A Mother’s Story, Mitogenome Relationships in the Genus Rupicapra
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Sequencing
2.2. Genome Assembly and Validation
2.3. Alignment, Post Processing, and Annotation of Protein Coding Genes
2.4. Phylogenetic Analyses
3. Results
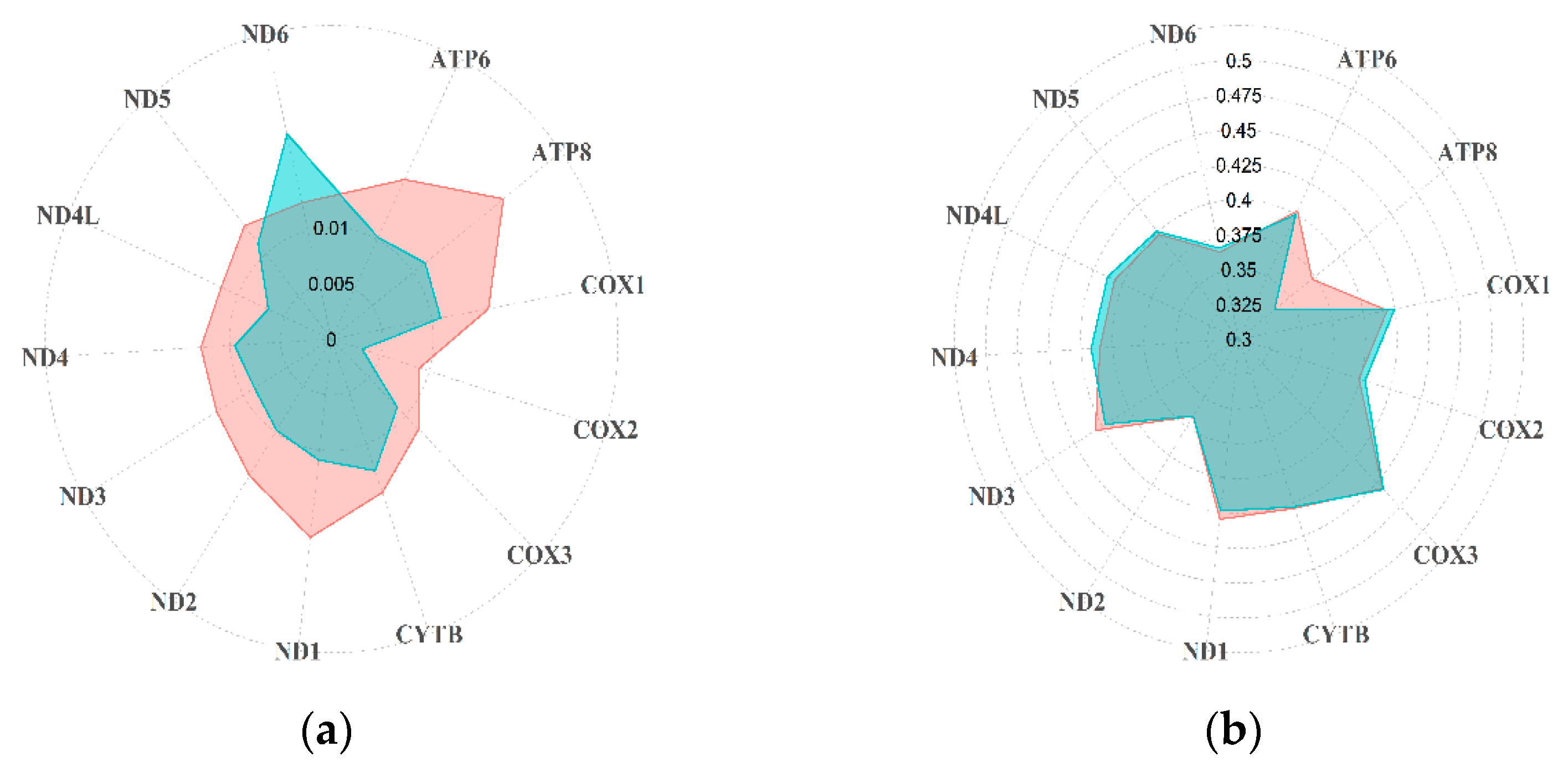
3.1. Diversity of Rupicapra Mitogenome
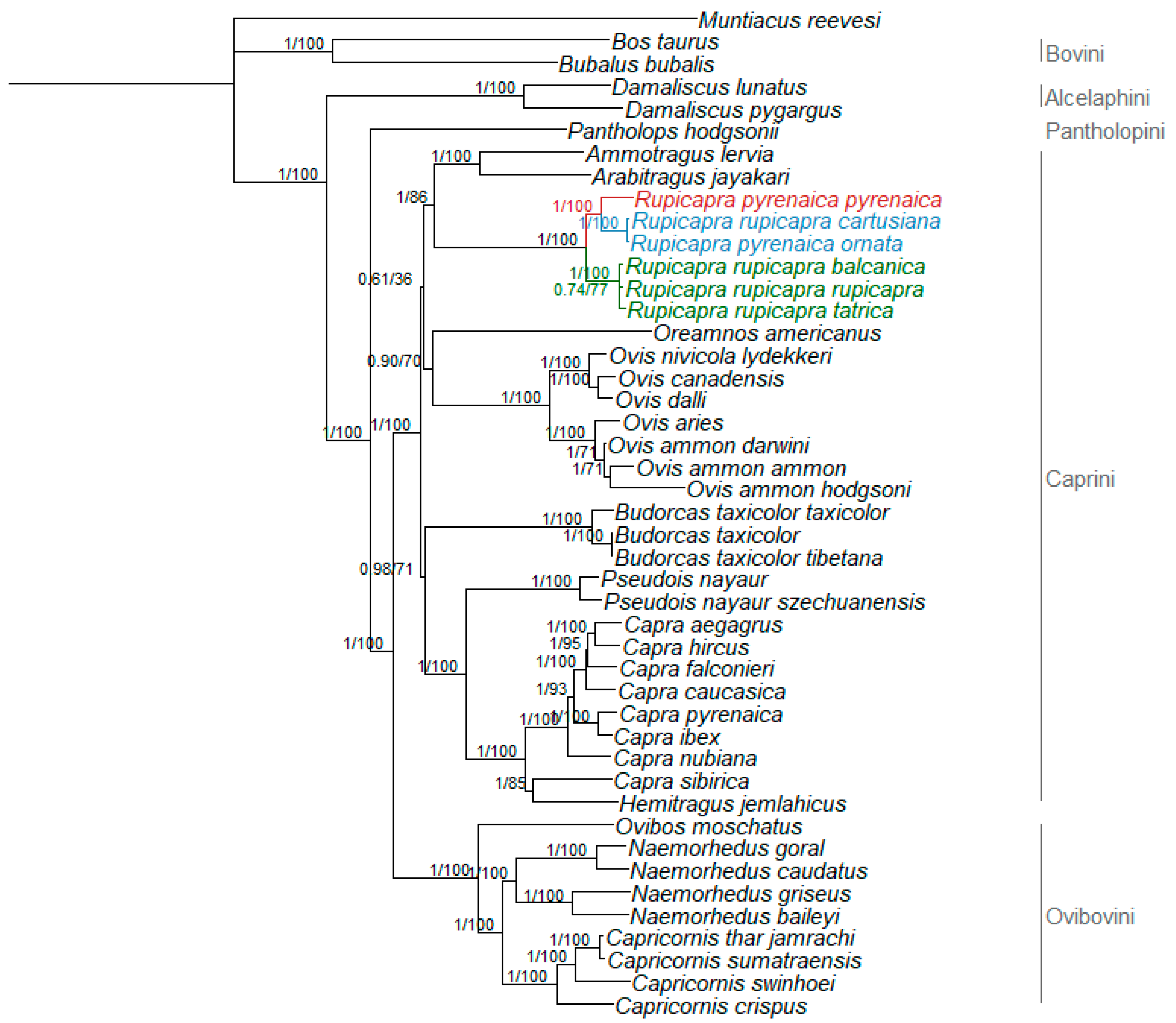
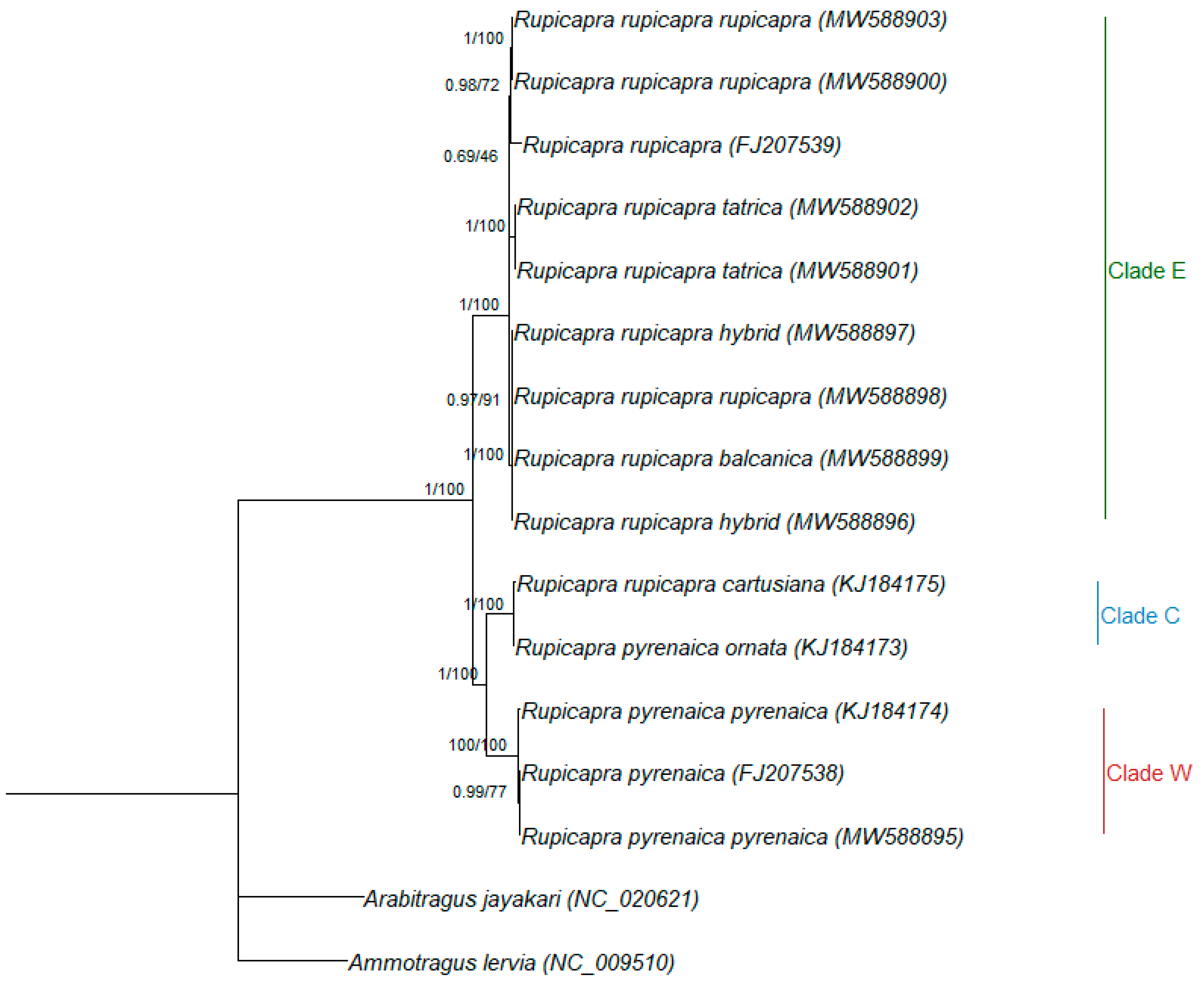
3.2. Phylogenetic Relationships
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lovari, S. Evolutionary aspects of the biology of chamois, Rupicapra spp. (Bovidae, Caprinae). In The Biology and Management of Capricornis and Related Mountain Antelopes; Springer: Dordrecht, The Netherlands, 1987; pp. 51–61. [Google Scholar]
- Corlatti, L.; Lorenzini, R.; Lovari, S. The conservation of the chamois Rupicapra spp. Mamm. Rev. 2011, 41, 163–174. [Google Scholar] [CrossRef]
- Herrero, J.; Lovari, S.; Nores, C.; Toigo, C. Rupicapra pyrenaica. IUCN Red List Threat. Species 2020, e.T19771A171131310. [Google Scholar] [CrossRef]
- Anderwald, P.; Ambarli, H.; Avramov, S.; Ciach, M.; Corlatti, L.; Farkas, A.; Jovanovic, M.; Papaioannou, H.; Peters, W.; Sarasa, M.; et al. Rupicapra rupicapra. IUCN Red List Threat. Species 2020, e.T39255A22149561. [Google Scholar] [CrossRef]
- Pérez, T.; Hammer, S.E.; Albornoz, J.; Domínguez, A. Y-chromosome phylogeny in the evolutionary net of chamois (genus Rupicapra). BMC Evol. Biol. 2011, 11, 272. [Google Scholar] [CrossRef]
- Rodríguez, F.; Hammer, S.; Pérez, T.; Suchentrunk, F.; Lorenzini, R.; Michallet, J.; Martinkova, N.; Albornoz, J.; Domínguez, A. Cytochrome b Phylogeography of Chamois (Rupicapra spp.). Population Contractions, Expansions and Hybridizations Governed the Diversification of the Genus. J. Hered. 2009, 100, 47–55. [Google Scholar] [CrossRef][Green Version]
- Rodríguez, F.; Pérez, T.; Hammer, S.E.; Albornoz, J.; Domínguez, A. Integrating phylogeographic patterns of microsatellite and mtDNA divergence to infer the evolutionary history of chamois (genus Rupicapra). BMC Evol. Biol. 2010, 10, 222. [Google Scholar] [CrossRef]
