The Concurrent Detection of Chelonid Alphaherpesvirus 5 and Chelonia mydas Papillomavirus 1 in Tumoured and Non-Tumoured Green Turtles
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction
2.3. Primer and Probe Design
2.4. Producing Standard Curves for ChHV5, CmPV1 and Green Turtle Genomic DNA
2.5. Calibration Curves and Real-Time Quantitative PCR
2.6. Viral and Host qPCR Reaction Conditions and Data Interpretation
2.7. Statistical Analysis
3. Results
3.1. qPCR Validation and Optimisation
3.2. Viral DNA Detection
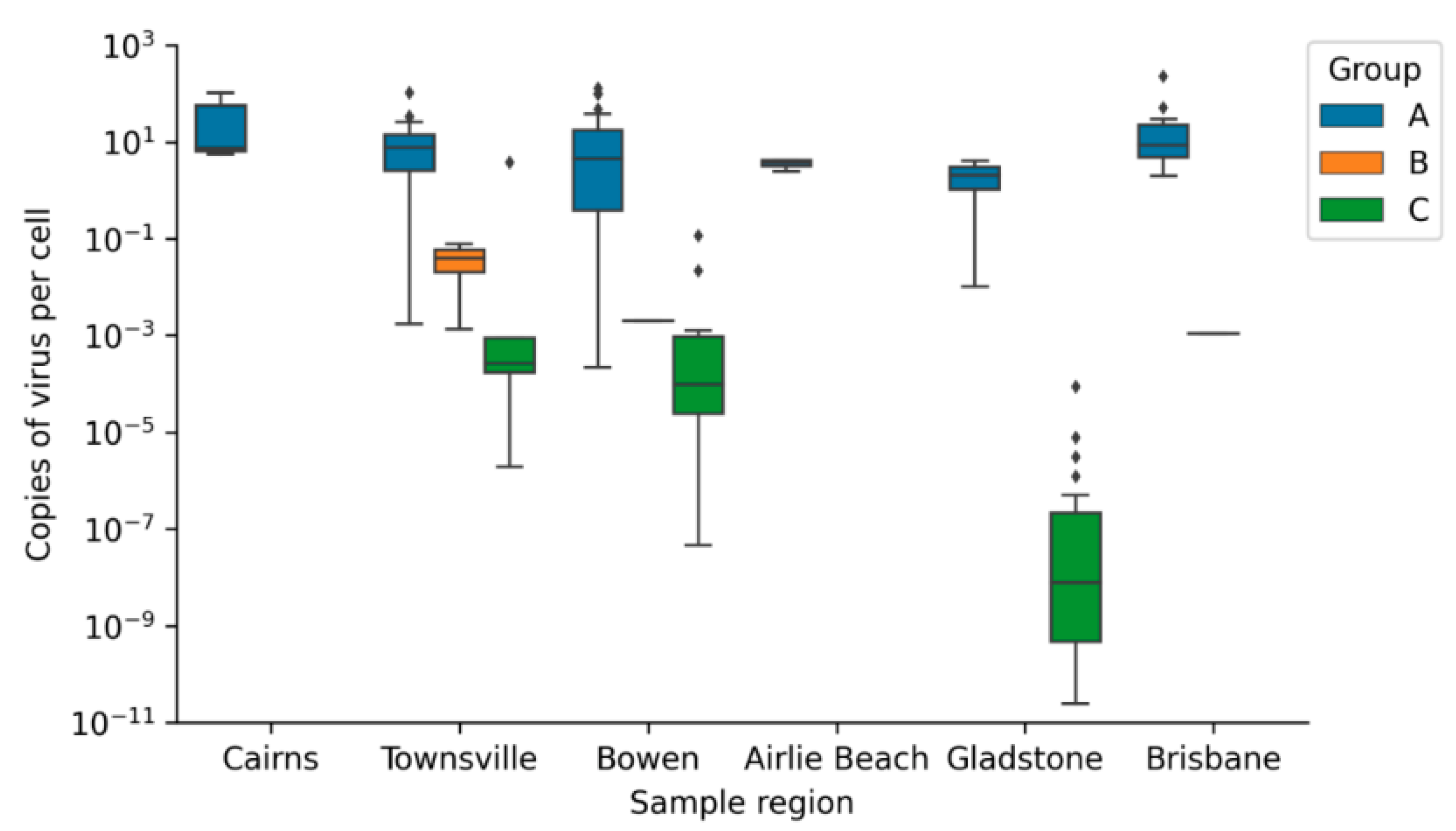
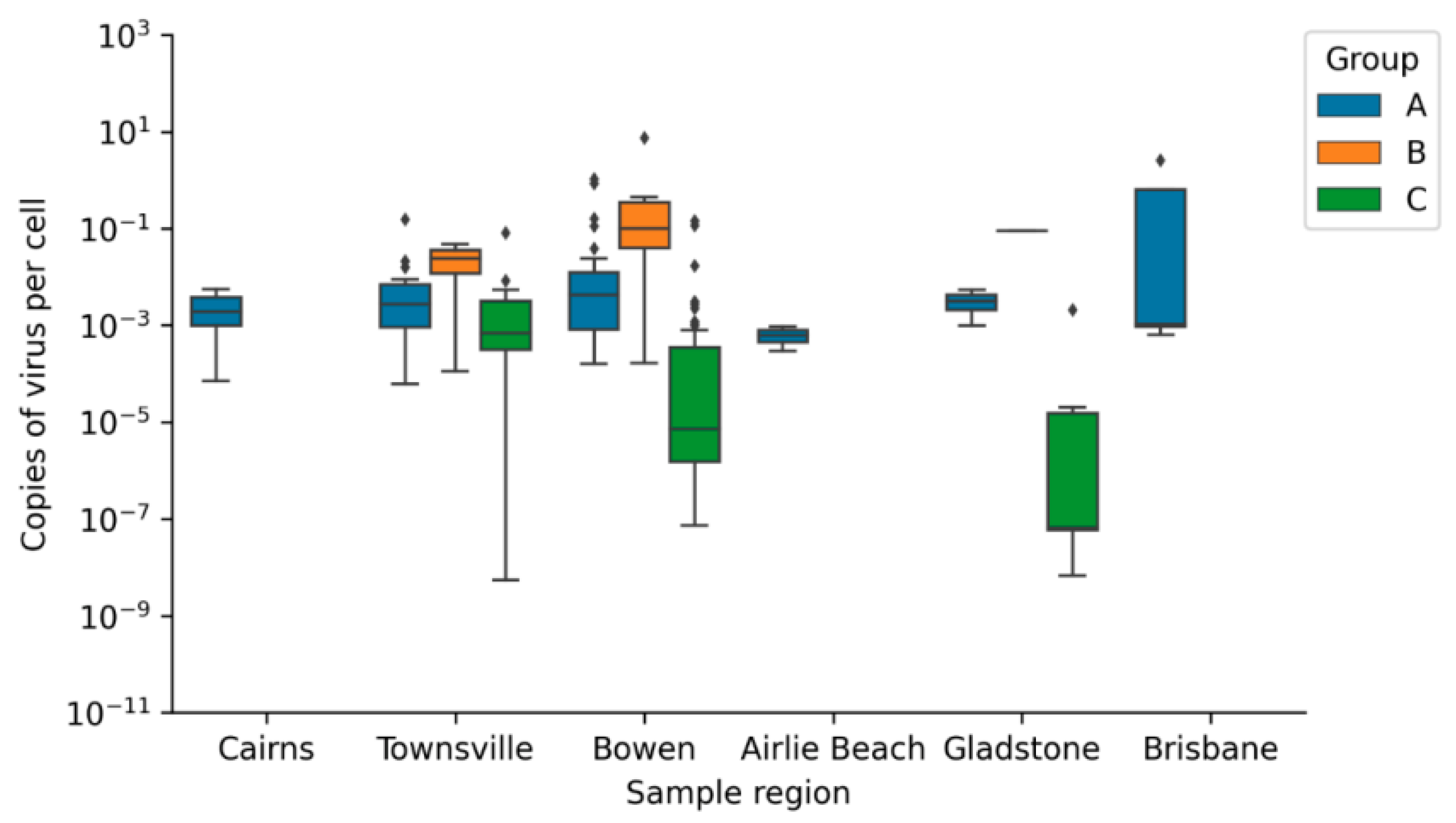
Viral Loads of CmPV1 and ChHV5
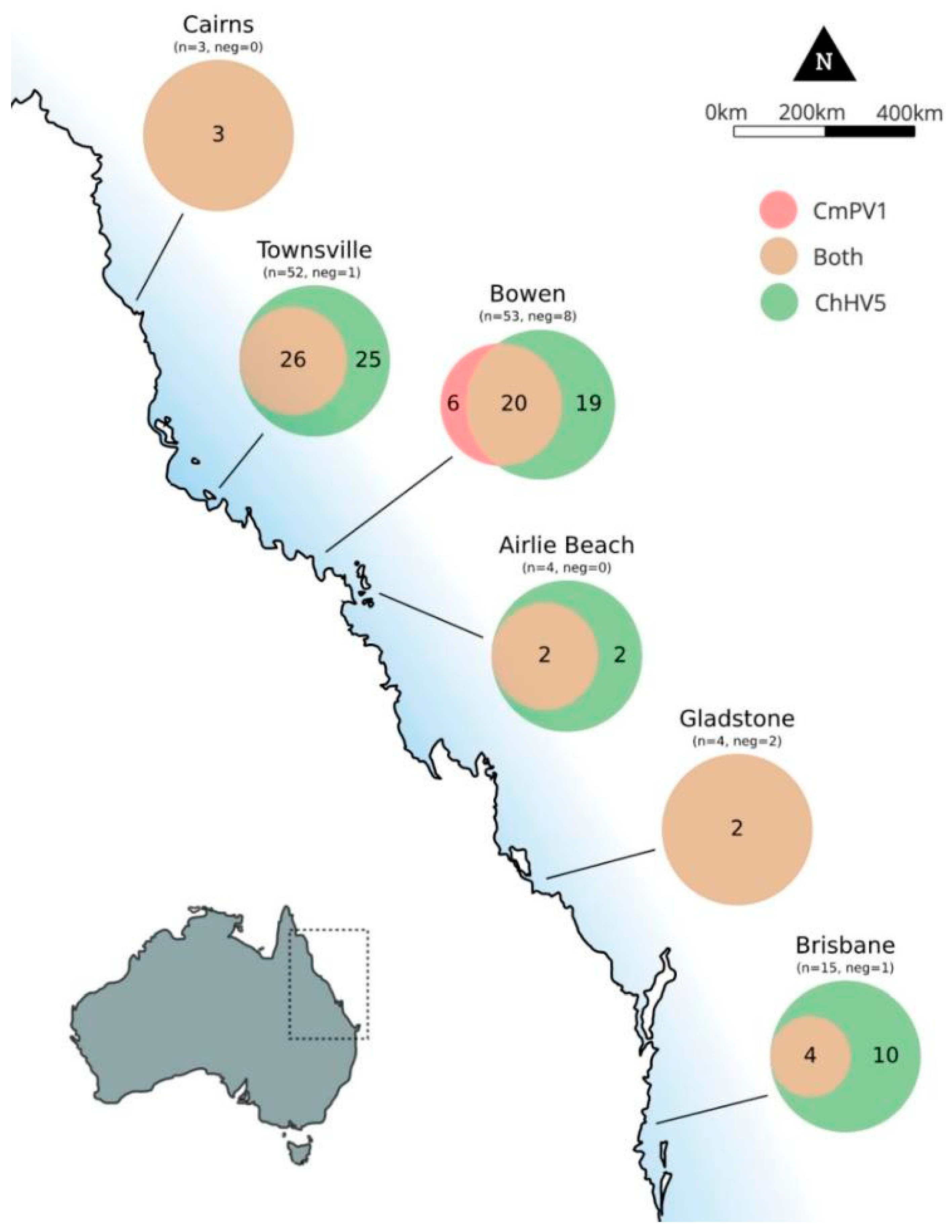
3.3. Geographic Distribution of ChHV5 and CmPV1
Geographic Distribution of Viral Loads
4. Discussion
4.1. Rate of Detection
4.2. Mean Viral Load
4.3. Concurrent Detection
4.4. Geographic Locations
4.5. Limitations
4.6. Future Perspective
4.7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mashkour, N.; Jones, K.; Kophamel, S.; Hipolito, T.; Ahasan, S.; Walker, G.; Jakob-Hoff, R.; Whittaker, M.; Hamann, M.; Bell, I.; et al. Disease risk analysis in sea turtles: A baseline study to inform conservation efforts. PLoS ONE 2020, 15, e0230760. [Google Scholar] [CrossRef]
- Jones, K.M.; Ariel, E.; Burgess, G.; Read, M. A review of fibropapillomatosis in Green turtles (Chelonia mydas). Vet. J. 2016, 212, 48–57. [Google Scholar] [CrossRef]
- Work, T.M.; Work, T.M.; Balazs, G.H.; Rameyer, R.A.; Morris, R.A. Retrospective pathology survey of green turtles Chelonia mydas with fibropapillomatosis in the Hawaiian Islands, 1993-2003. Dis. Aquat. Org. 2004, 62, 163–176. [Google Scholar] [CrossRef]
- Herbst, L.H. Fibropapillomatosis of marine turtles. Annu. Rev. Fish Dis. 1994, 4, 389–425. [Google Scholar] [CrossRef]
- Norton, T.M.; Jacobson, E.R.; Sundberg, J.P. Cutaneous Fibropapillomas and Renal Myxofibroma in a Green Turtle, Chelonia mydas. J. Wildl. Dis. 1990, 26, 265–270. [Google Scholar] [CrossRef]
