Comparative Microbial Profiles of Colonic Digesta between Ningxiang Pig and Large White Pig
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Housing and Experimental Design
2.2. Sample Collection and Preparation
2.3. Carcass and IMF
2.4. DNA Extraction, 16S rRNA Sequencing and Bioinformatics Analysis
2.5. Metagenomic Functional Predictions
2.6. Analysis of SCFAs
2.7. Statistical Analysis
3. Results
3.1. Growth Performance and Carcass Traits
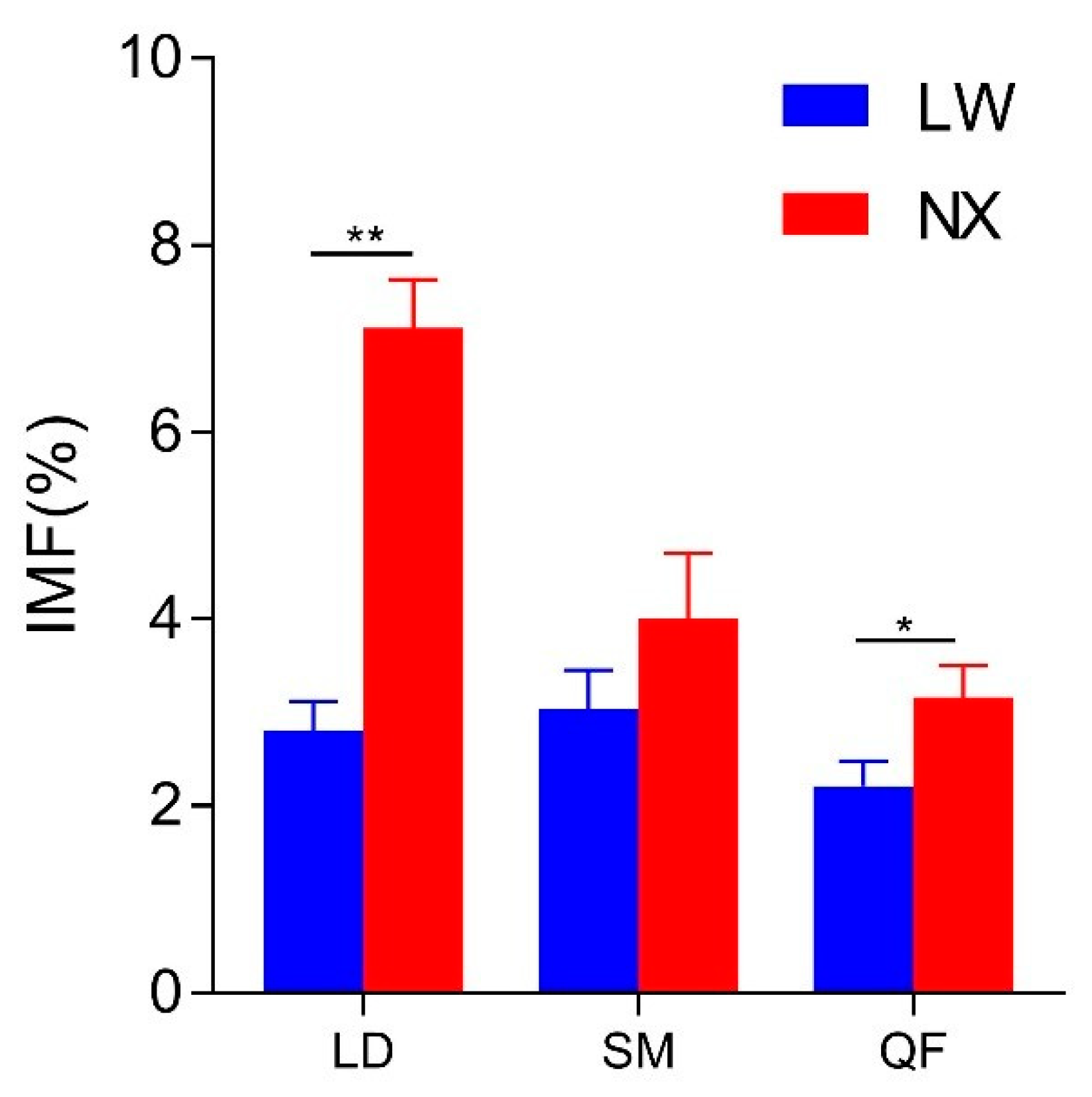
3.2. IMF Content
3.3. Microbiome Sequencing
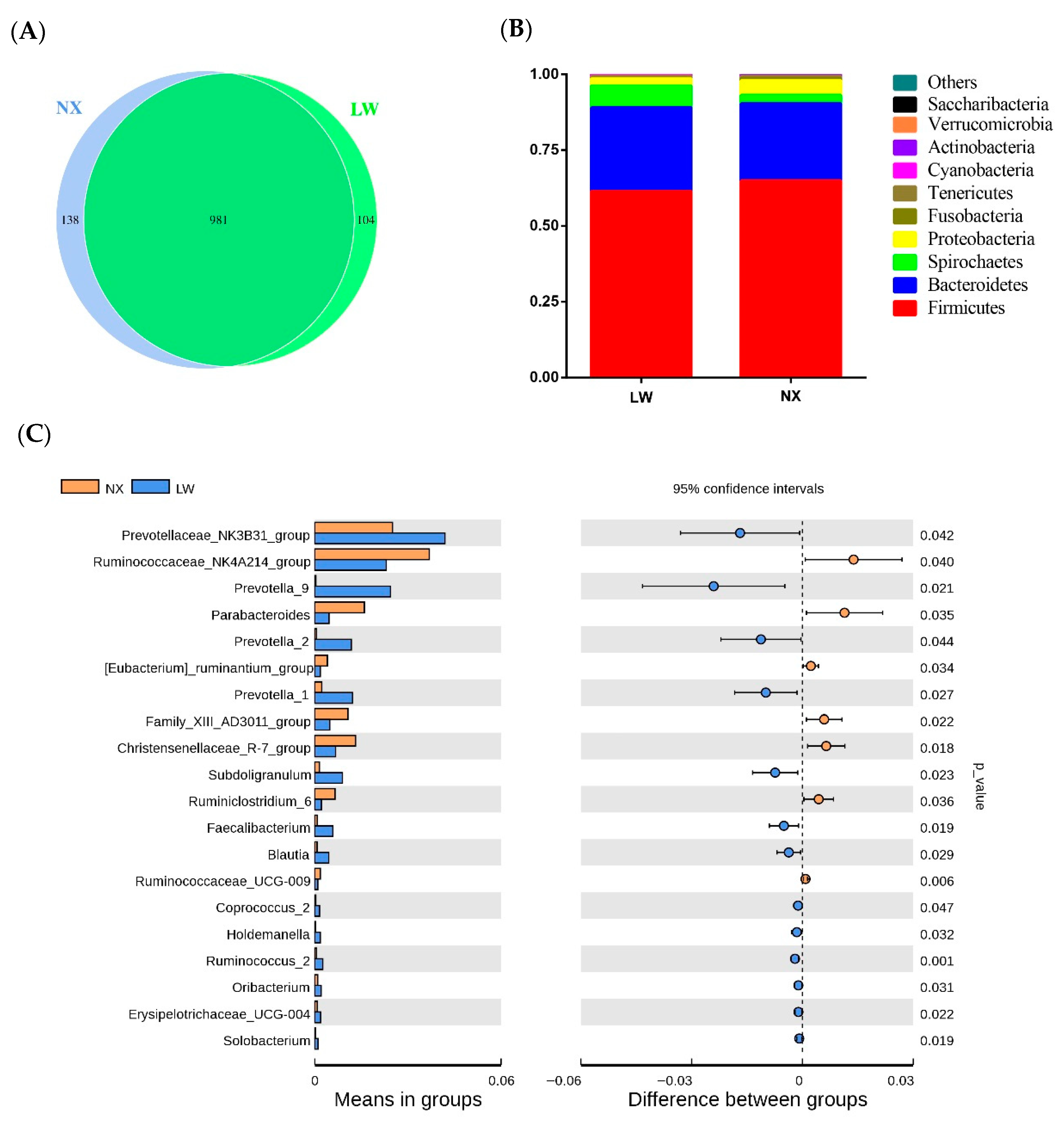
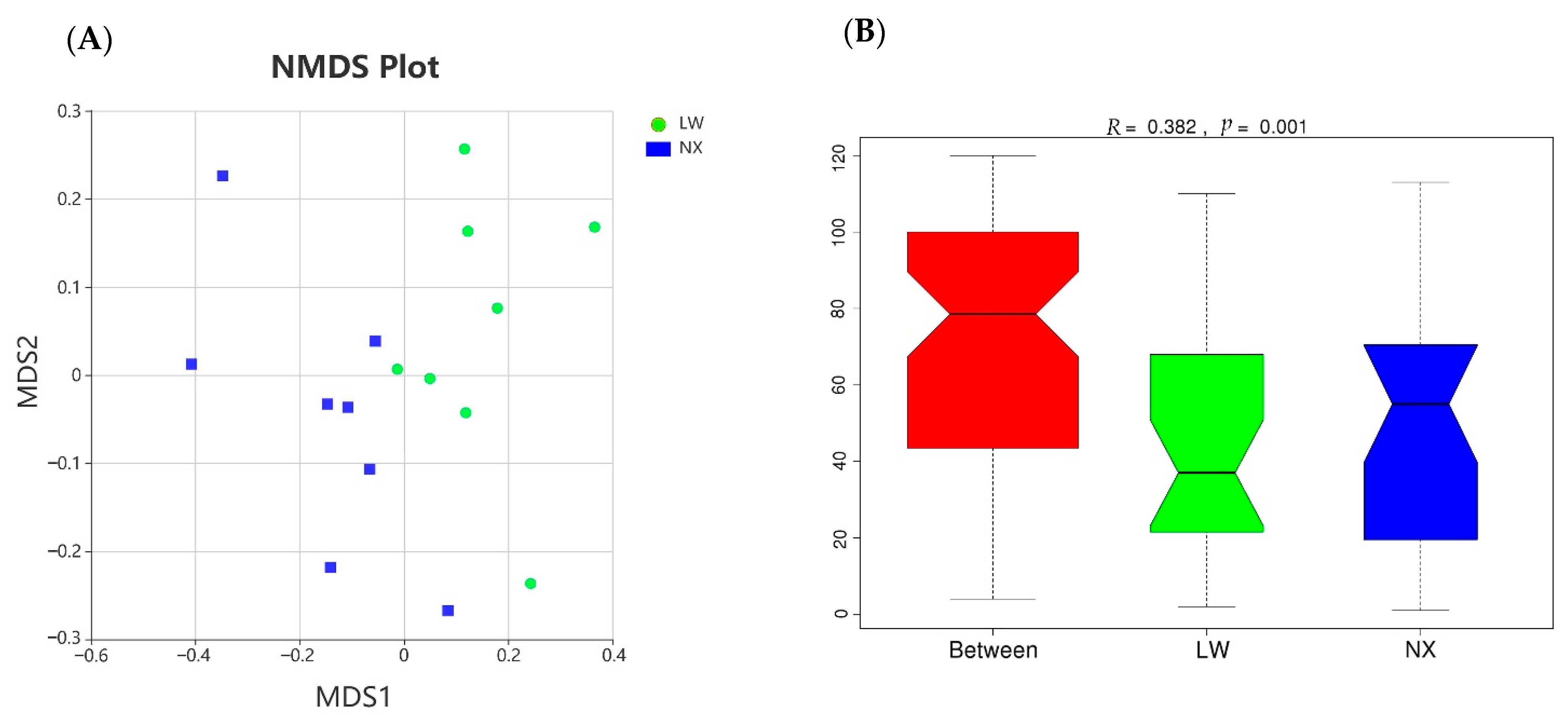
3.4. Bacterial Composition and Diversity
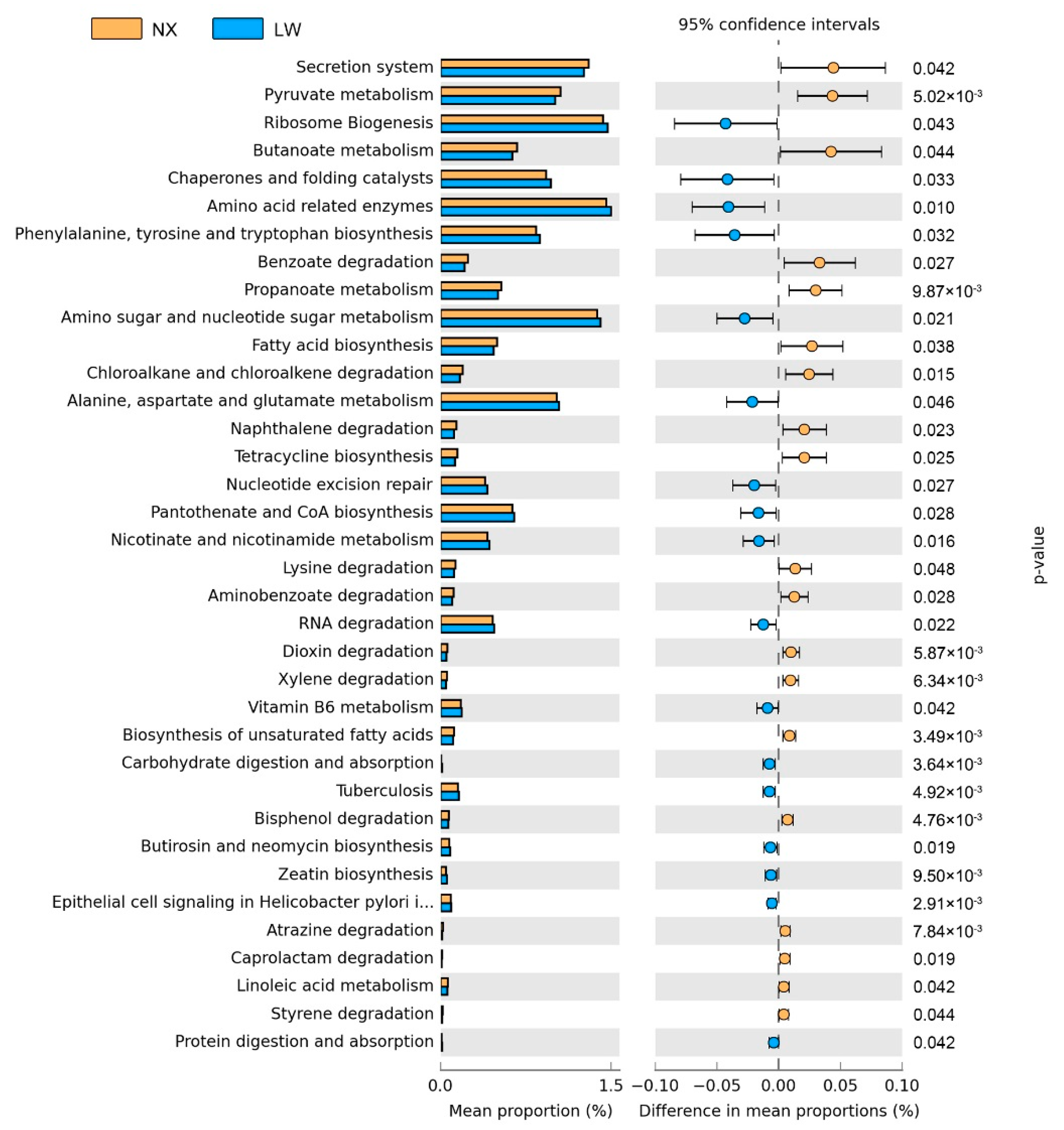
3.5. Functions of Colonic Microbiota
3.6. SCFAs Content in the Colon
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keenan, D.F. Pork meat quality, production and processing on. Encycl. Food Health 2016, 419–431. [Google Scholar] [CrossRef]
- Hausman, G.J.; Dodson, M.V.; Ajuwon, K.; Azain, M.; Barnes, K.M.; Guan, L.L.; Jiang, Z.; Poulos, S.P.; Sainz, R.D.; Smith, S. The biology and regulation of preadipocytes and adipocytes in meat animals. J. Anim. Sci. 2009, 87, 1218–1246. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Du, M.; Jiang, Z.; Hausman, G.J.; Zhang, L.; Dodson, M.V. Long noncoding RNAs in regulating adipogenesis: New rnas shed lights on obesity. Cell. Mol. Life. Sci. 2016, 73, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdes, A.M.; Jens, W.; Eran, S.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Ananthakrishnan, A.N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Portune, K.J.; Benítez-Páez, A.; del Pulgar, E.M.G.; Cerrudo, V.; Sanz, Y. Gut microbiota, diet, and obesity-related disorders-the good, the bad, and the future challenges. Mol. Nutr. Food Res. 2016, 61, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Kallus, S.J.; Brandt, L.J. The intestinal microbiota and obesity. J. Clin. Gastroenterol. 