- Pérez, T.; Rodríguez, F.; Fernández, M.; Albornoz, J.; Domínguez, A. Ancient mitochondrial pseudogenes reveal hybridization between distant lineages in the evolution of the Rupicapra genus. Gene 2017, 628, 63–71. [Google Scholar] [CrossRef]
- Pérez, T.; Fernández, M.; Hammer, S.E.; Domínguez, A. Multilocus Intron Trees Reveal Extensive Male-Biased Homogenization of Ancient Populations of Chamois (Rupicapra spp.) across Europe during Late Pleistocene. PLoS ONE 2017, 12, e0170392. [Google Scholar] [CrossRef] [PubMed]
- Aquadro, C.F.; Greenberg, B.D. Human mitochondrial DNA variation and evolution: Analysis of nucleottide sequences from seven individuals. Genetics 1983, 103, 287–312. [Google Scholar] [CrossRef]
- Cann, R.L.; Brown, W.M.; Wilson, A. Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics 1984, 106, 479–499. [Google Scholar] [CrossRef]
- Neupert, W. Mitochondrial Gene Expression: A Playground of Evolutionary Tinkering. Annu. Rev. Biochem. 2016, 85, 65–76. [Google Scholar] [CrossRef]
- Herrero, A.; Barja, G. H2O2 production of heart mitochondria and aging rate are slower in canaries and parakeets than in mice: Sites of free radical generation and mechanisms involved. Mech. Ageing Dev. 1998, 103, 133–146. [Google Scholar] [CrossRef]
- Shen, Y.-Y.; Liang, L.; Zhu, Z.-H.; Zhou, W.-P.; Irwin, D.M.; Zhang, Y.-P. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. USA 2010, 107, 8666–8671. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Huang, G.; Sun, J.; Wang, Z.; Teng, S.; Cao, Y.; Hanif, Q.; Chen, N.; Lei, C.; Liao, Y. Mitogenome Diversity and Maternal Origins of Guangxi Buffalo Breeds. Animals 2020, 10, 547. [Google Scholar] [CrossRef]
- Sun, C.-H.; Liu, H.-Y.; Min, X.; Lu, C.-H. Mitogenome of the little owl Athene noctua and phylogenetic analysis of Strigidae. Int. J. Biol. Macromol. 2020, 151, 924–931. [Google Scholar] [CrossRef]
