Simple Summary
Two species of chamois (Rupicapra rupicapra and R. pyrenaica) are currently recognized by taxonomy and further subdivided into seven and three subspecies, respectively. However, recent research based on molecular markers finds this classification questionable. We aim to increase the resolution of published research on chamois phylogeny by including mitogenomes of all available subspecies, including the previously unpublished mitogenomes of R. r. balcanica and R. r. tatrica subspecies. The inferred phylogeny based on the full mitogenomes confirms the previously reported genus subdivision in three clades and its monophyletic positioning within the Caprinae. Phylogeny and taxonomy of Rupicapra species thus remain controversial prompting for the inclusion of archeological remains to solve the controversy.
Abstract
Although the two species of chamois (Rupicapra rupicapra and R. pyrenaica) are currently classified as least-concern by the IUCN (International Union for Conservation of Nature), inconsistencies on the subspecies classification reported in literature make it challenging to assess the conservation status of the single subspecies. Previous studies relying on mitochondrial genes, sometimes in combination with nuclear or Y-chromosome markers, reported the presence of clusters corresponding to the geographic distribution but highlighting ambiguities in the genus phylogeny. Here we report novel de novo assembled sequences of the mitochondrial genome from nine individuals, including previously unpublished R. r. balcanica and R. r. tatrica subspecies, and use them to untangle the genus phylogeny. Our results based on the full mitogenome inferred phylogeny confirm the previously reported genus subdivision in three clades and its monophyletic positioning within the Caprinae. Phylogeny and taxonomy of Rupicapra species thus remain controversial prompting for the inclusion of archeological remains to solve the controversy.
1. Introduction
The chamois, genus Rupicapra, is the most abundant mountain-dwelling ungulate in Europe and the Near East, and is currently recognized to be divided into two species: Rupicapra rupicapra (Northern chamois) and Rupicapra pyrenaica (Southern chamois), further subdivided into seven (cartusiana, rupicapra, balcanica, tatrica, carpatica, caucasica, and asiatica) and three (parva, pyrenaica, and ornata) subspecies, respectively [1,2]. The conservation status of the species requires consideration. Neither of the species is threatened and both species are currently classified as least-concern in the IUCN Red List of Threatened Species [3,4]. Although some subspecies are protected at the national level in (part of) their distribution, detailed information on the conservation status of the different subspecies is patchy, if any, and chamois may be one of the most threatened European ungulates if considered at the subspecies level [2]. This picture is further complicated by molecular controversy concerning the subspecies subdivision based on morphological and behavioral characters. While some markers provided some support for this classification mitochondrial DNA (mtDNA) identified nominal species as paraphyletic [2,5] and references therein.
Previous studies relying on cytochrome b (cytb) identified three lineages geographically separated, a western clade in the Western Alps and Iberia, a central clade in the Apennines and the Chartreuse Massif, and an eastern clade covering all regions east of the Alps [6]. This subdivision, later confirmed by a combination of mitochondrial regions [7], differs from the pattern observed with microsatellites, mitochondrial pseudogenes, and introns [7,8,9]. The analyses using nuclear markers, although recognizing geographically structured clades, more closely resemble the subspecies subdivision based on morphological characteristics and highlight a complex population history strongly shaped by male-biased dispersal [7,8,9], as confirmed by Y-chromosome lineages [5]. However, mitochondrial genes evolution rates differ [10,11,12], with mutations accumulating faster in non-coding than in coding regions, and are strongly influenced by energetic demands [12,13,14], thus using the whole mitochondrial genome (mitogenome) could help to address different phylogenetic aspects [15,16]. So far, very few Rupicapra mitogenomes are available. Hassanin et al. [17] were the first to use the potential of mitogenomes to investigate phylogenetic relationships within the Caprini tribe but were unable to resolve the relationships of Rupicapra and a few other genera. In their study, the two Rupicapra species formed a separate branch in the tree [17] but represented only two of the three mitochondrial clades. The later addition of two mitogenomes of subspecies previously attributed to the central clade [18] confirmed the subdivision in three clades and the non-concordance with taxonomic classification.
To further complicate the picture, reintroductions and translocations—if performed without previous knowledge on the involved individuals’ taxonomic status [19,20]—might introduce an additional confounding effect. One of these examples is in the Northern Dinaric mountains, Mt. Velebit (Croatia), where R. r. rupicapra and R. r. balcanica were introduced for hunting purposes and led to a molecularly identified hybrid population [21]. However, such events might be difficult to trace due to a lack of historic records or illegal practices, thus leading to incorrect assumptions on the (sub)species being sampled and hampering phylogenetic and taxonomic reconstructions.
