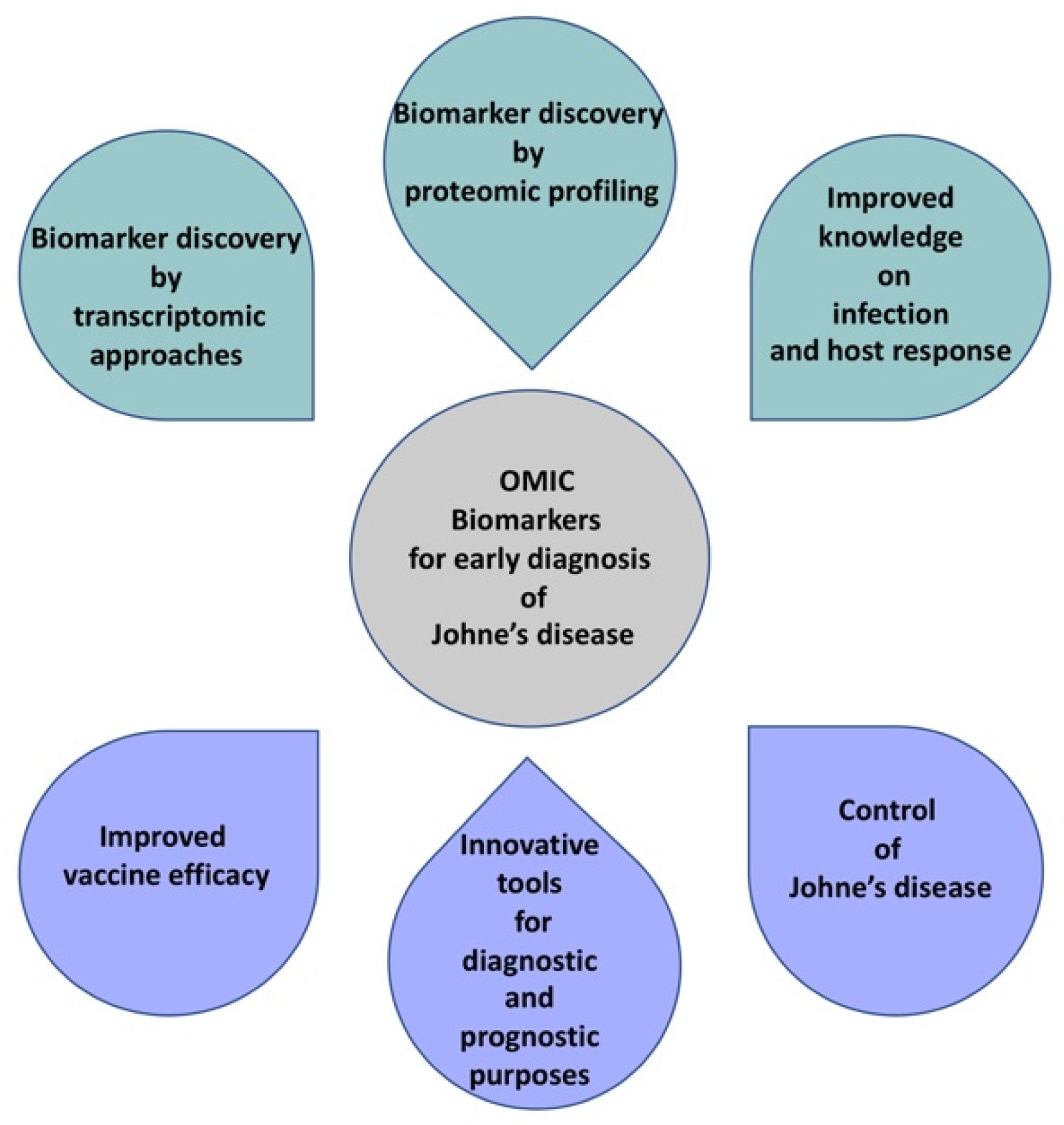
Using Omics Approaches in the Discovery of Biomarkers for Early Diagnosis of Johne’s Disease in Sheep and Goats
Abstract
:Simple Summary
Abstract
1. Introduction
2. Johne’s Disease All Over the World
3. Transmission and Signs
4. Mechanism of Infection and Host Response
5. Importance of Biomarkers
6. Biomarker Discovery by Transcriptomic Approaches
7. miRNA as Biomarkers
8. Biomarker Discovery by Proteomic Approaches
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lewis, C.R.; Ait-Ali, T.; Clapperton, M.; Archibald, A.L.; Bishop, S. Genetic perspectives on host responses to porcine reproductive and respiratory syndrome (PRRS). Viral Immunol. 2007, 20, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Harris, N.B.; Barletta, R.G. Mycobacterium avium. Clin. Microb. Rev. 2001, 14, 489–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salgado, M.; Monti, G.; Sevilla, I.; Manning, E. Association between cattle herd Mycobacterium avium subsp. paratuberculosis (MAP) infection and infection of a hare population. Trop. Anim. Health Prod. 2014, 46, 1313–1316. [Google Scholar] [CrossRef] [PubMed]
- Beard, P.M.; Daniels, M.J.; Henderson, D.; Pirie, A.; Rudge, K.; Buxton, D.; Rhind, S.; Greig, A.; Hutchings, M.R.; McKendrick, I.; et al. Paratuberculosis infection of nonruminant wildlife in Scotland. J. Clin. Microbiol. 2001, 39, 1517–1521. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, D. International efforts at paratuberculosis control. Vet. Clin. N. Am. Food Anim. Pract. 2011, 27, 647–654. [Google Scholar] [CrossRef]
- Lombard, J.E. Epidemiology and economics of paratuberculosis. Vet. Clin. N. Am. Food Anim. Pract. 2011, 27, 525–535. [Google Scholar] [CrossRef]
- Whittington, R.; Donat, K.; Weber, M.F.; Kelton, D.; Nielsen, S.S.; Eisenberg, S.; Arrigoni, N.; Juste, R.; Saez, J.L.; Dhand, N. Control of paratuberculosis: Who, why and how. A review of 48 countries. BMC Vet. Res. 2019, 15, 198. [Google Scholar] [CrossRef] [Green Version]
- Giannitti, F.; Fraga, M.; Caffarena, R.D.; Schild, C.O.; Banchero, G.; Armién, A.G.; Traveria, G.; Marthaler, D.; Wells, S.J.; Correa, F.R. Mycobacterium paratuberculosis sheep type strain in Uruguay: Evidence for a wider geographic distribution in South America. J. Infect. Dev. 2018, 12, 190–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, S.S.; Toft, N. A review of prevalences of paratuberculosis in farmed animals in Europe. Prev. Vet. Med. 2009, 88, 1–14. [Google Scholar] [CrossRef]
- Attili, A.R.; Ngu Ngwa, V.; Preziuso, S.; Pacifici, L.; Domesi, A.; Cuteri, V. Ovine paratuberculosis: A seroprevalence study in dairy flocks reared in the marche region, Italy. Vet. Med. Int. 2011, 2011, 782875. [Google Scholar]
- Iarussi, F.; Paradies, P.; Sardaro, R.; Rubino, G.; Scaltrito, D.; Pieragostini, E.; Petazzi, F. Epidemiology and risk factors of Mycobacterium avium subspecies paratuberculosis in semi-extensive dairy sheep and goat farms of Apulia, southern Italy. Small Rumin. Res. 2019, 177, 89–96. [Google Scholar] [CrossRef]
- Aduriz, J.J.; Juste, R.A.; Cortabarría, N. Lack of mycobactin dependence of mycobacteria isolated on Middlebrook 7H11 from clinical cases of ovine paratuberculosis. Vet. Microbiol. 1995, 45, 211–217. [Google Scholar] [CrossRef]
- Reviriego, F.J.; Moreno, M.A.; Domínguez, L. Soil type as a putative risk factor of ovine and caprine paratuberculosis seropositivity in Spain. Prev. Vet. Med. 2000, 43, 43–51. [Google Scholar] [CrossRef]
- Seaman, J.T.; Gardner, I.A.; Dent, C.H. Johne’s disease in sheep. Aust. Vet. J. 1981, 57, 102–103. [Google Scholar] [CrossRef] [PubMed]
- Sergeant, E.S.G.; Baldock, F.C. The estimated prevalence of Johne’s disease infected sheep flocks in Australia. Austr. Vet. J. 2002, 80, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Verdugo, C.; Jones, G.; Johnson, W.; Wilson, P.; Stringer, L.; Heuer, C. Estimation of flock/herd-level true Mycobacterium avium subspecies paratuberculosis prevalence on sheep, beef cattle and deer farms in New Zealand using a novel Bayesian model. Prev. Vet. Med. 2014, 117, 447–455. [Google Scholar] [CrossRef]
- Michel, A.L.; Bastianello, S.S. Paratuberculosis in sheep: An emerging disease in South Africa. Vet. Microb. 2000, 77, 299–307. [Google Scholar] [CrossRef]
- Fernández-Silva, J.A.; Correa-Valencia, N.M.; Ramírez, N.F. Systematic review of the prevalence of paratuberculosis in cattle, sheep, and goats in Latin America and the Caribbean. Trop. Anim. Health Produ. 2014, 46, 1321–1340. [Google Scholar] [CrossRef]
- Angelidou, E.; Kostoulas, P.; Leontides, L. Flock-level factors associated with the risk of Mycobacterium avium subsp. paratuberculosis (MAP) infection in Greek dairy goat flocks. Prev. Vet. Med. 2014, 117, 233–241. [Google Scholar] [CrossRef]
- Mercier, P.; Baudry, C.; Beaudeau, F.; Seegers, H.; Malher, X. Estimated prevalence of Mycobacterium avium subspecies paratuberculosis infection in herds of dairy goats in France. Vet. Rec. 2010, 167, 412–415. [Google Scholar] [CrossRef]
- Muehlherr, J.E.; Zweifel, C.; Corti, S.; Blanco, J.E.; Stephan, R. Microbiological quality of raw goat’s and ewe’s bulk-tank milk in Switzerland. J. Dairy Sci. 2003, 86, 3849–3856. [Google Scholar] [CrossRef]