- Hassanin, A.; Ropiquet, A.; Couloux, A.; Cruaud, C. Evolution of the Mitochondrial Genome in Mammals Living at High Altitude: New Insights from a Study of the Tribe Caprini (Bovidae, Antilopinae). J. Mol. Evol. 2009, 68, 293–310. [Google Scholar] [CrossRef] [PubMed]
- Pérez, T.; González, I.; Essler, S.E.; Fernández, M.; Domínguez, A. The shared mitochondrial genome of Rupicapra pyrenaica ornata and Rupicapra rupicapra cartusiana: Old remains of a common past. Mol. Phylogenet. Evol. 2014, 79, 375–379. [Google Scholar] [CrossRef]
- Iacolina, L.; Corlatti, L.; Buzan, E.; Safner, T.; Šprem, N. Hybridisation in European ungulates: An overview of the current status, causes, and consequences. Mamm. Rev. 2019, 49, 45–59. [Google Scholar] [CrossRef]
- Apollonio, M.; Scandura, M.; Šprem, N. Reintroductions as a management tool for European Ungulates. In Behavior and Management of European Ungulates; Putman, R., Apollonio, M., Eds.; Whittles Publishing: Dunbeath, UK, 2014; pp. 46–77. [Google Scholar]
- Šprem, N.; Buzan, E. The genetic impact of chamois management in the dinarides. J. Wildl. Manag. 2016, 80, 783–793. [Google Scholar] [CrossRef]
- Marinier, E.; Brown, D.G.; McConkey, B.J. Pollux: Platform independent error correction of single and mixed genomes. BMC Bioinform. 2015, 16, 10. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef] [PubMed]
- BLAST. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi. (accessed on 15 January 2021).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Conant, G.C.; Wolfe, K.H. GenomeVx: Simple web-based creation of editable circular chromosome maps. Bioinformatics 2008, 24, 861–862. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Pais, F.S.-M.; de Cássia Ruy, P.; Oliveira, G.; Coimbra, R.S. Assessing the efficiency of multiple sequence alignment programs. Algorithms Mol. Biol. 2014, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Achilli, A.; Olivieri, A.; Soares, P.; Lancioni, H.; Kashani, B.H.; Perego, U.A.; Nergadze, S.G.; Carossa, V.; Santagostino, M.; Capomaccio, S.; et al. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc. Natl. Acad. Sci. USA. 2012, 109, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Kumar, S. Evolutionary distance estimation under heterogeneous substitution pattern among lineages. Mol. Biol. Evol. 2002, 19, 1727–1736. [Google Scholar] [CrossRef]