In this study, we aimed to contribute to the unraveling of the complex genus phylogeny by increasing the number of individuals and subspecies investigated using the whole mitochondrial genome. Our results confirmed the grouping of individuals from Mt. Velebit with the Balkan subspecies, the subdivision of the genus into three mitochondrial clades and its monophyletic positioning within the Caprini, in a sister-group relationship with Ammotragus and Arabitragus, while prompting for additional multi-marker studies also involving historical samples to disentangle the genus phylogeny and taxonomy.
2. Materials and Methods
2.1. Sampling and DNA Sequencing
We collected samples from Rupicapra subspecies (rupicapra n = 4, balcanica n = 2, tatrica n = 2, balcanica x rupicapra putative hybrids n = 2, and pyrenaica n = 2). All samples were collected from dead animals during regular hunting seasons. Genomic DNA was extracted from tissue samples using a standard phenol-chloroform method. DNA fragments were then treated according to the Illumina DNA sample preparation protocol. Genome sequencing was performed on the Illumina HiSeq 2500 platform with paired-end adaptors and 100 bp reads length for libraries of 350 bp inserts. To investigate the phylogenetic relationships, we downloaded one mitogenome sequence for each of the 34 (sub)species from the subfamily Caprinae available in GenBank and five outgroups: Damaliscus pygarus, Damaliscus lunatus, Bos taurus, Bubalus bubalis, and Muntiacus reevesi (Table S1).
2.2. Genome Assembly and Validation
Raw sequences were corrected using the Pollux program [22] with default settings. Mitochondrial DNA, due to the presence of multiple copies per cell, had higher coverage and depth in whole-genome sequences which, if results are of sufficient quality, allows for de novo assembly. After screening with Pollux, we excluded two samples that did not pass the quality filtering. To avoid ambiguity of the assembled sequences, we used two approaches: a de novo assembly followed by an assembly relying on a reference genome. MitoZ [23] was used for the de novo approach with default settings. We then used NOVOPlasty [24] “seed-and-extend” method using the GenBank R. rupicapra sequence (FY207539) as seed. The resulting circular genomes were verified as belonging to the Genus in BLAST [25]. Obtained mitochondrial sequences were further validated by aligning them against GenBank R. rupicapra sequence using MEGA X [26], Bioedit [27], and BWA [28] software. Visualization of results was done using GenomeVx software [29].
2.3. Alignment, Post Processing, and Annotation of Protein Coding Genes
Newly assembled mitogenomes that passed the quality controls (Table 1) were aligned in Mega X [26] using Clustal W [30] considering the algorithm computational demand and the size of our file [31] and checked by eye. Downloaded sequences were also visually inspected for consistency and exclude the possible presence of erroneous rearrangements and nuclear pseudogenes [8,17,31]. To avoid erroneous hypothesis on homology, all indels, positions with ambiguity in the position of the gaps, and the portion of the control region presenting tandem repeats in the Caprini [17] were excluded. The final alignment consisted of 15,383 bp and 40 sequences. GeSeq [32] was used to identify both protein-coding and tRNA, with the embedded tRNAscan-SE v2.0.5 [33], regions setting the sequence source to linear mtDNA. DNAsp v6.12.03 [34] was used to infer the number of haplotypes, haplotype (h) and nucleotide (π) diversity, number of polymorphic sites (S), GC content, and Fu’s (Fs) statistic, for both the whole mitogenome and each individual protein-coding region for each Rupicapra subspecies. The ND6 region was reverse complemented to present the same reading direction as the other protein-coding sequences [35]. DNAsp was additionally used to identify the number of synonymous (SS) and non-synonymous (NSS) sites and to compute the ratio between synonymous and non-synonymous substitutions. The overall mean distance for synonymous substitutions per synonymous site (pS) and non-synonymous substitutions per non-synonymous site (pN) were calculated using Mega X according to the Nei-Gojobori method [36] with 1000 bootstraps.

Table 1.
Variability across the Rupicapra subspecies. N = number of individuals, N haplotypes = number of haplotypes, S = number of polymorphic sites, π = nucleotide diversity, h = haplotype diversity, SD = standard deviation, Fs = Fu’s Fs, TOT = total.