- Kang, K.; Torres-Velez, F.; Zhang, J.; Moore, P.; Moore, D.; Rivera, S.; Brown, C. Localization of Fibropapilloma-associated Turtle Herpesvirus in Green Turtles (Chelonia mydas) by In-Situ Hybridization. J. Comp. Pathol. 2008, 139, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Flint, M.; Limpus, C.; Patterson-Kane, J.; Murray, P.; Mills, P. Corneal Fibropapillomatosis in Green Sea Turtles (Chelonia mydas) in Australia. J. Comp. Pathol. 2010, 142, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.A.; Balazs, G.H.; Spraker, T.R.; Gross, T.S. Adrenal and Hematological Responses to Stress in Juvenile Green Turtles (Chelonia mydas) with and without Fibropapillomas. Physiol. Zool. 1995, 68, 831–854. [Google Scholar] [CrossRef]
- Work, T.M.; Rameyer, R.A.; Balazs, G.H.; Cray, C.; Chang, S.P. Immune Status of Free-Ranging Green Turtles with Fibropapillomatosis from Hawaii. J. Wildl. Dis. 2001, 37, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Work, T.M.; Dagenais, J.; Weatherby, T.M.; Balazs, G.H.; Ackermann, M. In Vitro Replication of Chelonid Herpesvirus 5 in Organotypic Skin Cultures from Hawaiian Green Turtles (Chelonia mydas). J. Virol. 2017, 91, e00404-17. [Google Scholar] [CrossRef]
- Herbst, L.H. The Etiology and Pathogenesis of Green Turtle Fibropapillomatosis. Ph.D. Thesis, University of Florida, Gainesville, FL, USA, 1995. [Google Scholar]
- Jacobson, E.; Mansell, J.; Sundberg, J.; Hajjar, L.; Reichmann, M.; Ehrhart, L.; Walsh, M.; Murru, F. Cutaneous fibropapillomas of green turtles (Chelonia mydas). J. Comp. Pathol. 1989, 101, 39–52. [Google Scholar] [CrossRef]
- Casey, R.; Quackenbush, S.; Work, T.; Balazs, G.; Bowser, P.; Casey, J. Evidence for retrovirus infections in green turtles Chelonia mydas from the Hawaiian islands. Dis. Aquat. Org. 1997, 31, 1–7. [Google Scholar] [CrossRef]
- Jacobson, E.; Buergelt, C.; Williams, B.; Harris, R. Herpesvirus in cutaneous fibropapillomas of the green turtle Chelonia mydas. Dis. Aquat. Org. 1991, 12, 1–6. [Google Scholar] [CrossRef]
- Quackenbush, S.L.; Work, T.M.; Balazs, G.H.; Casey, R.N.; Rovnaka, J.; Chavesd, A.; du Toit, L.; Baines, J.D.; Parrish, C.R.; Bowser, P.R.; et al. Three Closely Related Herpesviruses Are Associated with Fibropapillomatosis in Marine Turtles. Virology 1998, 246, 392–399. [Google Scholar] [CrossRef]
- Herbst, L.; Ene, A.; Su, M.; DeSalle, R.; Lenz, J. Tumor outbreaks in marine turtles are not due to recent herpesvirus mutations. Curr. Biol. 2004, 14, R697–R699. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Aguirre, A.A.; Work, T.M.; Balazs, G.H.; Nerurkar, V.R.; Yanagihara, R. Identification of a small, naked virus in tumor-like aggregates in cell lines derived from a green turtle, Chelonia mydas, with fibropapillomas. J. Virol. Methods 2000, 86, 25–33. [Google Scholar] [CrossRef]