- Hiendleder, S.; Lewalski, H.; Wassmuth, R.; Janke, A. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. J. Mol. Evol. 1998, 47, 441–448. [Google Scholar] [CrossRef]
- Mereu, P.; Palici di Suni, M.; Manca, L.; Masala, B. Complete nucleotide mtDNA sequence of Barbary sheep (Ammotragus lervia). DNA Seq. 2008, 19, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Nascetti, G.; Lovari, S.; Lanfranchi, P.; Berducou, C.; Mattiucci, S.; Rossi, L.; Bullini, L. Revision of Rupicapra genus. III. Electrophoretic studies demostrating species distintion of chamois populations of the Alps from those of the Apennines and Pyrenees. In The Biology and Management of Mountain Ungulates; Lovar, S., Ed.; Croom-Helm: London, UK, 1985; pp. 56–62. [Google Scholar]
- Lovari, S.; Scala, C. Revision of Rupicapra Genus. I. A statistical re-evaluation of Couturier’s data on the morphometry of six chamois subspecies. Boll. di Zool. 1980, 47, 113–124. [Google Scholar] [CrossRef]
- Zhou, M.; Yu, J.; Li, B.; Ouyang, B.; Yang, J. The complete mitochondrial genome of Budorcas taxicolor tibetana (Artiodactyla: Bovidae) and comparison with other Caprinae species: Insight into the phylogeny of the genus Budorcas. Int. J. Biol. Macromol. 2019, 121, 223–232. [Google Scholar] [CrossRef]
- Groves, P.; Shields, G.F. Phylogenetics of the Caprinae based on cytochrome b sequence. Mol. Phylogenet. Evol. 1996, 5, 467–476. [Google Scholar] [CrossRef]
- Groves, P.; Shields, G.F. Cytochrome B Sequences Suggest Convergent Evolution of the Asian Takin and Arctic Muskox. Mol. Phylogenet. Evol. 1997, 8, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Gorbachev, V. V Effect of random sample size on the accuracy of nucleotide diversity estimation. Russ. J. Genet. 2012, 48, 746–750. [Google Scholar] [CrossRef]