2.4. Phylogenetic Analyses
Phylogenetic relationships among Caprinae species were reconstructed using a reduced alignment containing a single sequence per Rupicapra subspecies, mitogenome sequences of Caprinae species available in GenBank, and five Bovidae as outgroups. The complete dataset used for this analysis consisted of 45 mitogenome sequences. Relationships within Rupicapra were investigated using all Rupicapra sequences available in GenBank (n = 5) and the nine new sequences obtained within this research. Mitogenome sequences of Ammotragus lervia and Arabitragus jakari were included as outgroups. PartitionFinder2 [37] was used to infer partitioning and corresponding evolutionary models, whereas ModelFinder in IQ-TREE [38] was used to select the most accurate evolutionary model for the complete mitogenome. Since partitioning the sequence did not lead to differences in tree topology (data not shown) we herewith specify evolutionary models only for the whole sequence. Two phylogenetic approaches were applied for tree inference: the maximum likelihood method in IQ-TREE [39] and Bayesian inference in MrBayes 3.2.7 [40]. Stochastic inference on both Caprinae and Rupicapra datasets was computed with IQ-TREE default parameters, genetic code set to vertebrate mitochondrial, substitution model TIM2+I+G4, and ultrafast bootstrap analysis UFBoot2 [41].
Bayesian inference on the Caprinae was performed with GTR+I+G4 model and Markov chain Monte Carlo simulation for 1.2 million generations. The trees were sampled every 500 generations until reaching an average standard deviation of split frequencies of 0.011. MrBayes parameters for Rupicapra were GTR+G4 and 1.7 million generations, sampled every 200 generations until reaching an average standard deviation of split frequencies of 0.006. For both analyses, the convergence of chains was additionally checked by verifying the stationarity of the log probability graphs [42]. For each run, 25% of the initial trees were discarded as burn-in.
Tree editing and visualization were performed using the R package ggtree [43]. Within and between group genetic distances of Rupicapra subspecies were calculated in Mega X using the Maximum Composite Likelihood model [44], rate variation among sites modeled with an uneven gamma distribution (shape parameter = 1) [45] and including all codon positions.
3. Results
3.1. Diversity of Rupicapra Mitogenome
After quality control screening, we obtained nine newly sequenced complete mitogenomes of Rupicapra, which showed the typical organization (Figure S1) of mammalian mitochondrial genomes, incomplete stop codons, overlapping coding regions, and different start codons [46,47]. Results of all methods used for validation of the alignment were concordant. The alignment with the five already available conspecific sequences and 39 from other species had an initial length of 16,850 bp and, after removal of sites with ambiguities, gaps, and the tandem repeat of the control region, the final alignment was 15,383 bp and presented 6075 variable sites (39.49%), out of which 4914 (31.94%) were parsimony informative and 1161 (7.55%) were singletons.
The 14 Rupicapra sequences included in the full dataset showed an overall high haplotype (h = 0.945; SD = 0.045) and nucleotide (π = 0.019; sd = 0.003) diversity, with high variability between the two species and among subspecies (Table 1). A similarly high variability between R. rupicapra and R. pyrenaica was observed in the variability of the coding regions, with R. rupicapra showing in general lower nucleotide and haplotype diversity, despite the larger sample size, with some exceptions (Figure 1a and Table S2). However, the GC content and the ratio between synonymous and non-synonymous substitutions were overall concordant between the two species (Figure 1b and Table S2). Although the number of NSS was higher than the SS (Table S2), the pS-pN difference showed an overall higher proportion of pS (R. pyrenaica 0.009, standard error –SE–0.000; R. rupicapra 0.008, SE 0.000).
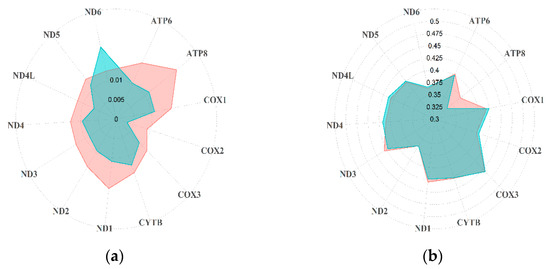
Figure 1.
Differences between R. rupicapra (green) and R. pyrenaica (red) in terms of (a) nucleotide diversity and (b) GC content.