2012, 46, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L. The gut microbiota and obesity: From correlation to causality. Nat. Rev. Microbiol. 2013, 11, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Wilson, I.D. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 2005, 3, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.S.; Bloom, S.R. The regulation of food intake by the gut-brain axis: Implications for obesity. Int. J. Obes. 2013, 37, 625–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bovo, S.; Mazzoni, G.; Galimberti, G.; Calò, D.G.; Fanelli, F.; Mezzullo, M.; Schiavo, G.; Manisi, A.; Trevisi, P.; Bosi, P. Metabolomics evidences plasma and serum biomarkers differentiating two heavy pig breeds. Animal 2016, 10, 1741–1748. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Zou, X.; Xia, W.; Wen, X.; Yang, H. Comparative metabolomic analysis of caecal digesta between Jinhua pig and Landrace pig. Czech. J. Anim. Sci. 2019, 64, 332–342. [Google Scholar] [CrossRef]
- Jiang, G.L.; Liu, Y.Y.; Oso, A.O.; Li, F.N.; Kong, X.F.; Geng, M.M.; Yang, H.S.; Yin, Y.L. The differences of bacteria and bacteria metabolites in the colon between fatty and lean pigs. J. Anim. Sci. 2016, 94, 349–353. [Google Scholar] [CrossRef]
- Ha, C.W.; Lam, Y.Y.; Holmes, A.J. Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health. World J. Gastroenterol. 2014, 20, 16498–16517. [Google Scholar] [CrossRef]