- Alfaro-Núñez, A.; Bertelsen, M.F.; Bojesen, A.M.; Rasmussen, I.; Zepeda-Mendoza, L.; Olsen, M.T.; Gilbert, M.T.P. Global distribution of Chelonid fibropapilloma-associated herpesvirus among clinically healthy sea turtles. BMC Evol. Biol. 2014, 14, 206. [Google Scholar] [CrossRef]
- Lackovich, J.K.; Brown, D.R.; Homer, B.L.; Garber, R.L.; Mader, D.R.; Moretti, R.H.; Patterson, A.D.; Herbst, L.H.; Orós, J.; Jacobson, E.R.; et al. Association of herpesvirus with fibropapillomatosis of the green turtle Chelonia mydas and the loggerhead turtle Caretta caretta in Florida. Dis. Aquat. Org. 1999, 37, 89–97. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, Y.; Aguirre, A.; Zhao, Z.; Liu, C.; Nerurkar, V.; Yanagihara, R. RT-PCR detection of the expression of the polymerase gene of a novel reptilian herpesvirus in tumor tissues of green turtles with fibropapilloma. Arch. Virol. 2003, 148, 1155–1163. [Google Scholar] [CrossRef]
- Nigro, O.; Aguirre, A.A.; Lu, Y. Nucleotide sequence of an ICP18. 5 assembly protein (UL28) gene of green turtle herpesvirus pathogenically associated with green turtle fibropapilloma. J. Virol. Methods 2004, 120, 107–112. [Google Scholar] [CrossRef]
- Nigro, O.; Yu, G.; Aguirre, A.A.; Lu, Y. Sequencing and characterization of the full-length gene encoding the single-stranded DNA binding protein of a novel Chelonian herpesvirus. Arch. Virol. 2004, 149, 337–347. [Google Scholar] [CrossRef]
- Page-Karjian, A.; Norton, T.M.; Ritchie, B.; Brown, C.; Mancia, C.; Jackwood, M.; Gottdenker, N.L. Quantifying chelonid herpesvirus 5 in symptomatic and asymptomatic rehabilitating green sea turtles. Endanger. Species Res. 2015, 28, 135–146. [Google Scholar] [CrossRef]
- Page-Karjian, A.; Torres, F.; Zhang, J.; Rivera, S.; Diez, C.; Moore, P.A.; Moore, D.; Brown, C. Presence of chelonid fibropapilloma-associated herpesvirus in tumored and non-tumored green turtles, as detected by polymerase chain reaction, in endemic and non-endemic aggregations, Puerto Rico. SpringerPlus 2012, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Quackenbush, S.L.; Aguirre, A.A.; Spraker, T.R.; Horrocks, J.A.; Vermeer, L.A.; Balazs, G.H.; Casey, J.W.; Casey, R.N.; Murcek, R.J.; Paul, T.A.; et al. Quantitative analysis of herpesvirus sequences from normal tissue and fibropapillomas of marine turtles with real-time PCR. Virology 2001, 287, 105–111. [Google Scholar] [CrossRef][Green Version]