3.2. Phylogenetic Relationships
The Bayesian phylogenetic tree of the Caprinae dataset identified the same topology as reported by Hassanin et al. [17], with tribes Bovini and Caprini being monophyletic, sister group relationship of Damaliscus and the Ovicaprini, the basal divergence of Pantholops within the Ovicaprini, and a well-supported separation of the Ovibovini. Within the Caprini tribe, Capra and Hemitragus form a goat-like clade and the more distant Pseudois and Budorcas as sister groups; Ovis and Oreamnos group together in a sheep-like clade; Ammotragus and Arabitragus showed a sister group relationship and finally, Rupicapra was monophyletic (Figure 2). The maximum likelihood analysis (tree not shown) was consistent with Bayesian results, showing both the same topology and the same high support for the nodes (bootstrap values provided in Figure 2).
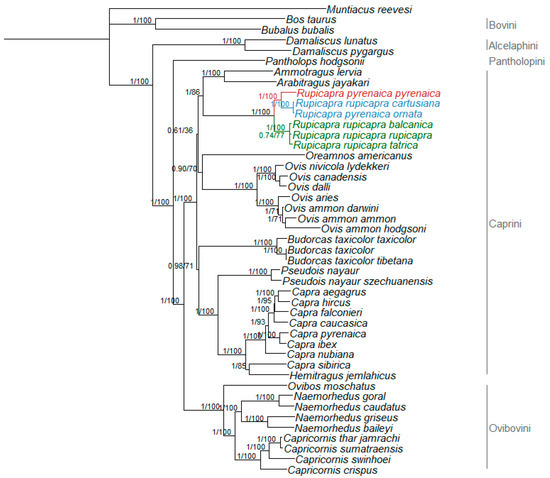
Figure 2.
Rooted phylogenetic tree obtained by Bayesian inference for the whole Caprinae. Nodal supports are indicated above the nodes (posterior probability for Bayesian inference and bootstrap for maximum likelihood, respectively). Colored branches represent Rupicapra clades: red—clade W; blue—clade C; and green—clade E.
The maximum likelihood and Bayesian analyses focusing on Rupicapra (Figure 3) showed the same highly supported subdivision in eastern, central, and western clades, with the western and central clade being closer (between group distance = 0.024 SE 0.002) to each other than to the eastern one (between group distance = 0.032 SE 0.003 and 0.030 SE 0.003, respectively). Within the eastern clade, R. r. balcanica was the most differentiated and included both sequences from the contact population from Mt. Velebit, whereas R. r. tatrica and R. r. rupicapra showed a sister group relationship (Figure 3).
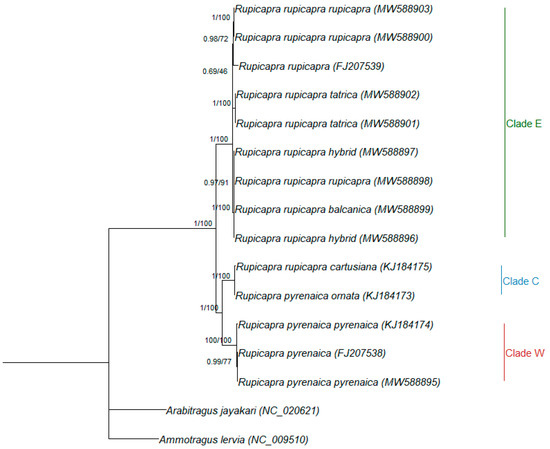
Figure 3.
Rooted phylogenetic tree obtained by Bayesian inference for Rupicapra only. Nodal supports are indicated above the nodes (posterior probability for Bayesian inference and bootstrap percentages for maximum likelihood, respectively). GenBank Accession numbers are presented in parentheses.
4. Discussion
The phylogeny and taxonomy of Rupicapra species remain controversial despite the improvements in molecular methods over the decades and are still an intriguing question, with important implications on both the evolution and conservation of this species complex [2,9]. Initial molecular investigations using electrophoresis in the 1980s supported the subdivision into an Alpine lineage separated from the Apennine and Pyrenean one [48]. Later studies, however, revealed a much more complex phylogenetic history with contrasting nuclear, Y-chromosome, and mitochondrial patterns [5,6,7,18], whereas more recent findings provide support to the hypothesis of ancient hybridization among lineages [8,9]. Although differences among markers in phylogenetic reconstructions are not unusual, due to the evolutionary mode of each marker and the fact that they represent either matrilineal (mitochondrial), patrilineal (Y-chromosome), or biparental (nuclear) evolutionary histories, what is intriguing with Rupicapra is that the same marker, in this case, mitochondrial DNA, led to contrasting conclusions on the number and composition of lineages at different times, depending on the methodological approach (restriction fragment length polymorphisms or sequencing) and the region analyzed (coding, non-coding, a combination or the whole mitogenome) [2,18]. Currently, the most accepted taxonomy recognizes two species (R. rupicapra and R. pyrenaica) and three mitochondrial clades corresponding to the geographic distribution of the populations (eastern, central, and western).
