRef]
- Jennen, D.G.J. Chicken Fatness: From QTL to Candidate Gene; Wageningen University: Wageningen, The Netherlands, 2004; ISBN 90-8504-069-8. [Google Scholar]
- Wood, J.D.; Enser, M.; Fisher, A.V.; Nute, G.R.; Sheard, P.R.; Richardson, R.I.; Hughes, S.I.; Whittington, F.M. Fat deposition, fatty acid composition and meat quality: A review. Meat Sci. 2008, 78, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Kang, L.; Hu, Y.; Xu, Q.; Zhang, N.; Jiang, Y. Effect of Dietary Betaine Supplementation on mRNA Expression and Promoter CpG Methylation of Lipoprotein Lipase Gene in Laying Hens. J. Poult. Sci. 2009, 46, 224–228. [Google Scholar] [CrossRef]
- Bai, S.; Wang, G.; Zhang, W.; Zhang, S.; Rice, B.B.; Cline, M.A.; Gilbert, E.R. Broiler chicken adipose tissue dynamics during the first two weeks post-hatch. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2015, 189, 115–123. [Google Scholar] [CrossRef]
- Nie, Q.; Fang, M.; Xie, L.; Peng, X.; Xu, H.; Luo, C.; Zhang, D.; Zhang, X. Molecular characterization of the ghrelin and ghrelin receptor genes and effects on fat deposition in chicken and duck. J. Biomed. Biotechnol. 2009, 2009, 567120. [Google Scholar] [CrossRef] [PubMed]
- Deaton, J.W.; Lott, B.D. Age and dietary energy effect on broiler abdominal fat deposition. Poult. Sci. 1985, 64, 2161–2164. [Google Scholar] [CrossRef]
- He, J.; Zheng, H.; Pan, D.; Liu, T.; Sun, Y.; Cao, J.; Wu, Z.; Zeng, X. Effects of aging on fat deposition and meat quality in Sheldrake duck. Poult. Sci. 2018, 97, 2005–2010. [Google Scholar] [CrossRef] [PubMed]
- Kotani, K.; Tokunaga, K.; Fujioka, S.; Kobatake, T.; Keno, Y.; Yoshida, S.; Shimomura, I.; Tarui, S.; Matsuzawa, Y. Sexual dimorphism of age-related changes in whole-body fat distribution in the obese. Int. J. Obes. Relat. Metab. Disord. J. Int. Assoc. Study Obes. 1994, 18, 202–207. [Google Scholar]
- Kuk, J.L.; Saunders, T.J.; Davidson, L.E.; Ross, R. Age-related changes in total and regional fat distribution. Ageing Res. Rev. 2009, 8, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential Expression of RNA-Seq Data at the Gene Level the DESeq Package; European Molecular Biology Laboratory: Heidelberg, Germany, 2012. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 27. [Google Scholar] [CrossRef]
- Deng, M.T.; Zhang, F.; Zhu, F.; Yang, Y.Z.; Yang, F.X.; Hao, J.P.; Hou, Z.C. Genome-wide association study reveals novel loci associated with fat-deposition and meat-quality traits in Pekin ducks. Anim. Genet. 2020, 51, 953–957. [Google Scholar] [CrossRef]
- Zhang, M.; Li, D.H.; Li, F.; Sun, J.W.; Jiang, R.R.; Li, Z.J.; Han, R.L.; Li, G.X.; Liu, X.J.; Kang, X.T.; et al. Integrated Analysis of MiRNA and Genes Associated with Meat Quality Reveals that Gga-MiR-140-5p Affects Intramuscular Fat Deposition in Chickens. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 46, 2421–2433. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Cui, H.; Zhao, G.; Liu, R.; Li, Q.; Zheng, M.; Guo, Y.; Wen, J. Intramuscular preadipocytes impede differentiation and promote lipid deposition of muscle satellite cells in chickens. BMC Genom. 2018, 19, 838. [Google Scholar] [CrossRef]