- Lombard, J.E.; Garry, F.B.; McCluskey, B.J.; Wagner, B.A. Risk of removal and effects on milk production associated with paratuberculosis status in dairy cows. J. Am. Vet. Med. Assoc. 2005, 227, 1975–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groenendaal, H.; Nielen, M.; Jalvingh, A.W.; Horst, S.H.; Galligan, D.T.; Hesselink, J.W. A simulation of Johne’s disease control. Prev. Vet. Med. 2002, 54, 225–245. [Google Scholar] [CrossRef]
- Ott, S.L.; Wells, S.J.; Wagner, B.A. Herd-level economic losses associated with Johne’s disease on US dairy operations. Prev. Vet. Med. 1999, 40, 179–192. [Google Scholar] [CrossRef]
- Bush, R.D.; Windsor, P.A.; Toribio, J.A. Losses of adult sheep due to ovine Johne’s disease in 12 infected flocks over a 3-year period. Aust. Vet. J. 2006, 84, 246–253. [Google Scholar] [CrossRef]
- Kampen, A.H.; Mork, J.; Klevar, S. The surveillance and control programme for Brucella melitensis in small ruminants in Norway 2011. In Surveillance and Control Programmes for Terrestrial and Aquatic Animals in Norway; Annual report; Norwegian Veterinary Institute: Oslo, Norway, 2011. [Google Scholar]
- Yayo Ayele, W.; Macháčková, M.; Pavlík, I. The transmission and impact of paratuberculosis infection in domestic and wild ruminants. Vet. Med. 2001, 46, 205–224. [Google Scholar] [CrossRef] [Green Version]
- Delgado, L.; Juste, R.A.; Muñoz, M.; Morales, S.; Benavides, J.; Ferreras, M.C.; Marín, J.F.; Pérez, V. Differences in the peripheral immune response between lambs and adult ewes experimentally infected with Mycobacterium avium subspecies paratuberculosis. Vet. Immunol. Immunopathol. 2012, 145, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Whittington, R.J.; Marsh, I.B.; Taylor, P.J.; Marshall, D.J.; Taragel, C.; Reddacliff, L.A. Isolation of Mycobacterium avium subsp paratuberculosis from environmental samples collected from farms before and after destocking sheep with paratuberculosis. Aust. Vet. J. 2003, 81, 559–563. [Google Scholar] [CrossRef]
- Morris, C.A.; Hickey, S.M.; Henderson, H.V. The effect of Johne’s disease on production traits in Romney, Merino and Merino x Romney-cross ewes. N. Z. Vet. J. 2006, 54, 204–209. [Google Scholar] [CrossRef]
- Lugton, I.W. Cross-sectional study of risk factors Johne’ s disease on New South Wales. Aust. Vet. J. 2004, 82, 355–365. [Google Scholar] [CrossRef]
- Cocito, C.; Gilot, P.; Coene, M.; de Kesel, M.; Poupart, P.; Vannuffel, P. Paratuberculosis. Clin. Microbiol. Rev. 1994, 7, 328–345. [Google Scholar] [CrossRef] [PubMed]
- Carrigan, M.J.; Seaman, J.T. The pathology of Johne’s disease in sheep. Aust. Vet. J. 1990, 67, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Robbe-Austerman, S. Control of paratuberculosis in small ruminants. Vet. Clin. N. Am. Food Anim. Pract. 2011, 27, 609–620. [Google Scholar] [CrossRef]
- Gezon, H.M.; Bither, H.D.; Gibbs, H.C.; Acker, E.J.; Hanson, L.A.; Thompson, J.K.; Jorgenson, R.D. Identification and control of paratuberculosis in a large goat herd. Am. J. Vet. Res. 1988, 49, 1817–1823. [Google Scholar]
- Pérez, V.; García Marín, J.F.; Badiola, J.J. Description and classification of different types of lesion associated with natural paratuberculosis infection in sheep. J. Comp. Pathol. 1996, 114, 107–122. [Google Scholar] [CrossRef]
- Corpa, J.M.; Garrido, J.; García Marín, J.F.; Pérez, V. Classification of lesions observed in natural cases of paratuberculosis in goats. J. Comp. Pathol. 2000, 122, 255–265. [Google Scholar] [CrossRef]
- Valheim, M.; Storset, A.K.; Aleksersen, M.; Brun-Hansen, H.; McL Press, C. Lesions in subclinical paratuberculosis of goats are associated with persistent gut-associated lymphoid tissue. J. Comp. Pathol. 2002, 127, 194–202. [Google Scholar] [CrossRef]
- Lybeck, K.R.; Løvoll, M.; Johansen, T.B.; Olsen, I.; Storset, A.K.; Valheim, M. Intestinal strictures, fibrous adhesions and high local interleukin-10 levels in goats infected naturally with Mycobacterium avium subsp. paratuberculosis. J. Comp. Pathol. 2013, 148, 157–172. [Google Scholar] [CrossRef]
- Wood, P.L.; Erol, E.; Hoffsis, G.F.; Steinman, M.; De Buck, J. Serum lipidomics of bovine paratuberculosis: Disruption of choline-containing glycerophospholipids and sphingolipids. SAGE Open Med. 2018, 6, 2050312118775302. [Google Scholar] [CrossRef] [Green Version]
- Smeed, J.A.; Watkins, C.A.; Rhind, S.M.; Hopkins, J. Differential cytokine gene expression profiles in the three pathological forms of sheep paratuberculosis. BMC Vet. Res. 2007, 3, 18. [Google Scholar] [CrossRef] [Green Version]
- Whittington, R.J.; Reddacliff, L.A.; Marsh, I.; Mc Allister, S.; Saunders, V. Temporal patterns and quantification of excretion of Mycobacterium avium subsp paratuberculosis in sheep with Johne’s disease. Aust. Vet. J. 2000, 78, 34–37. [Google Scholar] [CrossRef]
- Bannantine, J.P.; Stabel, J.R. Killing of Mycobacterium avium subspecies paratuberculosis within macrophages. BMC Microbiol. 2002, 2, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, D.J.; Vaughan, J.A.; Stiles, P.L.; Noske, P.J.; Tizard, M.L.; Prowse, S.J.; Michalski, W.P.; Butler, K.L.; Jones, S.L. A long-term study in Merino sheep experimentally infected with Mycobacterium avium subsp. paratuberculosis: Clinical disease, faecal culture and immunological studies. Vet. Microbiol. 2004, 104, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.J.; Vaughan, J.A.; Stiles, P.L.; Noske, P.J.; Tizard, M.L.; Prowse, S.J.; Michalski, W.P.; Butler, K.L.; Jones, S.L. A long-term study in Angora goats experimentally infected with Mycobacterium avium subsp. paratuberculosis: Clinical disease, faecal culture and immunological studies. Vet. Microbiol. 2006, 113, 13–24. [Google Scholar] [CrossRef]