- Rodenbusch, C.R.; Baptistotte, C.; Werneck, M.R.; Pires, T.T.; Melo, M.T.D.; De Ataíde, M.W.; Dos Reis, K.; Testa, P.; Alieve, M.M.; Canal, C.W. Fibropapillomatosis in green turtles Chelonia mydas in Brazil: Characteristics of tumors and virus. Dis. Aquat. Org. 2014, 111, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Hu, N.; Lu, Y.; Nerurkar, V.R.; Yanagihara, R. Rapid acquisition of entire DNA polymerase gene of a novel herpesvirus from green turtle fibropapilloma by a genomic walking technique. J. Virol. Methods 2001, 91, 183–195. [Google Scholar] [CrossRef]
- Yu, Q.; Lu, Y.; Nerurkar, V.R.; Yanagihara, R. Amplification and analysis of DNA flanking known sequences of a novel herpesvirus from green turtles with fibropapilloma. Arch. Virol. 2000, 145, 2669–2676. [Google Scholar] [CrossRef]
- Jones, K.; Burgess, G.; Budd, A.M.; Huerlimann, R.; Mashkour, N.; Ariel, E. Molecular evidence for horizontal transmission of chelonid alphaherpesvirus 5 at green turtle (Chelonia mydas) foraging grounds in Queensland, Australia. PLoS ONE 2020, 15, e0227268. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wang, Y.; Yu, Q.; Aguirre, A.A.; Balazs, G.H.; Nerurkar, V.R.; Yanagihara, R. Detection of herpesviral sequences in tissues of green turtles with fibropapilloma by polymerase chain reaction. Arch. Virol. 2000, 145, 1885–1893. [Google Scholar] [CrossRef] [PubMed]
- McGeoch, D.J.; Gatherer, D. Integrating Reptilian Herpesviruses into the Family Herpesviridae. J. Virol. 2005, 79, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Page-Karjian, A.; Gottdenker, N.L.; Whitfield, J.; Herbst, L.; Norton, T.M.; Ritchie, B. Potential Noncutaneous Sites of Chelonid Herpesvirus 5 Persistence and Shedding in Green Sea Turtles Chelonia mydas. J. Aquat. Anim. Health 2017, 29, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Alfaro-Núñez, A.; Gilbert, M.T.P. Validation of a sensitive PCR assay for the detection of Chelonid fibropapilloma-associated herpesvirus in latent turtle infections. J. Virol. Methods 2014, 206, 38–41. [Google Scholar] [CrossRef]