- Liu, Y.; Jia, Y.; Liu, C.; Ding, L.; Xia, Z. RNA-Seq transcriptome analysis of breast muscle in Pekin ducks supplemented with the dietary probiotic Clostridium butyricum. BMC Genom. 2018, 19, 844. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Zhang, L.P.; Li, J.J.; Wang, D.Q.; Tian, Y.; Lu, L.Z. De novo assembly and characterization of Muscovy duck liver transcriptome and analysis of differentially regulated genes in response to heat stress. Cell Stress Chaperones 2015, 20, 483–493. [Google Scholar] [CrossRef]
- Wójcik, E.; Smalec, E. Description of the Muscovy duck (Cairina moschata) karyotype. Folia Biol. 2008, 56, 243–248. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Z.; Ren, X.; Wang, L.; Li, C.; Sun, Y.; Wang, M.; Tong, Q.; Sun, H.; Pu, J. Infection of chicken H9N2 influenza viruses in different species of domestic ducks. Vet. Microbiol. 2019, 233, 1–4. [Google Scholar] [CrossRef]
- Xu, T.S.; Gu, L.H.; Schachtschneider, K.M.; Liu, X.L.; Huang, W.; Xie, M.; Hou, S.S. Identification of Differentially Expressed Genes in Breast Muscle and Skin Fat of Postnatal Pekin Duck. PLoS ONE 2014, 9, 13. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xin, Y.; Li, C.; Guo, Y.; Xiao, R.; Zhang, H.; Zhou, G. RNA-Seq analysis reveals a negative role of MSMO1 with a synergized NSDHL expression during adipogenesis of 3T3-L1. Biosci. Biotechnol. Biochem. 2019, 83, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Seth, G.; Mcivor, R.S.; Hu, W.S. 17Beta-hydroxysteroid dehydrogenase type 7 (Hsd17b7) reverts cholesterol auxotrophy in NS0 cells. J. Biotechnol. 2006, 121, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.M.; Li, L.O.; Wu, P.C.; Koves, T.R.; Ilkayeva, O.; Stevens, R.D.; Watkins, S.M.; Muoio, D.M.; Coleman, R.A. Adipose Acyl-CoA Synthetase-1 Directs Fatty Acids toward beta-Oxidation and Is Required for Cold Thermogenesis. Cell Metab. 2010, 12, 53–64. [Google Scholar] [CrossRef]
- Burgess, S.C.; He, T.; Yan, Z.; Lindner, J.; Sherry, A.D.; Malloy, C.R.; Browning, J.D.; Magnuson, M.A. Cytosolic Phosphoenolpyruvate Carboxykinase Does Not Solely Control the Rate of Hepatic Gluconeogenesis in the Intact Mouse Liver. Cell Metab. 2007, 5, 313–320. [Google Scholar] [CrossRef]
- Latorre, P.; Burgos, C.; Hidalgo, J.; Varona, L.; Carrodeguas, J.A.; López-Buesa, P. c.A2456C-substitution in Pck1 changes the enzyme kinetic and functional properties modifying fat distribution in pigs. Sci. Rep. 2016, 6, 19617. [Google Scholar] [CrossRef]
- Sirwi, A.; Hussain, M.M. Lipid transfer proteins in the assembly of apoB-containing lipoproteins. J. Lipid Res. 2018, 59, 1094–1102. [Google Scholar] [CrossRef]