- Stewart, D.J.; Vaughan, J.A.; Stiles, P.L.; Noske, P.J.; Tizard, M.L.; Prowse, S.J.; Michalski, W.P.; Butler, K.L.; Jones, S.L. A long-term bacteriological and immunological study in Holstein-Friesian cattle experimentally infected with Mycobacterium avium subsp. paratuberculosis and necropsy culture results for Holstein-Friesian cattle, Merino sheep and Angora goats. Vet. Microbiol. 2007, 122, 83–96. [Google Scholar] [CrossRef]
- Collins, D.M.; Gabric, D.M.; De Lisle, G.W. Identification of two groups of Mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J. Clin. Microbiol. 1990, 28, 1591–1596. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, K.; Hughes, V.M.; De Juan, L.; Inglis, N.F.; Wright, F.; Sharp, J.M. Molecular characterization of pigmented and non pigmented isolates of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 2002, 40, 1798–1804. [Google Scholar] [CrossRef] [Green Version]
- De Juan, L.; Mateos, A.; Domínguez, L.; Sharp, J.M.; Stevenson, K. Genetic diversity of Mycobacterium avium subspecies paratuberculosis isolates from goats detected by pulsed-field gel electrophoresis. Vet. Microbiol. 2005, 106, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Moloney, B.J.; Whittington, R.J. Cross species transmission of ovine Johne’s disease from sheep to cattle: An estimate of prevalence in exposed susceptible cattle. Aust. Vet. J. 2008, 86, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.; Benavides, J.; Sevilla, I.A.; Fuertes, M.; Castaño, P.; Delgado, L.; García Marín, J.F.; Garrido, J.M.; Ferreras, M.C.; Pérez, V. Experimental infection of lambs with C and S-type strains of Mycobacterium avium subspecies paratuberculosis: Immunological and pathological findings. Vet. Res. 2014, 45, 5. [Google Scholar] [CrossRef] [Green Version]
- Momotani, E.; Whipple, D.L.; Thiermann, A.B.; Cheville, N.F. Role of M cells and macrophages in the entrance of Mycobacterium paratuberculosis into domes of ileal Peyer’s patches in calves. Vet. Pathol. 1988, 25, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Sigur-Dardóttir, O.G.; Press, C.M.; Evensen, O. Uptake of Mycobacterium avium subsp. paratuberculosis through the distal small intestinal mucosa in goats: An ultrastructural study. Vet. Pathol. 2001, 38, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, L.E.; Petrofsky, M.; Sommer, S.; Barletta, R.G. Peyer’s patch-deficient mice demonstrate that Mycobacterium avium subsp. paratuberculosis translocates across the mucosal barrier via both M cells and enterocytes but has inefficient dissemination. Infect. Immun. 2010, 78, 3570–3577. [Google Scholar] [CrossRef] [Green Version]
- Sigurethardóttir, O.G.; Valheim, M.; Press, C.M. Establishment of Mycobacterium avium subsp. paratuberculosis infection in the intestine of ruminants. Adv. Drug Deliv Rev. 2004, 56, 819–834. [Google Scholar] [CrossRef]
- Arsenault, R.J.; Maattanen, P.; Daigle, J.; Potter, A.; Griebel, P.; Napper, S. From mouth to macrophage: Mechanisms of innate immune subversion by Mycobacterium avium subsp. paratuberculosis. Vet. Res. 2014, 45, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Secott, T.E.; Lin, T.L.; Wu, C.C. Mycobacterium avium subsp. paratuberculosis fibronectin attachment protein facilitates M-cell targeting and invasion through a fibronectin bridge with host integrins. Infect. Immun. 2004, 72, 3724–3732. [Google Scholar] [CrossRef] [Green Version]
- Khare, S.; Nunes, J.S.; Figueiredo, J.F.; Lawhon, S.D.; Rossetti, C.A.; Gull, T.; Rice-Ficht, A.C.; Adams, L.G. Early phase morphological lesions and transcriptional responses of bovine ileum infected with Mycobacterium avium subsp. paratuberculosis. Vet. Pathol. 2009, 46, 717–728. [Google Scholar] [CrossRef]
- Ponnusamy, D.; Periasamy, S.; Tripathi, B.N.; Pal, A. Mycobacterium avium subsp. paratuberculosis invades through M cells and enterocytes across ileal and jejunal mucosa of lambs. Res. Vet. Sci. 2013, 94, 306–312. [Google Scholar] [CrossRef]
- Whittington, R.J.; Begg, D.J.; De Silva, K.; Plain, K.M.; Purdie, A.C. Comparative immunological and microbiological aspects of paratuberculosis as a model mycobacterial infection. Vet. Immunol. Immunopathol. 2012, 148, 29–47. [Google Scholar] [CrossRef]
- Ben-Ali, M.; Barbouche, M.R.; Bousnina, S.; Chabbou, A.; Dellagi, K. Toll-like receptor 2 Arg677Trp polymorphism is associated with susceptibility to tuberculosis in Tunisian patients. Clin. Diagn Lab. Immunol. 2004, 11, 625–626. [Google Scholar] [CrossRef] [Green Version]