- Xiao, Y.; Li, K.; Xiang, Y.; Zhou, W.; Yang, H. The fecal microbiota composition of boar Duroc, Yorkshire, Landrace and Hampshire pigs. Asian Austral. J. Anim. 2017, 30, 1456–1463. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Wu, X.; Xie, C.; Xiao, D.; Zhang, B. Meat quality and fatty acid profiles of chinese ningxiang pigs following supplementation with n-carbamylglutamate. Animals 2020, 10, 88. [Google Scholar] [CrossRef] [Green Version]
- Velotto, S.; Claudia, V.; Antonio, C. Muscle fibre types, fat deposition and fatty acid profile of casertana versus large white pig. Anim. Sci. Pap. Rep. 2012, 30, 35–44. [Google Scholar] [CrossRef]
- Miao, Z.G.; Wang, L.J.; Xu, Z.R.; Huang, J.F.; Wang, Y.R. Developmental changes of carcass composition, meat quality and organs in the Jinhua pig and Landrace. Animal 2009, 3, 468–473. [Google Scholar] [CrossRef] [Green Version]
- Miao, Z.G.; Wei, P.; Khan, M.A.; Zhang, J.; Wang, S. Transcriptome analysis reveals differential gene expression in intramuscular adipose tissues of Jinhua and Landrace pigs. J. Vet. Med. Sci. 2018, 80, 953–959. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Jiang, Z.Y.; Lin, Y.C.; Zheng, C.T.; Jiang, S.Q.; Ma, X.Y. Effects of dibutyryl cAMP on growth performance and carcass traits in finishing pigs. Livest. Sci. 2012, 146, 67–72. [Google Scholar] [CrossRef]
- Du, F.; Zhang, X.; Gu, H.; Song, J.; Gao, X. Dynamic changes in the bacterial community during the fermentation of traditional chinese fish sauce (TCFS) and their correlation with TCFS quality. Microorganisms 2019, 7, 371. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high throughput sequencing reads. Embnet J. 2011, 17. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Christopher, Q.; Rob, K. Uchime improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2011, 41, 590–596. [Google Scholar] [CrossRef]