- Linton, M.F.; Yancey, P.G.; Davies, S.S.; Jerome, W.G.; Linton, E.F.; Song, W.L.; Doran, A.C.; Vickers, K.C. The Role of Lipids and Lipoproteins in Atherosclerosis. In Endotext; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., De Herder, W.W., Dhatariya, K., Dungan, K., Grossman, A., Hershman, J.M., Hofland, J., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Leduc, M.S.; Shimmin, L.C.; Klos, K.L.; Hanis, C.; Boerwinkle, E.; Hixson, J.E. Comprehensive evaluation of apolipoprotein H gene (APOH) variation identifies novel associations with measures of lipid metabolism in GENOA. J. Lipid Res. 2008, 49, 2648–2656. [Google Scholar] [CrossRef] [PubMed]
- Cassader, M.; Ruiu, G.; Gambino, R.; Guzzon, F.; Pagano, A.; Veglia, F.; Pagni, R.; Pagano, G. Influence of apolipoprotein H polymorphism on levels of triglycerides. Atherosclerosis 1994, 110, 45–51. [Google Scholar] [CrossRef]
- Takada, D.; Ezura, Y.; Ono, S.; Iino, Y.; Katayama, Y.; Xin, Y.; Wu, L.L.; Larringa-Shum, S.; Stephenson, S.H.; Hunt, S.C.; et al. Apolipoprotein H variant modifies plasma triglyceride phenotype in familial hypercholesterolemia: A molecular study in an eight-generation hyperlipidemic family. J. Atheroscler. Thromb. 2003, 10, 79–84. [Google Scholar] [CrossRef]
- Rajkumar, A.; Liaghati, A.; Chan, J.; Lamothe, G.; Dent, R.; Doucet, É.; Rabasa-Lhoret, R.; Prud’homme, D.; Harper, M.E.; Tesson, F. ACSL5 genotype influence on fatty acid metabolism: A cellular, tissue, and whole-body study. Metab. Clin. Exp. 2018, 83, 271–279. [Google Scholar] [CrossRef]
- Bowman, T.A.; O’keeffe, K.R.; D’aquila, T.; Yan, Q.W.; Griffin, J.D.; Killion, E.A.; Salter, D.M.; Mashek, D.G.; Buhman, K.K.; Greenberg, A.S. Acyl CoA synthetase 5 (ACSL5) ablation in mice increases energy expenditure and insulin sensitivity and delays fat absorption. Mol. Metab. 2016, 5, 210–220. [Google Scholar] [CrossRef]
- Savastano, S.; Barrea, L.; Savanelli, M.C.; Nappi, F.; Di Somma, C.; Orio, F.; Colao, A. Low vitamin D status and obesity: Role of nutritionist. Rev. Endocr. Metab. Disord. 2017, 18, 215–225. [Google Scholar] [CrossRef]
- Dastorani, M.; Aghadavod, E.; Mirhosseini, N.; Foroozanfard, F.; Zadeh Modarres, S.; Amiri Siavashani, M.; Asemi, Z. The effects of vitamin D supplementation on metabolic profiles and gene expression of insulin and lipid metabolism in infertile polycystic ovary syndrome candidates for in vitro fertilization. Reprod. Biol. Endocrinol. RBE 2018, 16, 94. [Google Scholar] [CrossRef]
- Chanet, A.; Salles, J.; Guillet, C.; Giraudet, C.; Berry, A.; Patrac, V.; Domingues-Faria, C.; Tagliaferri, C.; Bouton, K.; Bertrand-Michel, J.; et al. Vitamin D supplementation restores the blunted muscle protein synthesis response in deficient old rats through an impact on ectopic fat deposition. J. Nutr. Biochem. 2017, 46, 30–38. [Google Scholar] [CrossRef]
- Grave, N.; Tovo-Rodrigues, L.; Da Silveira, J.; Rovaris, D.L.; Dal Bosco, S.M.; Contini, V.; Genro, J.P. A vitamin D pathway gene-gene interaction affects low-density lipoprotein cholesterol levels. J. Nutr. Biochem. 2016, 38, 12–17. [Google Scholar] [CrossRef]
- Zhou, J.C.; Zhu, Y.; Gong, C.; Liang, X.; Zhou, X.; Xu, Y.; Lyu, D.; Mo, J.; Xu, J.; Song, J.; et al. The GC2 haplotype of the vitamin D binding protein is a risk factor for a low plasma 25-hydroxyvitamin D concentration in a Han Chinese population. Nutr. Metab. 2019, 16, 5. [Google Scholar] [CrossRef]