To disentangle these controversies, we integrated previous knowledge on the topic with newly sequenced mitogenomes. So far only two studies used whole mitogenomes of the genus [17,18], and our results, based on a larger dataset, identified the same subdivision of the Caprinae tree (Figure 2) into Pantholopini, Caprini, and Ovibovini as previously reported [17,18], and the same eastern, central and western clades described elsewhere [5,6,7,18] for the Rupicapra tree (Figure 3), with the central clade being more closely related to the western than to the eastern one [18]. This observation agrees with the intermediate morphological phenotype of R. r. cartusiana compared to R. pyrenaica and R. rupicapra [18,49]. Additionally, concordantly with what was reported in mitogenome publications [17,50], but contrary to previous studies based on cytb [51,52], Budorcas sequences did not group with Ovis but were closer to Capra and Pseudois, highlighting the increased resolution provided by whole mitogenomes compared to single mitochondrial genes.
The other point worth mentioning is the presence of a sequence attributed to R. r. rupicapra within the R. r. balcanica clade. This result is particularly intriguing as it taps into an interesting management issue. The sample comes from the Northern Dinaric Mountains. The southern part of this mountain chain was identified as inhabited by a human-induced hybrid population based on the comparison between nuclear and mitochondrial markers [21]. The same study reported no shared haplotypes between the Northern and Southern Velebit populations. Although based on a single individual, our result prompts further investigation of this area to validate the extension of the contact area between the two subspecies and the potential implications for management.
The clustering of R. r. cartusiana sequence with R. p. ornata (Figure 2 and Figure 3) and its grouping in the same clade as other subspecies of R. pyrenaica confirms previous findings based on morphological [25] and genetic data [6,7,18], and further increases disconcordance between current chamois systematics and the results of genetic analyses.
Quite interestingly, Pérez and colleagues [18] reported that in the central clade COX genes were quite conserved while ATP genes were more variable. Our analyses at the species level observed a similar pattern but with some important differences between species. Nucleotide diversity was generally lower in R. rupicapra but with a trend similar to R. pyrenaica, with the exception of ND4L, where a slight decrease (0.007) was observed compared to the Iberian chamois (0.012), and unexpectedly high variation at the ND6 gene (0.019 and 0.013, respectively). GC content was more homogeneous between the two species, with a single noticeable difference at the ATP8 gene, with the Alpine chamois showing lower variability (0.335 and 0.372, respectively). While estimates of nucleotide diversity on small samples are found to be underestimated [53], it is interesting to notice that greater variability was observed in the species with a lower sample size, opening questions on the population history of both species. All mitochondrial genes are involved in cellular respiration and are thus extremely important for efficient energy production and aging [12,13]. However, variation in mitochondrial genes can lead to a number of pathological disorders [54,55,56]. In particular, in humans, mutations at ND genes have been associated with progressive loss of central vision [57], epilepsy [58], and muscular dystonia [59], whereas ATP8 has been associated with autoimmune skin diseases [60], epilepsy [58] and cancer [61], making the differences of variability between species worth further investigation, considering the potential implications for conservation.
5. Conclusions
Overall, our results provide increased resolution compared to the available literature, confirm the phylogeny previously reported for both whole and partial mitochondrial sequences and the discrepancy among markers in phylogenetic reconstruction of the genus. Pérez and colleagues [9] explained the differences among nuclear, mitochondrial, and Y-chromosome phylogenies with male-mediated introgression and female philopatry. Hybridization is a particularly controversial aspect for this genus, since both natural and human-mediated hybridization have been repeatedly hypothesized or anecdotally reported on several occasions, but confirmed on few [19]. However, the taxonomic subdivision of the genus was recently questioned again, since female philopatry might bias mitochondrial-based phylogenies and the extremely low variation of nuclear markers observed suggests a single species [9]. Further studies based on multiple markers, and possibly including archeological remains, are thus needed to solve the controversy.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/article/10.3390/ani11041065/s1, Figure S1: Structural organization of Rupicapra mitochondrial genome; Table S1: List of sequences and accession codes; Table S2: Variability among Rupicapra subspecies for each gene.