- Kong, X.F.; Zhang, Y.Z.; Wu, X.; Yin, Y.L.; Tan, Z.L.; Feng, Y.; Yan, F.Y.; Bo, M.J.; Huang, R.L.; Li, T.J. Fermentation characterization of Chinese yam polysaccharide and its effects on the gut microbiota of rats. Int. J. Microbiol. 2009, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latorre, M.A.; Pomar, C.; Faucitano, L.; Gariépy, C.; Méthot, S. The relationship within and between production performance and meat quality characteristics in pigs from three different genetic lines. Livest. Sci. 2008, 115, 258–267. [Google Scholar] [CrossRef]
- Maiorano, G.; Gambacorta, M.; Tavaniello, S.; D’Andrea, M.; Stefanon, B.; Piila, F. Growth, carcass and meat quality of Casertana, Italian Large White and Duroc × (Landrace x Italian Large White) pigs reared outdoors. Ital. J. Anim. Sci. 2013, 12, e69. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.M.; Guo, J.P.; Kong, X.F.; Wang, W.C.; Yin, Y.L. Molecular cloning and expression profiling of g protein coupled receptor 120 in landrace pig and different chinese indigenous pig breeds. J. Food Agric. Environ. 2012, 10, 809–814. [Google Scholar]
- Guo, J.; Shan, T.Z.; Wu, T.; Zhu, L.N.; Wang, Y.Z. Comparisons of different muscle metabolic enzymes and muscle fiber types in Jinhua and Landrace pigs. J. Anim. Sci. 2010, 89, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Aura, A.M. Microbial metabolism of dietary phenolic compounds in the colon. Phytochem. Rev. 2008, 7, 407–429. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2008, 457, 480–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Kong, F.; Xiang, Y.; Zhou, W.; Wang, J.; Yang, H.; Zhang, G.; Zhao, J. Comparative biogeography of the gut microbiome between jinhua and landrace pigs. Sci. Rep. 2018, 8, 5985. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Liu, Y.; Le, B.; Qin, B.; Liu, M.; Zhao, Y.; Guo, X.; Cao, G.; Liu, J.; Li, B. A comparison of dynamic distributions of intestinal microbiota between Large White and Chinese Shanxi Black pigs. Arch. Microbiol. 2019, 201, 357–367. [Google Scholar] [CrossRef]
- Gordon, J. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 1518–1523. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Bian, G.; Su, Y.; Zhu, W. Comparison of faecal microbial community of lantang, bama, erhualian, meishan, xiaomeishan, duroc, landrace, and yorkshire sows. Asian Austral. J. Anim. Sci. 2014, 27, 898–906. [Google Scholar] [CrossRef]
- Heinritz, S.N.; Weiss, E.; Eklund, M.; Aumiller, T.; Mosenthin, R. Intestinal Microbiota and Microbial Metabolites Are Changed in a Pig Model Fed a High-Fat/Low-Fiber or a Low-Fat/High-Fiber Diet. PLoS ONE 2016, 11, e0154329. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Xia, X.; Tang, R.; Wang, K. Real-time PCR quantification of the predominant bacterial divisions in the distal gut of Meishan and Landrace pigs. Anaerobe 2008, 14, 224–228. [Google Scholar] [CrossRef]
- Biddle, A.; Stewart, L.; Blanchard, J.; Leschine, S. Untangling the genetic basis of fibrolytic specialization by lachnospiraceae and ruminococcaceae in diverse gut communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- Vital, M.; Howe, C.; Tiedje, M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. Mbio 2014, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Segain, J.P.; Raingeard de la Blétière, D.; Bourreille, A.; Leray, V.; Gervois, N.; Rosales, C.; Ferrier, L.; Bonnet, C.; Blottière, H.M.; Galmiche, J.P. Butyrate inhibits inflammatory responses through nfκb inhibition: Implications for crohn’s disease. Gut 2000, 47, 397–403. [Google Scholar] [CrossRef] [Green Version]