- Hatakeyama, H.; Goto, Y. Concise Review: Heteroplasmic Mitochondrial DNA Mutations and Mitochondrial Diseases: Toward iPSC-Based Disease Modeling, Drug Discovery, and Regenerative Therapeutics. Stem Cells 2016, 34, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.L.; Sue, C.M. The Genetics of Mitochondrial Disease. Semin. Neurol. 2011, 31, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Craven, L.; Alston, C.L.; Taylor, R.W.; Turnbull, D.M. Recent Advances in Mitochondrial Disease. Annu. Rev. Genomics Hum. Genet. 2017, 18, 257–275. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.A.; Govindaraj, P.; Soumittra, N.; Srilekha, S.; Ambika, S.; Vanniarajan, A.; Meena, A.K.; Uppin, M.S.; Sundaram, C.; Taly, A.B.; et al. Haplogroup Heterogeneity of LHON Patients Carrying the m.14484T>C Mutation in India. Investig. Opthalmol. Vis. Sci. 2013, 54, 3999. [Google Scholar] [CrossRef]
- Gurses, C.; Azakli, H.; Alptekin, A.; Cakiris, A.; Abaci, N.; Arikan, M.; Kursun, O.; Gokyigit, A.; Ustek, D. Mitochondrial DNA profiling via genomic analysis in mesial temporal lobe epilepsy patients with hippocampal sclerosis. Gene 2014, 538, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.-S.; Ki, C.-S.; Park, K.-J. Pediatric-Onset Dystonia Associated with Bilateral Striatal Necrosis and G14459A Mutation in a Korean Family: A Case Report. J. Korean Med. Sci. 2010, 25, 180. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hirose, M.; Schilf, P.; Benoit, S.; Eming, R.; Gläser, R.; Homey, B.; Kunz, M.; Nebel, A.; Peitsch, W.K.; Pföhler, C.; et al. Polymorphisms in the mitochondrially encoded ATP synthase 8 gene are associated with susceptibility to bullous pemphigoid in the German population. Exp. Dermatol. 2015, 24, 715–717. [Google Scholar] [CrossRef] [PubMed]
- Thapa, S.; Lalrohlui, F.; Ghatak, S.; Zohmingthanga, J.; Lallawmzuali, D.; Pautu, J.L.; Senthil Kumar, N. Mitochondrial complex I and V gene polymorphisms associated with breast cancer in mizo-mongloid population. Breast Cancer 2016, 23, 607–616. [Google Scholar] [CrossRef]