- Thuong, N.T.; Hawn, T.R.; Thwaites, G.E.; Chau, T.T.; Lan, N.T.; Quy, H.T.; Hieu, N.T.; Aderem, A.; Hien, T.T.; Farrar, J.J. A polymorphism in human TLR2 is associated with increased susceptibility to tuberculous meningitis. Genes Immun. 2007, 8, 422–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bochud, P.Y.; Hawn, T.R.; Siddiqui, M.R.; Saunderson, P.; Britton, S.; Abraham, I.; Argaw, A.T.; Janer, M.; Zhao, L.P.; Kaplan, G. Toll-like receptor 2 (TLR2) polymorphisms are associated with reversal reaction in leprosy. J. Infect. Dis. 2008, 197, 253–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khor, C.C.; Chapman, S.J.; Vannberg, F.O.; Dunne, A.; Murphy, C.; Ling, Y.E.; Frodsham, A.J.; Walley, A.J.; Kyrieleis, O.; Khan, A. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat. Genet. 2007, 39, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Tabel, Y.; Berdeli, A.; Mir, S. Association of TLR2 gene Arg753Gln polymorphism with urinary tract infection in children. Int. J. Immunogenet. 2007, 34, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.L.; Zhong, L.; Begg, D.J.; De Silva, K.; Whittington, R.J. Toll-like receptor genes are differentially expressed at the sites of infection during the progression of Johne’s disease in outbred sheep. Vet. Immunol. Immunopathol. 2008, 124, 132–151. [Google Scholar] [CrossRef]
- De Silva, K.; Begg, D.; Whittington, R. The interleukin 10 response in ovine Johne’s disease. Vet. Immunol. Immunopathol. 2011, 139, 10–16. [Google Scholar] [CrossRef]
- Sohal, J.S.; Singh, S.V.; Tyagi, P.; Subhodh, S.; Singh, P.K.; Singh, A.V.; Narayanasamy, K.; Sheoran, N.; Sandhu, K.S. Immunology of mycobacterial infections: With special reference to Mycobacterium avium subspecies paratuberculosis. Immunobiology 2008, 213, 585–598. [Google Scholar] [CrossRef]
- Periasamy, S.; Tripathi, B.N.; Singh, N. Mechanisms of Mycobacterium avium subsp. paratuberculosis induced apoptosis and necrosis in bovine macrophages. Vet. Microb. 2013, 165, 392–401. [Google Scholar] [CrossRef]
- Allen, S.; Sotos, J.; Sylte, M.J.; Czuprynski, C.J. Use of Hoechst 33342 staining to detect apoptotic changes in bovine mononuclear phagocytes infected with Mycobacterium avium subsp. paratuberculosis. Clin. Diagn Lab. Immunol. 2001, 8, 460–464. [Google Scholar] [CrossRef] [Green Version]
- Kabara, E.; Coussens, P.M. Infection of primary bovine macrophages with Mycobacterium avium subspecies paratuberculosis suppresses host cell apoptosis. Front. Microb. 2012, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Coussens, P.M.; Jeffers, A.; Colvin, C. Rapid and transient activation of gene expression in peripheral blood mononuclear cells from Johne’s disease positive cows exposed to Mycobacterium paratuberculosis in vitro. Microb Pathog. 2004, 36, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Sommer, S.; Pudrith, C.B.; Colvin, C.J.; Coussens, P.M. Mycobacterium avium subspecies paratuberculosis suppresses expression of IL-12p40 and iNOS genes induced by signalling through CD40 in bovine monocyte-derived macrophages. Vet. Immunol. Immunopathol. 2009, 128, 44–52. [Google Scholar] [CrossRef]
- Begg, D.J.; De Silva, K.; Carter, N.; Plain, K.M.; Purdie, A.; Whittington, R.J. Does a Th1 over Th2 dominancy really exist in the early stages of Mycobacterium avium subspecies paratuberculosis infections? Immunobiology 2011, 216, 840–846. [Google Scholar] [CrossRef]
- Gillan, S.; O’Brien, R.; Hughes, A.D.; Griffin, J.F. Identification of immune parameters to differentiate disease states among sheep infected with Mycobacterium avium subsp. paratuberculosis. Clin. Vaccine Immunol. 2010, 17, 108–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddacliff, L.A.; McClure, S.J.; Whittington, R.J. Immunoperoxidase studies of cell mediated immune effector cell populations in early Mycobacterium avium subsp. paratuberculosis infection in sheep. Vet. Immunol. Immunopathol. 2004, 97, 149–162. [Google Scholar] [CrossRef]
- Pérez, V.; Tellechea, J.; Corpa, J.M.; Gutiérrez, M.; García Marín, J.F. Relation between pathologic findings and cellular immune responses in sheep with naturally acquired paratuberculosis. Am. J. Vet. Res. 1999, 60, 123–127. [Google Scholar]
- Storset, A.K.; Berg, I.; Djønne, B. Evaluation of the gamma interferon test for diagnosis of paratuberculosis in goats. Vet. Immunol. Immunopathol. 2005, 107, 87–94. [Google Scholar] [CrossRef]
- Lybeck, K.R.; Storset, A.K.; Olsen, I. Neutralization of interleukin-10 from CD14(+) monocytes enhances gamma interferon production in peripheral blood mononuclear cells from Mycobacterium avium subsp. paratuberculosis-infected goats. Clin. Vaccine Immunol. 2009, 16, 1003–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coussens, P.M.; Sipkovsky, S.; Murphy, B.; Roussey, J.; Colvin, C.J. Regulatory T cells in cattle and their potential role in bovine paratuberculosis. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 233–239. [Google Scholar] [CrossRef]
- Buczinski, S.; Arsenault, J.; Kostoulas, P.; Corbière, F.; Fecteau, G.; Dendukuri, N. Accuracy of paratuberculosis diagnostic tests in small ruminants: Protocol for a systematic review and meta-analysis. Anim. Health Res. Rev. 2019, 20, 98–102. [Google Scholar] [CrossRef]
- Bauman, C.A.; Jones-Bitton, A.; Menzies, P.; Toft, N.; Jansen, J.; Kelton, D. Prevalence of paratuberculosis in the dairy goat and dairy sheep industries in Ontario, Canada. Can. Vet. J. 2016, 57, 169–175. [Google Scholar]
- Gwozdz, J.M.; Thompson, K.G.; Murray, A.; West, D.M.; Manktelow, B.W. Use of the polymerase chain reaction assay for the detection of Mycobacterium avium subspecies paratuberculosis in blood and liver biopsies from experimentally infected sheep. Aust. Vet. J. 2000, 78, 622–624. [Google Scholar] [CrossRef]
- Sergeant, E.S.; Marshall, D.J.; Eamens, G.J.; Kearns, C.; Whittington, R.J. Evaluation of an absorbed ELISA and an agar-gel immuno-diffusion test for ovine paratuberculosis in sheep in Australia. Prev. Vet. Med. 2003, 61, 235–248. [Google Scholar] [CrossRef]