Author Contributions
Conceptualization, N.Š. and E.B.; software, N.B., U.G. and T.T.; formal analysis, L.I.; investigation, J.C. and J.H.; resources, P.L., M.C.A., E.B. and N.Š.; writing—original draft preparation, L.I.; writing—review and editing, T.S., E.B., T.T., U.G. and N.Š.; visualization, T.S., U.G., T.T.; supervision, E.B. and N.Š.; project administration, T.S. and N.Š.; funding acquisition, N.Š. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Croatian Science Foundation, project IP 2016-06-5751, “DNA as evidence of distribution and vitality of endangered Balkan chamois”; the RESBIOS European Union’s Horizon 2020 Research and Innovation Program (No. 872146) and the Open Access Publication Fund of the University of Zagreb Faculty of Agriculture.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The newly assembled DNA sequences presented in this study are openly available in GenBank, with accession number of each sequence stated in the manuscript.
Acknowledgments
The authors thank Ante Turudić for advice and technical help with handling of sequencing data. The authors thank Sandro Lovari, and Luca Corlatti, for information and discussion about chamois taxonomy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lovari, S. Evolutionary aspects of the biology of chamois, Rupicapra spp. (Bovidae, Caprinae). In The Biology and Management of Capricornis and Related Mountain Antelopes; Springer: Dordrecht, The Netherlands, 1987; pp. 51–61. [Google Scholar]
- Corlatti, L.; Lorenzini, R.; Lovari, S. The conservation of the chamois Rupicapra spp. Mamm. Rev. 2011, 41, 163–174. [Google Scholar] [CrossRef]
- Herrero, J.; Lovari, S.; Nores, C.; Toigo, C. Rupicapra pyrenaica. IUCN Red List Threat. Species 2020, e.T19771A171131310. [Google Scholar] [CrossRef]
- Anderwald, P.; Ambarli, H.; Avramov, S.; Ciach, M.; Corlatti, L.; Farkas, A.; Jovanovic, M.; Papaioannou, H.; Peters, W.; Sarasa, M.; et al. Rupicapra rupicapra. IUCN Red List Threat. Species 2020, e.T39255A22149561. [Google Scholar] [CrossRef]
- Pérez, T.; Hammer, S.E.; Albornoz, J.; Domínguez, A. Y-chromosome phylogeny in the evolutionary net of chamois (genus Rupicapra). BMC Evol. Biol. 2011, 11, 272. [Google Scholar] [CrossRef]
- Rodríguez, F.; Hammer, S.; Pérez, T.; Suchentrunk, F.; Lorenzini, R.; Michallet, J.; Martinkova, N.; Albornoz, J.; Domínguez, A. Cytochrome b Phylogeography of Chamois (Rupicapra spp.). Population Contractions, Expansions and Hybridizations Governed the Diversification of the Genus. J. Hered. 2009, 100, 47–55. [Google Scholar] [CrossRef][Green Version]
- Rodríguez, F.; Pérez, T.; Hammer, S.E.; Albornoz, J.; Domínguez, A. Integrating phylogeographic patterns of microsatellite and mtDNA divergence to infer the evolutionary history of chamois (genus Rupicapra). BMC Evol. Biol. 2010, 10, 222. [Google Scholar] [CrossRef]
- Pérez, T.; Rodríguez, F.; Fernández, M.; Albornoz, J.; Domínguez, A. Ancient mitochondrial pseudogenes reveal hybridization between distant lineages in the evolution of the Rupicapra genus. Gene 2017, 628, 63–71. [Google Scholar] [CrossRef]
- Pérez, T.; Fernández, M.; Hammer, S.E.; Domínguez, A. Multilocus Intron Trees Reveal Extensive Male-Biased Homogenization of Ancient Populations of Chamois (Rupicapra spp.) across Europe during Late Pleistocene. PLoS ONE 2017, 12, e0170392. [Google Scholar] [CrossRef] [PubMed]
- Aquadro, C.F.; Greenberg, B.D. Human mitochondrial DNA variation and evolution: Analysis of nucleottide sequences from seven individuals. Genetics 1983, 103, 287–312. [Google Scholar] [CrossRef]
- Cann, R.L.; Brown, W.M.; Wilson, A. Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics 1984, 106, 479–499. [Google Scholar] [CrossRef]