- Peng, L.; Li, Z.R.; Green, R.S.; Holzman, I.R.; Lin, J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 2009, 139, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Brummer, R.J. Review article: The role of butyrate on colonic function. Aliment. Pharm. Ther. 2010, 27, 104–119. [Google Scholar] [CrossRef]
- Maslowski, K.M.; Mackay, C.R. Diet, gut microbiota and immune responses. Nat. Immunol. 2011, 12, 5–9. [Google Scholar] [CrossRef]
- Payne, A.N.; Chassard, C.; Zimmermann, M.; Müller, P.; Stinca, S.; Lacroix, C. The metabolic activity of gut microbiota in obese children is increased compared with normal-weight children and exhibits more exhaustive substrate utilization. Nutr. Diabetes 2011, 1, e12. [Google Scholar] [CrossRef] [Green Version]
- Berndt, J.; Kovacs, P.; Ruschke, K.; Kloting, N.; Fasshauer, M.; Schon, M.R.; Körner, A.; Stumvoll, M.; Blüher, M. Fatty acid synthase gene expression in human adipose tissue: Association with obesity and type 2 diabetes. Diabetologia 2007, 50, 1472–1480. [Google Scholar] [CrossRef] [Green Version]
- Bajzer, M.; Seeley, R.J. Physiology: Obesity and gut flora. Nature 2006, 444, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Tsai, F.; Coyle, W.J. The microbiome and obesity: Is obesity linked to our gut flora? Curr. Gastroenterol. Rep. 2009, 11, 307–313. [Google Scholar] [CrossRef] [PubMed]
| Items 1 | Genotype 2 | SEM 3 | p-Value | |
|---|---|---|---|---|
| LW | NX | |||
| ADG (g/day) | 842.26 a | 392.50 b | 11.84 | 0.025 |
| ADFI (g/day) | 1953 a | 1402 b | 35.03 | 0.037 |
| F/G (g/g) | 2.32 b | 3.60 a | 0.04 | 0.054 |
| Final weight (kg) | 99.34 a | 49.39 b | 1.35 | 0.018 |