- Hope, A.F.; Kluver, P.F.; Jones, S.L.; Condron, R.J. Sensitivity and specificity of two serological tests for the detection of ovine paratuberculosis. Aust. Vet. J. 2000, 78, 850–856. [Google Scholar] [CrossRef]
- Vazquez, P.; Garrido, J.M.; Juste, R.A. Specific antibody and interferon-gamma responses associated with immunopathological forms of bovine paratuberculosis in slaughtered Friesian cattle. PLoS ONE 2013, 8, e64568. [Google Scholar] [CrossRef] [PubMed]
- Stabel, J.R.; Bannantine, J.P. Development of a Nested PCR Method Targeting a Unique Multicopy Element, ISMap 02, for Detection of Mycobacterium avium subsp. paratuberculosis in Fecal Samples. J. Clin. Microb. 2005, 43, 4744–4750. [Google Scholar] [CrossRef] [Green Version]
- Marquetoux, N.; Mitchell, R.; Ridler, A.; Heuer, C.; Wilson, P. A synthesis of the patho-physiology of Mycobacterium avium subspecies paratuberculosis infection in sheep to inform mathematical modelling of ovine paratuberculosis. Vet. Res. 2018, 49, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nascimento, E.J.; Braga-Neto, U.; Calzavara-Silva, C.E.; Gomes, A.L.; Abath, F.G.; Brito, C.A.A.; Cordeiro, M.T.; Silva, A.M.; Magalhães, C.; Andrade, R. Gene expression profiling during early acute febrile stage of dengue infection can predict the disease outcome. PLoS ONE 2009, 4, e7892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, P.; Peter, S.; Jung, M.; Lewin, A.; Hemmrich-Stanisak, G.; Franke, A.; von Kleist, M.; Schütte, C.; Einspanier, R.; Sharbati, S. Analysis of long non-coding RNA and mRNA expression in bovine macrophages brings up novel aspects of Mycobacterium avium subspecies paratuberculosis infections. Sci. Rep. 2019, 9, 1571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, M.D.; de Silva, K.; Plain, K.M.; Whittington, R.J.; Purdie, A.C. Mycobacterium avium subspecies paratuberculosis is able to manipulate host lipid metabolism and accumulate cholesterol within macrophages. Microb. Pathog. 2019, 130, 44–53. [Google Scholar] [CrossRef]
- Cha, S.B.; Yoo, A.; Park, H.T.; Sung, K.Y.; Shin, M.K.; Yoo, H.S. Analysis of transcriptional profiles to discover biomarker candidates in Mycobacterium avium subsp. paratuberculosis-infected macrophages, RAW 264.7. J. Microbiol. Biotechnol. 2013, 23, 1167–1175. [Google Scholar] [CrossRef]
- Weiss, D.J.; Evanson, O.A.; Deng, M.; Abrahamsen, M.S. Gene expression and antimicrobial activity of bovine macrophages in response to Mycobacterium avium subsp. paratuberculosis. Vet. Pathol. 2004, 41, 326–337. [Google Scholar] [CrossRef] [Green Version]
- Coussens, P.M.; Colvin, C.J.; Wiersma, K.; Abouzied, A.; Sipkovsky, S. Gene expression profiling of peripheral blood mononuclear cells from cattle infected with Mycobacterium paratuberculosis. Infect. Immun. 2002, 70, 5494–5502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thirunavukkarasu, S.; Plain, K.M.; de Silva, K.; Begg, D.; Whittington, R.J.; Purdie, A.C. Expression of genes associated with cholesterol and lipid metabolism identified as a novel pathway in the early pathogenesis of Mycobacterium avium subspecies paratuberculosis-infection. Vet. Immunol. Immunopathol. 2014, 160, 147–157. [Google Scholar] [CrossRef]
- Thirunavukkarasu, S.; De Silva, K.; Whittington, R.J.; Plain, K.M. In vivo and in vitro expression pattern of Toll-like receptors in Mycobacterium avium subspecies paratuberculosis infection. Vet. Immunol. Immunopathol. 2013, 156, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Smeed, J.A.; Watkins, C.A.; Gossner, A.G.; Hopkins, J. Expression profiling reveals differences in immuno-inflammatory gene expression between the two disease forms of sheep paratuberculosis. Vet. Immunol. Immunopathol. 2010, 135, 218–225. [Google Scholar] [CrossRef] [Green Version]
- Malvisi, M.; Curti, N.; Remondini, D.; De Iorio, M.G.; Palazzo, F.; Gandini, G.; Vitali, S.; Polli, M.; Williams, J.L.; Minozzi, G. Combinatorial Discriminant Analysis Applied to RNAseq Data Reveals a Set of 10 Transcripts as Signatures of Exposure of Cattle to Mycobacterium avium subsp. paratuberculosis. Animals 2020, 10, 253. [Google Scholar] [CrossRef] [Green Version]
- Plain, K.M.; Purdie, A.C.; Begg, D.J.; de Silva, K.; Whittington, R.J. Toll-like receptor (TLR) 6 and TLR1 differentiation in gene expression studies of Johne’s disease. Vet. Immunol. Immunopathol. 2010, 137, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Nalubamba, K.; Smeed, J.; Gossner, A.; Watkins, C.; Dalziel, R.; Hopkins, J. Differential expression of pattern recognition receptors in the three pathological forms of sheep paratuberculosis. Microbes Infect. 2008, 10, 598–604. [Google Scholar] [CrossRef] [Green Version]
- Arsenault, R.J.; Li, Y.; Maattanen, P.; Scruten, E.; Doig, K.; Potter, A.; Griebel, P.; Kusalik, A.; Napper, S. Altered Toll-like receptor 9 signaling in Mycobacterium avium subsp. paratuberculosis-infected bovine monocytes reveals potential therapeutic targets. Infect. Immun. 2013, 81, 226–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, E.K.; Yang, C.S.; Choi, C.H.; Harding, C.V. Intracellular signalling cascades regulating innate immune responses to Mycobacteria: Branching out from Toll-like receptors. Cell Microbiol. 2007, 9, 1087–1098. [Google Scholar] [CrossRef]