- Neupert, W. Mitochondrial Gene Expression: A Playground of Evolutionary Tinkering. Annu. Rev. Biochem. 2016, 85, 65–76. [Google Scholar] [CrossRef]
- Herrero, A.; Barja, G. H2O2 production of heart mitochondria and aging rate are slower in canaries and parakeets than in mice: Sites of free radical generation and mechanisms involved. Mech. Ageing Dev. 1998, 103, 133–146. [Google Scholar] [CrossRef]
- Shen, Y.-Y.; Liang, L.; Zhu, Z.-H.; Zhou, W.-P.; Irwin, D.M.; Zhang, Y.-P. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. USA 2010, 107, 8666–8671. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Huang, G.; Sun, J.; Wang, Z.; Teng, S.; Cao, Y.; Hanif, Q.; Chen, N.; Lei, C.; Liao, Y. Mitogenome Diversity and Maternal Origins of Guangxi Buffalo Breeds. Animals 2020, 10, 547. [Google Scholar] [CrossRef]
- Sun, C.-H.; Liu, H.-Y.; Min, X.; Lu, C.-H. Mitogenome of the little owl Athene noctua and phylogenetic analysis of Strigidae. Int. J. Biol. Macromol. 2020, 151, 924–931. [Google Scholar] [CrossRef]
- Hassanin, A.; Ropiquet, A.; Couloux, A.; Cruaud, C. Evolution of the Mitochondrial Genome in Mammals Living at High Altitude: New Insights from a Study of the Tribe Caprini (Bovidae, Antilopinae). J. Mol. Evol. 2009, 68, 293–310. [Google Scholar] [CrossRef] [PubMed]
- Pérez, T.; González, I.; Essler, S.E.; Fernández, M.; Domínguez, A. The shared mitochondrial genome of Rupicapra pyrenaica ornata and Rupicapra rupicapra cartusiana: Old remains of a common past. Mol. Phylogenet. Evol. 2014, 79, 375–379. [Google Scholar] [CrossRef]
- Iacolina, L.; Corlatti, L.; Buzan, E.; Safner, T.; Šprem, N. Hybridisation in European ungulates: An overview of the current status, causes, and consequences. Mamm. Rev. 2019, 49, 45–59. [Google Scholar] [CrossRef]
- Apollonio, M.; Scandura, M.; Šprem, N. Reintroductions as a management tool for European Ungulates. In Behavior and Management of European Ungulates; Putman, R., Apollonio, M., Eds.; Whittles Publishing: Dunbeath, UK, 2014; pp. 46–77. [Google Scholar]
- Šprem, N.; Buzan, E. The genetic impact of chamois management in the dinarides. J. Wildl. Manag. 2016, 80, 783–793. [Google Scholar] [CrossRef]
- Marinier, E.; Brown, D.G.; McConkey, B.J. Pollux: Platform independent error correction of single and mixed genomes. BMC Bioinform. 2015, 16, 10. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef] [PubMed]
- BLAST. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi. (accessed on 15 January 2021).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Conant, G.C.; Wolfe, K.H. GenomeVx: Simple web-based creation of editable circular chromosome maps. Bioinformatics 2008, 24, 861–862. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Pais, F.S.-M.; de Cássia Ruy, P.; Oliveira, G.; Coimbra, R.S. Assessing the efficiency of multiple sequence alignment programs. Algorithms Mol. Biol. 2014, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Achilli, A.; Olivieri, A.; Soares, P.; Lancioni, H.; Kashani, B.H.; Perego, U.A.; Nergadze, S.G.; Carossa, V.; Santagostino, M.; Capomaccio, S.; et al. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc. Natl. Acad. Sci. USA. 2012, 109, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Kumar, S. Evolutionary distance estimation under heterogeneous substitution pattern among lineages. Mol. Biol. Evol. 2002, 19, 1727–1736. [Google Scholar] [CrossRef]
- Hiendleder, S.; Lewalski, H.; Wassmuth, R.; Janke, A. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. J. Mol. Evol. 1998, 47, 441–448. [Google Scholar] [CrossRef]