| Carcass weight (kg) | 67.17 a | 30.98 b | 0.99 | 0.014 |
| Dressing percentage (%) | 67.57 a | 63.38 b | 0.63 | 0.048 |
| Average backfat thickness (cm) | 2.61 b | 4.54 a | 0.08 | 0.032 |
| Drip loss (%) | 5.21 a | 3.25 b | 0.06 | 0.029 |
| Items 1 | Genotype 2 | SEM 3 | p-Value | |
|---|---|---|---|---|
| NX | LW | |||
| Acetate | 9.19 | 12.65 | 0.820 | 0.107 |
| Propionate | 4.30 b | 7.02 a | 0.478 | 0.036 |
| Butyrate | 2.41 b | 4.88 a | 0.340 | 0.014 |
| Isobutyrate | 0.56 | 0.63 | 0.035 | 0.450 |
| Valerate | 0.67 | 0.95 | 0.064 | 0.094 |
| Isovalerate | 0.86 | 0.94 | 0.061 | 0.593 |
| Total SCFAs | 17.98 b | 27.07 a | 1.670 | 0.044 |
| A/P 4 | 2.14 | 1.84 | 0.090 | 0.189 |
| Total BCFAs | 1.42 | 1.57 | 0.098 | 0.538 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, L.; Wang, Z.; Li, J.; Yang, H.; Yin, Y.; Tan, B.; Chen, J. Comparative Microbial Profiles of Colonic Digesta between Ningxiang Pig and Large White Pig. Animals 2021, 11, 1862. https://doi.org/10.3390/ani11071862
Lei L, Wang Z, Li J, Yang H, Yin Y, Tan B, Chen J. Comparative Microbial Profiles of Colonic Digesta between Ningxiang Pig and Large White Pig. Animals. 2021; 11(7):1862. https://doi.org/10.3390/ani11071862
Chicago/Turabian StyleLei, Linfeng, Zhaobin Wang, Jianzhong Li, Huansheng Yang, Yulong Yin, Bie Tan, and Jiashun Chen. 2021. "Comparative Microbial Profiles of Colonic Digesta between Ningxiang Pig and Large White Pig" Animals 11, no. 7: 1862. https://doi.org/10.3390/ani11071862
APA StyleLei, L., Wang, Z., Li, J., Yang, H., Yin, Y., Tan, B., & Chen, J. (2021). Comparative Microbial Profiles of Colonic Digesta between Ningxiang Pig and Large White Pig. Animals, 11(7), 1862. https://doi.org/10.3390/ani11071862