- Basu, J.; Shin, D.M.; Jo, E.K. Mycobacterial signaling through toll-like receptors. Front. Cell Infect. Microbiol. 2012, 2, 145. [Google Scholar] [CrossRef] [Green Version]
- Purdie, A.C.; Plain, K.M.; Begg, D.J.; de Silva, K.; Whittington, R.J. Expression of genes associated with the antigen presentation and processing pathway are consistently regulated in early Mycobacterium avium subsp. paratuberculosis infection. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Plain, K.M.; Begg, D.J.; de Silva, K.; Purdie, A.C.; Whittington, R.J. Enhancement of the interferon gamma assay to detect paratuberculosis using interleukin-7 and interleukin-12 potentiation. Vet. Immunol. Immunopathol. 2012, 149, 28–37. [Google Scholar] [CrossRef]
- Stabel, J.R.; Bannantine, J.P.; Hostetter, J.M. Comparison of Sheep, Goats, and Calves as Infection Models for Mycobacterium avium subsp. paratuberculosis. Vet. Immunol. Immunopathol. 2020, 225, 110060. [Google Scholar] [CrossRef]
- Gossner, A.G.; Venturina, V.M.; Peers, A.; Watkins, C.A.; Hopkins, J. Expression of sheep interleukin 23 (IL23A, alpha subunit p19) in two distinct gastrointestinal diseases. Vet. Immunol. Immunopathol. 2012, 150, 118–122. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, Y.; Suda, T.; Asada, K.; Miwa, S.; Suzuki, M.; Fujie, M.; Furuhashi, K.; Nakamura, Y.; Inui, N.; Shirai, T. Serum indoleamine 2,3-dioxygenase activity predicts prognosis of pulmonary tuberculosis. Clin. Vaccine Immunol. 2012, 19, 436–442. [Google Scholar] [CrossRef]
- Plain, K.M.; de Silva, K.; Earl, J.; Begg, D.J.; Purdie, A.C.; Whittington, R.J. Indoleamine 2,3-dioxygenase, tryptophan catabolism, and Mycobacterium avium subsp. paratuberculosis: A model for chronic mycobacterial infections. Infect. Immun. 2011, 79, 3821–3832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.E.; Park, H.T.; Jung, Y.H.; Yoo, H.S. Gene expression profiles of immune-regulatory genes in whole blood of cattle with a subclinical infection of Mycobacterium avium subsp. paratuberculosis. PLoS ONE 2018, 13, e0196502. [Google Scholar]
- Singh, P.K.; Singh, A.V.; Chauhan, D.S. Current understanding on micro RNAs and its regulation in response to Mycobacterial infections. J. Biomed. Sci. 2013, 20, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, P.K.; Singh, S.V.; Saxena, V.K.; Singh, M.K.; Singh, A.V.; Sohal, J.S. Expression profiles of different cytokine genes in peripheral blood mononuclear cells of goats infected experimentally with native strain of Mycobacterium avium subsp. paratuberculosis. Anim. Biotechnol. 2013, 24, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Singh, S.V.; Kumar, H.; Sohal, J.S.; Singh, A.V. Diagnostic Application of IS900 PCR Using Blood as a Source Sample for the Detection of Mycobacterium avium Subspecies Paratuberculosis in Early and Subclinical Cases of Caprine Paratuberculosis. Vet. Med. Int. 2010, 2010, 748621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velu, V.K.; Ramesh, R.; Srinivasan, A.R. Circulating MicroRNAs as Biomarkers in Health and Disease. J. Clin. Diagn Res. 2012, 6, 1791–1795. [Google Scholar] [CrossRef] [PubMed]
- Lima, R.T.; Busacca, S.; Almeida, G.M.; Gaudino, G.; Fennell, D.A.; Vasconcelos, M.H. MicroRNA regulation of core apoptosis pathways in cancer. Eur. J. Cancer 2011, 47, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Rajewsky, N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007, 8, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Swarup, V.; Rajeswari, M.R. Circulating (cell-free) nucleic acids—A promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007, 581, 795–799. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Gu, Z.; Jiang, H. MicroRNAs in farm animals. Animal 2013, 7, 1567–1575. [Google Scholar] [CrossRef] [Green Version]
- McBride, D.; Carré, W.; Sontakke, S.D.; Hogg, C.O.; Law, A.; Donadeu, F.X.; Clinton, M. Identification of miRNAs associated with the follicular-luteal transition in the ruminant ovary. Reproduction 2012, 144, 221–233. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Zhao, F.; Wei, C.; Sheng, X.; Ren, H.; Xu, L.; Lu, J.; Liu, J.; Zhang, L.; Du, L. Identification and characterization of the miRNA transcriptome of Ovis aries. PLoS ONE 2013, 8, e58905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, C.; Fang, X.; Zhao, Y.; Chen, X.; Sun, J.; Zhou, Y.; Wang, J.; Wang, Y.; Lan, X. Identification and profiling of microRNAs and their target genes from developing caprine skeletal Muscle. PLoS ONE 2014, 9, e96857. [Google Scholar] [CrossRef] [Green Version]
- Yuan, C.; Wang, X.; Geng, R.; He, X.; Qu, L.; Chen, Y. Discovery of cashmere goat (Capra hircus) microRNAs in skin and hair follicles by Solexa sequencing. BMC Genom. 2013, 14, 511. [Google Scholar] [CrossRef] [Green Version]
- Wenguang, Z.; Jianghong, W.; Jinquan, L.; Yashizawa, M. A subset of skin-expressed microRNAs with possible roles in goat and sheep hair growth based on expression profiling of mammalian microRNAs. OMICS 2007, 11, 385–396. [Google Scholar] [CrossRef]
- Hossain, M.M.; Sohel, M.M.; Schellander, K.; Tesfaye, D. Characterization and importance of microRNAs in mammalian gonadal functions. Cell Tissue Res. 2012, 349, 679–690. [Google Scholar] [CrossRef]
- Baril, P.; Ezzine, S.; Pichon, C. Monitoring the spatiotemporal activities of miRNAs in small animal models using molecular imaging modalities. Int. J. Mol. Sci. 2015, 16, 4947–4972. [Google Scholar] [CrossRef] [Green Version]