- Ariel, E.; Nainu, F.; Jones, K.; Juntunen, K.; Bell, I.; Gaston, J.; Scott, J.; Trocini, S.; Burgess, G. Phylogenetic variation of chelonid alphaherpesvirus 5 in green turtle (Chelonia mydas) populations along the Queensland Coast, Australia. J. Aquat. Anim. Health 2017. [Google Scholar] [CrossRef]
- Lawrance, M.F.; Mansfield, K.L.; Sutton, E.; Savage, A.E. Molecular evolution of fibropapilloma-associated herpesviruses infecting juvenile green and loggerhead sea turtles. Virology 2018, 521, 190–197. [Google Scholar] [CrossRef]
- Ugochukwu, I.C.I.; Aneke, C.I.; Idoko, I.S.; Sani, N.A.; Amoche, A.J.; Mshiela, W.P.; Ede, R.E.; Ibrahim, N.D.G.; Njoku, C.I.O.; Sackey, A.K.B. Bovine papilloma: Aetiology, pathology, immunology, disease status, diagnosis, control, prevention and treatment: A review. Comp. Haematol. Int. 2019, 28, 737–745. [Google Scholar] [CrossRef]
- Garcês, A.; Pires, I.; Savini, F.; Scagliarini, A.; Gallina, L. Cutaneous Fibropapilloma in a Red Deer (Cervus elaphus) Associated with Cervus elaphus Papillomavirus in Portugal. J. Wildl. Dis. 2020, 56, 636. [Google Scholar] [CrossRef]
- Munday, J.S.; Cullum, A.A.; Thomson, N.A.; Bestbier, M.; McCormack, T.; Julian, A.F. Anal fibropapillomas containing bovine papillomavirus type 2 DNA in two groups of heifers. N. Z. Vet. J. 2018, 66, 267–271. [Google Scholar] [CrossRef]
- Brown, D.R.; Lackovich, J.K.; Klein, P.A. Further evidence for the absence of papillomaviruses from sea turtle fibropapillomas. Vet. Rec. 1999, 145, 616–617. [Google Scholar] [CrossRef]
- Mashkour, N.; MacLaine, A.; Burgess, G.W.; Ariel, E. Discovery of an Australian Chelonia mydas papillomavirus via green turtle primary cell culture and qPCR. J. Virol. Methods 2018, 258, 13–23. [Google Scholar] [CrossRef]
- Limpus, C.J.; Reed, P.C. The green turtle, Chelonia mydas, in Queensland: Population structure in a coral reef feeding area. In Biology of Australian Frogs and Reptiles; Grigg, G., Shine, R., Ehmann, H., Eds.; Surrey Beatty and Sons: Sydney, Australia, 1985; pp. 47–52. [Google Scholar]