- Mereu, P.; Palici di Suni, M.; Manca, L.; Masala, B. Complete nucleotide mtDNA sequence of Barbary sheep (Ammotragus lervia). DNA Seq. 2008, 19, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Nascetti, G.; Lovari, S.; Lanfranchi, P.; Berducou, C.; Mattiucci, S.; Rossi, L.; Bullini, L. Revision of Rupicapra genus. III. Electrophoretic studies demostrating species distintion of chamois populations of the Alps from those of the Apennines and Pyrenees. In The Biology and Management of Mountain Ungulates; Lovar, S., Ed.; Croom-Helm: London, UK, 1985; pp. 56–62. [Google Scholar]
- Lovari, S.; Scala, C. Revision of Rupicapra Genus. I. A statistical re-evaluation of Couturier’s data on the morphometry of six chamois subspecies. Boll. di Zool. 1980, 47, 113–124. [Google Scholar] [CrossRef]
- Zhou, M.; Yu, J.; Li, B.; Ouyang, B.; Yang, J. The complete mitochondrial genome of Budorcas taxicolor tibetana (Artiodactyla: Bovidae) and comparison with other Caprinae species: Insight into the phylogeny of the genus Budorcas. Int. J. Biol. Macromol. 2019, 121, 223–232. [Google Scholar] [CrossRef]
- Groves, P.; Shields, G.F. Phylogenetics of the Caprinae based on cytochrome b sequence. Mol. Phylogenet. Evol. 1996, 5, 467–476. [Google Scholar] [CrossRef]
- Groves, P.; Shields, G.F. Cytochrome B Sequences Suggest Convergent Evolution of the Asian Takin and Arctic Muskox. Mol. Phylogenet. Evol. 1997, 8, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Gorbachev, V. V Effect of random sample size on the accuracy of nucleotide diversity estimation. Russ. J. Genet. 2012, 48, 746–750. [Google Scholar] [CrossRef]
- Hatakeyama, H.; Goto, Y. Concise Review: Heteroplasmic Mitochondrial DNA Mutations and Mitochondrial Diseases: Toward iPSC-Based Disease Modeling, Drug Discovery, and Regenerative Therapeutics. Stem Cells 2016, 34, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.L.; Sue, C.M. The Genetics of Mitochondrial Disease. Semin. Neurol. 2011, 31, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Craven, L.; Alston, C.L.; Taylor, R.W.; Turnbull, D.M. Recent Advances in Mitochondrial Disease. Annu. Rev. Genomics Hum. Genet. 2017, 18, 257–275. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.A.; Govindaraj, P.; Soumittra, N.; Srilekha, S.; Ambika, S.; Vanniarajan, A.; Meena, A.K.; Uppin, M.S.; Sundaram, C.; Taly, A.B.; et al. Haplogroup Heterogeneity of LHON Patients Carrying the m.14484T>C Mutation in India. Investig. Opthalmol. Vis. Sci. 2013, 54, 3999. [Google Scholar] [CrossRef]
- Gurses, C.; Azakli, H.; Alptekin, A.; Cakiris, A.; Abaci, N.; Arikan, M.; Kursun, O.; Gokyigit, A.; Ustek, D. Mitochondrial DNA profiling via genomic analysis in mesial temporal lobe epilepsy patients with hippocampal sclerosis. Gene 2014, 538, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.-S.; Ki, C.-S.; Park, K.-J. Pediatric-Onset Dystonia Associated with Bilateral Striatal Necrosis and G14459A Mutation in a Korean Family: A Case Report. J. Korean Med. Sci. 2010, 25, 180. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hirose, M.; Schilf, P.; Benoit, S.; Eming, R.; Gläser, R.; Homey, B.; Kunz, M.; Nebel, A.; Peitsch, W.K.; Pföhler, C.; et al. Polymorphisms in the mitochondrially encoded ATP synthase 8 gene are associated with susceptibility to bullous pemphigoid in the German population. Exp. Dermatol. 2015, 24, 715–717. [Google Scholar] [CrossRef] [PubMed]
- Thapa, S.; Lalrohlui, F.; Ghatak, S.; Zohmingthanga, J.; Lallawmzuali, D.; Pautu, J.L.; Senthil Kumar, N. Mitochondrial complex I and V gene polymorphisms associated with breast cancer in mizo-mongloid population. Breast Cancer 2016, 23, 607–616. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).