- Stefan, N.; Hennige, A.M.; Staiger, H.; Machann, J.; Schick, F.; Kröber, S.M.; Machicao, F.; Fritsche, A.; Häring, H.-U. Alpha2-Heremans-Schmid glycoprotein/fetuin-A is associated with insulin resistance and fat accumulation in the liver in humans. Diabetes Care 2006, 29, 853–857. [Google Scholar] [CrossRef] [PubMed]
- Lavebratt, C.; Dungner, E.; Hoffstedt, J. Polymorphism of the AHSG gene is associated with increased adipocyte beta2-adrenoceptor function. J. Lipid Res. 2005, 46, 2278–2281. [Google Scholar] [CrossRef]
- Zhou, M.C.; Yu, P.; Sun, Q.; Li, Y.X. Expression profiling analysis: Uncoupling protein2 deficiency improves hepatic glucose, lipid profiles and insulin sensitivity in high-fat diet-fed mice by modulating expression of genes in peroxisome proliferator-activated receptor signaling pathway. J. Diabetes Investig. 2016, 7, 179–189. [Google Scholar] [CrossRef]
- Fujii, H. Nuclear Receptor PPARs and magnesium. Clin. Calcium 2005, 15, 52–64. [Google Scholar]
- Royan, M.; Navidshad, B. Peroxisome proliferator-activated receptor gamma (PPAR gamma), a key regulatory gene of lipid metabolism in chicken. Worlds Poult. Sci. J. 2016, 72, 773–784. [Google Scholar] [CrossRef]


| Sample | Raw Reads | Q20 Value | Clean Reads | Clean Reads Ratio | Mapped Reads | Mapping Ratio |
|---|---|---|---|---|---|---|
| 35w-3 | 42,850,794 | 89.89% | 40,327,910 | 94.11% | 16,701,309 | 41.46% |
| 35w-2 | 48,548,724 | 90.02% | 45,637,136 | 94.00% | 19,810,653 | 43.44% |
| 35w-1 | 43,708,580 | 89.88% | 41,001,746 | 93.81% | 17,815,651 | 43.48% |
| 66w-3 | 41,752,056 | 88.66% | 38,826,006 | 92.99% | 13,653,232 | 35.19% |
| 66w-2 | 39,392,088 | 91.83% | 37,119,504 | 94.23% | 15,430,183 | 41.60% |
| 66w-1 | 43,543,486 | 91.58% | 40,983,242 | 94.12% | 17,051,338 | 41.64% |
| 12w-3 | 44,862,056 | 91.83% | 42,440,164 | 94.60% | 15,662,882 | 36.93% |
| 12w-2 | 50,191,846 | 93.64% | 47,533,720 | 94.70% | 20,748,045 | 43.69% |
| 12w-1 | 60,636,404 | 93.22% | 57,268,052 | 94.45% | 24,977,931 | 43.68% |
| Gene Name | FC | p Value | Description | Trend |
|---|---|---|---|---|
| PCK1 | 0.03 | 0.03 | Phosphoenolpyruvate carboxykinase | Down |
| MX1 | 0.16 | 0.04 | Interferon-induced GTP-binding protein Mx isoform X1 | Down |
| TMPRSS7 | 0.19 | 0.00 | Transmembrane protease serine 7 isoform X1 | Down |
| EPSTI1 | 0.20 | 0.01 | Epithelial-stromal interaction protein 1 | Down |
| ANGPTL5 | 0.21 | 0.01 | Angiopoietin-related protein 5 isoform X4 | Down |
| NRXN1 | 0.22 | 0.03 | Neurexin-1 isoform X6 | Down |
| C18orf21 | 0.24 | 0.01 | Uncharacterized protein c18orf21-like protein, partial | Down |
| ABHD6 | 0.26 | 0.00 | Monoacylglycerol lipase ABHD6 | Down |
| SYN3 | 0.27 | 0.04 | Synapsin-3 | Down |
| TRH | 0.27 | 0.01 | Prothyroliberin, partial | Down |
| NELL2 | 16.93 | 0.00 | Protein kinase C-binding protein NELL2, partial | Up |
| MLKL | 15.35 | 0.00 | Mixed lineage kinase domain-like protein, partial | Up |