- Hou, Y.; Zhang, H.; Miranda, L.; Lin, S. Serious Overestimation in Quantitative PCR by Circular (Supercoiled) Plasmid Standard: Microalgal pcna as the Model Gene. PLoS ONE 2010, 5, e9545. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Greenblatt, R.J.; Work, T.M.; Dutton, P.; Sutton, C.A.; Spraker, T.R.; Casey, R.N.; Diez, C.E.; Parker, D.; Leger, J.S.; Balazs, G.H.; et al. Geographic Variation in Marine Turtle Fibropapillomatosis. J. Zoo Wildl. Med. 2005, 36, 527–530. [Google Scholar] [CrossRef]
- Alfaro-Núñez, A.; Bojesen, A.M.; Bertelsen, M.F.; Wales, N.; Balazs, G.H.; Gilbert, M.T.P. Further evidence of Chelonid herpesvirus 5 (ChHV5) latency: High levels of ChHV5 DNA detected in clinically healthy marine turtles. PeerJ 2016, 4, e2274. [Google Scholar] [CrossRef]
- Díaz, F. OIE Standard on principles and methods of validation of diagnostic assays for infectious diseases. In Proceedings of the OIE Regional Workshop for OIE National Focal Points for Veterinary Products, Tokyo, Japan, 3–5 December 2014. [Google Scholar]
- Sun, Z.; Zhang, R.; Liu, Z.; Liu, C.; Li, X.; Zhou, W.; Yang, L.; Ruan, Q.; Zhang, X. Development of a fluorescence–based multiplex genotyping method for simultaneous determination of human papillomavirus infections and viral loads. BMC Cancer 2015, 15, 860. [Google Scholar] [CrossRef]
- Oyervides-Muñoz, M.A.; Pérez-Maya, A.A.; Sánchez-Domínguez, C.N.; Berlanga-Garza, A.; Antonio-Macedo, M.; Valdéz-Chapa, L.D.; Cerda-Flores, R.M.; Trevino, V.; Barrera-Saldaña, H.A.; Garza-Rodríguez, M.L. Multiple HPV Infections and Viral Load Association in Persistent Cervical Lesions in Mexican Women. Viruses 2020, 12, 380. [Google Scholar] [CrossRef]
- Duan, L.; Du, H.; Wang, C.; Huang, X.; Qu, X.; Shi, B.; Liu, Y.; Zhang, W.; Duan, X.; Wei, L.; et al. The application of BMRT-HPV viral load to secondary screening strategies for cervical cancer. PLoS ONE 2020, 15, e0232117. [Google Scholar] [CrossRef]
- Manire, C.A.; Stacy, B.A.; Kinsel, M.J.; Daniel, H.T.; Anderson, E.T.; Wellehan, J.F. Proliferative dermatitis in a loggerhead turtle, Caretta caretta, and a green turtle, Chelonia mydas, associated with novel papillomaviruses. Vet. Microbiol. 2008, 130, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Herbst, L.H.; Lenz, J.; Van Doorslaer, K.; Chen, Z.; Stacy, B.A.; Wellehan, J.F.; Manire, C.A.; Burk, R.D. Genomic characterization of two novel reptilian papillomaviruses, Chelonia mydas papillomavirus 1 and Caretta caretta papillomavirus 1. Virology 2009, 383, 131–135. [Google Scholar] [CrossRef]