| Subspecies | N | N Haplotypes | S | π (SD) | h (SD) | Fs | Accession Number |
|---|---|---|---|---|---|---|---|
| R. rupicapra | 1 | 1 | FJ207539 1 | ||||
| R. r. rupicapra | 3 | 2 | 52 | 0.002 (0.001) | 0.667 (0.314) | 6.481 | MW588898 2 MW588900 2 MW588903 2 |
| R. r. balcanica | 1 | 1 | MW588899 2 | ||||
| R. r. rupicapra x R. r. balcanica putative hybrid | 2 | 2 | 1 | 0.000 (0.000) | 1.000 (0.500) | 0.000 | MW588896 2 MW588897 2 |
| R. r. tatrica | 2 | 1 | MW588901 2 MW588902 2 | ||||
| R. r. cartusiana | 1 | 1 | KJ184175 3 | ||||
| R. rupicapra TOT | 10 | 6 | 573 | 0.009 (0.004) | 0.889 (0.075) | 12.454 | |
| R. pyrenaica | 1 | 1 | FJ207538 1 | ||||
| R. p. ornata | 1 | 1 | KJ184173 3 | ||||
| R. p. pyrenaica | 2 | 2 | 14 | 0.001 (0.000) | 1.000 (0.500) | 2.639 | KJ184174 3 MW588895 2 |
| R. pyrenaica TOT | 4 | 4 | 371 | 0.012 (0.006) | 1.000 (0.177) | 3.426 | |
| TOT | 14 | 10 | 771 | 0.019 (0.003) | 0.945 (0.045) | 11.944 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacolina, L.; Buzan, E.; Safner, T.; Bašić, N.; Geric, U.; Tesija, T.; Lazar, P.; Arnal, M.C.; Chen, J.; Han, J.; et al. A Mother’s Story, Mitogenome Relationships in the Genus Rupicapra. Animals 2021, 11, 1065. https://doi.org/10.3390/ani11041065
Iacolina L, Buzan E, Safner T, Bašić N, Geric U, Tesija T, Lazar P, Arnal MC, Chen J, Han J, et al. A Mother’s Story, Mitogenome Relationships in the Genus Rupicapra. Animals. 2021; 11(4):1065. https://doi.org/10.3390/ani11041065
Chicago/Turabian StyleIacolina, Laura, Elena Buzan, Toni Safner, Nino Bašić, Urska Geric, Toni Tesija, Peter Lazar, María Cruz Arnal, Jianhai Chen, Jianlin Han, and et al. 2021. "A Mother’s Story, Mitogenome Relationships in the Genus Rupicapra" Animals 11, no. 4: 1065. https://doi.org/10.3390/ani11041065
APA StyleIacolina, L., Buzan, E., Safner, T., Bašić, N., Geric, U., Tesija, T., Lazar, P., Arnal, M. C., Chen, J., Han, J., & Šprem, N. (2021). A Mother’s Story, Mitogenome Relationships in the Genus Rupicapra. Animals, 11(4), 1065. https://doi.org/10.3390/ani11041065