- Sontakke, S.D.; Mohammed, B.T.; McNeilly, A.S.; Donadeu, F.X. Characterization of microRNAs differentially expressed during bovine follicle development. Reproduction 2014, 148, 271–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Linher-Melville, K.; Yang, B.B.; Wu, D.; Li, J. Micro-RNA378 (miR-378) regulates ovarian estradiol production by targeting aromatase. Endocrinology 2011, 152, 3941–3951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melar-New, M.; Laimins, L.A. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J. Virol. 2010, 84, 5212–5221. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef]
- Quinn, S.R.; O’Neill, L.A. A trio of microRNAs that control Toll-like receptor signalling. Int. Immunol. 2011, 23, 421–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, C.P.; He, L.; Tsai, Y.C.; Peng, S.; Kang, T.H.; Pang, X.; Monie, A.; Hung, C.F.; Wu, T.C. In vivo microRNA-155 expression influences antigen-specific T cell-mediated immune responses generated by DNA vaccination. Cell Biosci. 2011, 1, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lind, E.F.; Elford, A.R.; Ohashi, P.S. Micro-RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J. Immunol. 2013, 190, 1210–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedersen, I.; David, M. MicroRNAs in the immune response. Cytokine 2008, 43, 391–394. [Google Scholar] [CrossRef] [Green Version]
- Russo, A.; Potenza, N. Antiviral effects of human microRNAs and conservation of their target sites. FEBS Lett. 2011, 585, 2551–2555. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Schulte, L.; Vogel, J. The mammalian microRNA response to bacterial infections. RNA Biol. 2012, 9, 742–750. [Google Scholar] [CrossRef]
- Vegh, P.; Foroushani, A.B.; Magee, D.A.; McCabe, M.S.; Browne, J.A.; Nalpas, N.C.; Conlon, K.M.; Gordon, S.V.; Bradley, D.G.; MacHugh, D.E. Profiling microRNA expression in bovine alveolar macrophages using RNA-seq. Vet. Immunol. Immunopathol. 2013, 155, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.T.; Wheelwright, M.; Teles, R.; Komisopoulou, E.; Edfeldt, K.; Ferguson, B.; Mehta, M.D.; Vazimia, A.; Rea, T.H.; Samo, E.N. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat. Med. 2012, 18, 267–273. [Google Scholar] [CrossRef]
- Gupta, S.K.; Maclean, P.H.; Ganesh, S.; Shu, D.; Buddle, B.M.; Wedlock, D.N.; Heiser, A. Detection of microRNA in cattle serum and their potential use to diagnose severity of Johne’s disease. J. Dairy Sci. 2018, 101, 10259–10270. [Google Scholar] [CrossRef] [PubMed]
- Farrell, D.; Shaughnessy, R.G.; Britton, L.; MacHugh, D.E.; Markey, B.; Gordon, S.V. The identification of circulating MiRNA in bovine serum and their potential as novel biomarkers of early Mycobacterium avium subsp paratuberculosis infection. PLoS ONE 2015, 10, e0134310. [Google Scholar] [CrossRef] [Green Version]
- Malvisi, M.; Palazzo, F.; Morandi, N.; Lazzari, B.; Williams, J.L.; Pagnacco, G.; Minozzi, G. Responses of Bovine Innate Immunity to Mycobacterium avium subsp. paratuberculosis Infection Revealed by Changes in Gene Expression and Levels of MicroRNA. PLoS ONE 2016, 11, e0164461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purdie, A.C.; Plain, K.M.; Begg, D.J.; de Silva, K.; Whittington, R.J. Gene expression profiles during subclinical Mycobacterium avium subspecies paratuberculosis infection in sheep can predict disease outcome. Sci. Rep. 2019, 9, 8245. [Google Scholar] [CrossRef]
- Berry, A.; Wu, C.W.; Venturino, A.J.; Talaat, A.M. Biomarkers for Early Stages of Johne’s Disease Infection and Immunization in Goats. Front. Microbiol. 2018, 9, 2284. [Google Scholar] [CrossRef]
- Van den Esker, M.H.; Koets, A.P. Application of Transcriptomics to Enhance Early Diagnostics of Mycobacterial Infections, with an Emphasis on Mycobacterium avium ssp. paratuberculosis. Vet. Sci. 2019, 6, 59. [Google Scholar] [CrossRef] [Green Version]
- Röcken, C.; Ebert, M.P.; Roessner, A. Proteomics in pathology, research and practice. Path Res. Pract. 2004, 200, 69–82. [Google Scholar] [CrossRef]
- Lee, P.Y.; Osman, J.; Low, T.Y.; Jamal, R. Plasma/serum proteomics: Depletion strategies for reducing high-abundance proteins for biomarker discovery. Bioanalysis 2019, 11, 1799–1812. [Google Scholar] [CrossRef]
- Liumbruno, G.; D’Alessandro, A.; Grazzini, G.; Zolla, L. Blood-related proteomics. J. Proteomics. 2010, 73, 483–507. [Google Scholar] [CrossRef]
- Petricoin, E.F.; Belluco, C.; Araujo, R.P.; Liotta, L.A. The blood peptidome: A higher dimension of information content for cancer biomarker discovery. Nat. Rev. Cancer 2006, 6, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Liotta, L.A.; Ferrari, M.; Petricoin, E. Clinical proteomics: Written in blood. Nature 2003, 425, 905. [Google Scholar] [CrossRef] [Green Version]
- Rioux, M.C.; Carmona, C.; Acosta, D.; Ward, B.; Ndao, M.; Gibbs, B.F.; Bennett, H.P.; Spithill, T.W. Discovery and validation of serum biomarkers expressed over the first twelve weeks of Fasciola hepatica infection in sheep. Int. J. Parasitol. 2008, 38, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Taylor, D.L.; Whittington, R.J. Proteomic profiling of ovine serum by SELDI-TOF MS: Optimisation, reproducibility and feasibility of biomarker discovery using routinely collected samples. Comp. Immunol. Microbiol. Infect. Dis. 2010, 33, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Taylor, D.; Begg, D.J.; Whittington, R.J. Biomarker discovery for ovine paratuberculosis (Johne’s disease) by proteomic serum profiling. Comp. Immunol. Microbiol. Infect. Dis. 2011, 34, 315–326. [Google Scholar] [CrossRef]