- Jones, K. Environmental Influences on the Epidemiology of Fibropapillomatosis in Green Turtles (Chelonia mydas) and Consequences for Management of Inshore Areas of the great Barrier Reef. Ph.D. Thesis, James Cook University, Townsville, Australia, 2019. [Google Scholar]
- Greenblatt, R.J.; Work, T.M.; Balazs, G.H.; Sutton, C.A.; Casey, R.N.; Casey, J.W. The Ozobranchus leech is a candidate mechanical vector for the fibropapilloma-associated turtle herpesvirus found latently infecting skin tumors on Hawaiian green turtles (Chelonia mydas). Virology 2004, 321, 101–110. [Google Scholar] [CrossRef]
- Smith, C.J.; Osborn, A.M. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol. Ecol. 2009, 67, 6–20. [Google Scholar] [CrossRef]
- Page-Karjian, A.; Serrano, M.E.; Cartzendafner, J.; Morgan, A.; Ritchie, B.W.; Gregory, C.R.; McNeill, J.B.; Perrault, J.R.; Christiansen, E.F.; Harms, C.A. Molecular Assessment of Chelonid Alphaherpesvirus 5 Infection in Tumor-Free Green (Chelonia mydas) and Loggerhead (Caretta caretta) Sea Turtles in North Carolina, USA, 2015–2019. Animals 2020, 10, 1964. [Google Scholar] [CrossRef]
- Work, T.M.; Dagenais, J.; Willimann, A.; Balazs, G.; Mansfield, K.; Ackermann, M. Differences in Antibody Responses against Chelonid Alphaherpesvirus 5 (ChHV5) Suggest Differences in Virus Biology in ChHV5-Seropositive Green Turtles from Hawaii and ChHV5-Seropositive Green Turtles from Florida. J. Virol. 2020, 94, 01658-19. [Google Scholar] [CrossRef]
- Blackburn, N.B.; Leandro, A.C.; Nahvi, N.; Devlin, M.A.; Leandro, M.; Escobedo, I.M.; Peralta, J.M.; George, J.; Stacy, B.A.; deMaar, T.W.; et al. Transcriptomic profiling of fibropapillomatosis in green sea turtles (Chelonia mydas) from South Texas. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yetsko, K.; Farrell, J.; Stammnitz, M.R.; Whitmore, L.; Whilde, J.; Eastman, C.B.; Ramia, D.R.; Thomas, R.; Krstic, A.; Linser, P.; et al. Mutational, transcriptional and viral shedding dynamics of the marine turtle fibropapillomatosis tumor epizootic. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yetsko, K.; Farrell, J.A.; Blackburn, N.B.; Whitmore, L.; Stammnitz, M.R.; Whilde, J.; Eastman, C.B.; Ramia, D.R.; Thomas, R.; Krstic, A.; et al. Molecular characterization of a marine turtle tumor epizootic, profiling external, internal and postsurgical regrowth tumors. Commun. Biol. 2021, 4, 1–16. [Google Scholar] [CrossRef]