| HYDIN | 14.59 | 0.03 | Belongs to the cytochrome P450 family | Up |
| ASTN1 | 10.16 | 0.04 | Astrotactin-1 | Up |
| KIAA1211 | 7.74 | 0.04 | Uncharacterized protein KIAA1211 homolog isoform X2 | Up |
| GDF1 | 5.25 | 0.02 | Embryonic growth/differentiation factor 1 | Up |
| LCN15 | 4.72 | 0.01 | LOW QUALITY PROTEIN: lipocalin-15-like | Up |
| PLS1 | 4.60 | 0.04 | Plastin-1 | Up |
| CD38 | 4.55 | 0.04 | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 2 | Up |
| FGF12 | 4.02 | 0.02 | Fibroblast growth factor 12 isoform X3 | Up |
| Gene Name | FC | p Value | Description | Trend |
|---|---|---|---|---|
| MB | 0.00 | 0.01 | Myoglobin, partial | Down |
| APOBEC2 | 0.00 | 0.02 | Apolipoprotein B mRNA editing enzyme catalytic subunit 2 | Down |
| HPD | 0.00 | 0.02 | 4-hydroxyphenylpyruvate dioxygenase | Down |
| SPP2 | 0.00 | 0.03 | secreted phosphoprotein 24 | Down |
| FTCD | 0.00 | 0.03 | Formimidoyltransferase-cyclodeaminase | Down |
| CYP7A1 | 0.00 | 0.03 | Cytochrome P450 7A1, partial | Down |
| TM4SF4 | 0.00 | 0.04 | transmembrane 4 L6 family member 4 | Down |
| SLC13A3 | 0.00 | 0.04 | Solute carrier family 13 member 3, partial | Down |
| CPB2 | 0.00 | 0.04 | carboxypeptidase B2 | Down |
| SLC6A12 | 0.00 | 0.05 | sodium- and chloride-dependent betaine transporter | Down |
| FGA | 0.00 | 0.01 | fibrinogen alpha chain | Down |
| FGB | 0.00 | 0.01 | fibrinogen beta chain | Down |
| APOB | 0.00 | 0.01 | apolipoprotein B-100 isoform X1 | Down |
| ALB | 0.00 | 0.02 | albumin | Down |
| TCN2 | 5.00 | 0.03 | transcobalamin-2, partial | Up |
| SYT12 | 4.63 | 0.04 | Synaptotagmin-12, partial | Up |
| GNA14 | 3.90 | 0.03 | G protein subunit alpha 14 | Up |
| PAMR1 | 3.44 | 0.03 | inactive serine protease PAMR1 isoform X2 | Up |
| TNIP2 | 3.26 | 0.02 | TNFAIP3-interacting protein 2 | Up |
| NET1 | 2.15 | 0.05 | neuroepithelial cell-transforming gene 1 protein | Up |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, L.; Wei, C.; Yi, L.; Yang, W.; Geng, Z.; Chen, X. Transcriptional Insights into Key Genes and Pathways Underlying Muscovy Duck Subcutaneous Fat Deposition at Different Developmental Stages. Animals 2021, 11, 2099. https://doi.org/10.3390/ani11072099
Guo L, Wei C, Yi L, Yang W, Geng Z, Chen X. Transcriptional Insights into Key Genes and Pathways Underlying Muscovy Duck Subcutaneous Fat Deposition at Different Developmental Stages. Animals. 2021; 11(7):2099. https://doi.org/10.3390/ani11072099
Chicago/Turabian StyleGuo, Liping, Congcong Wei, Li Yi, Wanli Yang, Zhaoyu Geng, and Xingyong Chen. 2021. "Transcriptional Insights into Key Genes and Pathways Underlying Muscovy Duck Subcutaneous Fat Deposition at Different Developmental Stages" Animals 11, no. 7: 2099. https://doi.org/10.3390/ani11072099
APA StyleGuo, L., Wei, C., Yi, L., Yang, W., Geng, Z., & Chen, X. (2021). Transcriptional Insights into Key Genes and Pathways Underlying Muscovy Duck Subcutaneous Fat Deposition at Different Developmental Stages. Animals, 11(7), 2099. https://doi.org/10.3390/ani11072099