- Power, D.M.; Elias, N.P.; Richardson, S.J.; Mendes, J.; Soares, C.M.; Santos, C.R. Evolution of the thyroid hormone-binding protein, transthyretin. Gen. Comp. Endocrinol. 2000, 119, 241–255. [Google Scholar]
- Seth, M.; Lamont, E.A.; Janagama, H.K.; Widdel, A.; Vulchanova, L.; Stabel, J.R.; Waters, W.R.; Palmer, M.V.; Sreevatsan, S. Biomarker discovery in subclinical mycobacterial infections of cattle. PLoS ONE 2009, 4, e5478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agranoff, D.; Fernandez-Reyes, D.; Papadopoulos, M.C.; Rojas, S.A.; Herbster, M.; Loosemore, A.; Tarelli, E.; Sheldon, J.; Schwenk, A.; Pollok, R. Identification of diagnostic markers for tuberculosis by proteomic fingerprinting of serum. Lancet 2006, 368, 1012–1021. [Google Scholar] [CrossRef]
- Crowle, A.J.; Ross, E.J. Inhibition by Retinoic Acid of Multiplication of Virulent Tubercle Bacilli in Cultured Human Macrophages. Infect. Immun. 1989, 57, 840–844. [Google Scholar] [CrossRef] [Green Version]
- Yamada, H.; Mizuno, S.; Ross, A.C.; Sugawara, I. Retinoic acid therapy attenuates the severity of tuberculosis while altering lymphocyte and macrophage numbers and cytokine expression in rats infected with Mycobacterium tuberculosis. J. Nutr. 2007, 137, 2696–2700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, Q.; Verschoor, C.P.; Pant, S.D.; Macri, J.; Kirby, G.M.; Karrow, N.A. Proteomic analysis of plasma from Holstein cows testing positive for Mycobacterium avium subsp. paratuberculosis (MAP). Vet. Immunol. Immunopathol. 2012, 148, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Gomollón, F.; Gisbert, J.P. Anemia and inflammatory bowel diseases. World J. Gastroenterol. 2009, 15, 4659–4665. [Google Scholar] [CrossRef] [PubMed]
- Owczarek, D.; Cibor, D.; Głowacki, M.K.; Rodacki, T.; Mach, T. Inflammatory bowel disease: Epidemiology, pathology and risk factors for hypercoagulability. World J. Gastroenterol. 2014, 20, 53–63. [Google Scholar] [CrossRef]
- Piras, C.; Soggiu, A.; Bonizzi, L.; Greco, V.; Ricchi, M.; Arrigoni, N.; Bassols, A.; Urbani, A.; Roncada, P. Identification of immunoreactive proteins of Mycobacterium avium subsp. paratuberculosis. Proteomics 2015, 15, 813–823. [Google Scholar] [CrossRef]
- Nagabhushanam, V.; Praszkier, J.; Cheers, C. Molecular and immunological characterization of Mycobacterium avium 65 kDa heat shock protein (Hsp65). Immunol. Cell Biol. 2001, 79, 454–461. [Google Scholar] [CrossRef]
- Miyata, M.; Kogure, A.; Sato, H.; Kodama, E.; Watanabe, H.; Ohira, H.; Kuroda, M.; Takagi, T.; Sato, Y.; Kasukawa, R. Detection of antibodies to 65 KD heat shock protein and to human superoxide dismutase in autoimmune hepatitis-molecular mimicry between 65 KD heat shock protein and superoxide dismutase. Clin. Rheumatol. 1995, 14, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Rajaiah, R.; Moudgil, K.D. Heat shock protein can promote as well as regulate autoimmunity. Autoimmun. Rev. 2010, 8, 388–393. [Google Scholar] [CrossRef] [Green Version]
- Dow, C.T.M. paratuberculosis Heat Shock Protein 65 and Human Diseases: Bridging Infection and Autoimmunity. Autoimmune Dis. 2012, 2012, 150824. [Google Scholar] [PubMed] [Green Version]
- Phillips, I.L.; Danelishvili, L.; Bermudez, L.E. Macrophage Proteome Analysis at Different Stages of Mycobacterium avium Subspecies paratuberculosis Infection Reveals a Mechanism of Pathogen Dissemination. Proteomes 2021, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Pisanu, S.; Cubeddu, T.; Uzzau, S.; Rocca, S.; Addis, M.F. Proteomic changes in the ileum of sheep infected with Mycobacterium avium subspecies paratuberculosis. Vet. J. 2017, 219, 1–3. [Google Scholar] [CrossRef]
- Pisanu, S.; Cubeddu, T.; Cacciotto, C.; Pilicchi, Y.; Pagnozzi, D.; Uzzau, S.; Rocca, S.; Addis, M.F. Characterization of paucibacillary ileal lesions in sheep with subclinical active infection by Mycobacterium avium subsp. paratuberculosis. Vet. Res. 2018, 49, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiorentina, P.; Martino, C.; Mancini, Y.; De Iorio, M.G.; Williams, J.L.; Minozzi, G. Using Omics Approaches in the Discovery of Biomarkers for Early Diagnosis of Johne’s Disease in Sheep and Goats. Animals 2021, 11, 1912. https://doi.org/10.3390/ani11071912
Fiorentina P, Martino C, Mancini Y, De Iorio MG, Williams JL, Minozzi G. Using Omics Approaches in the Discovery of Biomarkers for Early Diagnosis of Johne’s Disease in Sheep and Goats. Animals. 2021; 11(7):1912. https://doi.org/10.3390/ani11071912
Chicago/Turabian StyleFiorentina, Palazzo, Camillo Martino, Ylenia Mancini, Maria Grazia De Iorio, John L. Williams, and Giulietta Minozzi. 2021. "Using Omics Approaches in the Discovery of Biomarkers for Early Diagnosis of Johne’s Disease in Sheep and Goats" Animals 11, no. 7: 1912. https://doi.org/10.3390/ani11071912
APA StyleFiorentina, P., Martino, C., Mancini, Y., De Iorio, M. G., Williams, J. L., & Minozzi, G. (2021). Using Omics Approaches in the Discovery of Biomarkers for Early Diagnosis of Johne’s Disease in Sheep and Goats. Animals, 11(7), 1912. https://doi.org/10.3390/ani11071912