- Guidry, J.T.; Scott, R.S. The interaction between human papillomavirus and other viruses. Virus Res. 2017, 231, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Cruz, D.; Rodríquez, M.; Kouri, V.; Soto, Y.; Zamora, L.; Rodríguez, D.; Barrera, M.; Rehtanz, M. Concurrent papillomavirus- and herpesvirus-infection in Atlantic bottlenose dolphins (Tursiops truncatus) inhabiting the Cuban coast. Mar. Mammal Sci. 2014, 30, 1564–1572. [Google Scholar] [CrossRef]
- Kullander, J.; Handisurya, A.; Forslund, O.; Geusau, A.; Kirnbauer, R.; Dillner, J. Cutaneous human papillomavirus 88: Remarkable differences in viral load. Int. J. Cancer 2008, 122, 477–480. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Antonsson, A.; Forslund, O.; Ekberg, H.; Sterner, G.; Hansson, B.G. The Ubiquity and Impressive Genomic Diversity of Human Skin Papillomaviruses Suggest a Commensalic Nature of These Viruses. J. Virol. 2000, 74, 11636–11641. [Google Scholar] [CrossRef]
- Shimada, T.; Jones, R.; Limpus, C.; Groom, R.; Hamann, M. Long-term and seasonal patterns of sea turtle home ranges in warm coastal foraging habitats: Implications for conservation. Mar. Ecol. Prog. Ser. 2016, 562, 163–179. [Google Scholar] [CrossRef]
- Duffy, D.J.; Schnitzler, C.; Karpinski, L.; Thomas, R.; Whilde, J.; Eastman, C.; Yang, C.; Krstic, A.; Rollinson, D.; Zirkelbach, B.; et al. Sea turtle fibropapilloma tumors share genomic drivers and therapeutic vulnerabilities with human cancers. Commun. Biol. 2018, 1, 1–13. [Google Scholar] [CrossRef]
- Shamblin, B.M.; Dodd, M.G.; Williams, K.L.; Frick, M.G.; Bell, R.; Nairn, C.J. Loggerhead turtle eggshells as a source of maternal nuclear genomic DNA for population genetic studies. Mol. Ecol. Resour. 2011, 11, 110–115. [Google Scholar] [CrossRef]
| Number of Samples | |||||
|---|---|---|---|---|---|
| Study Site | Total Number of Turtles | Number of Turtles with FP Tumours | Group A (FP Tumours) | Group B (Non-Tumoured Skin from Turtles with FP Tumours) | Group C (Skin from Non-Tumoured Turtles) |
| Cairns | 2 | 2 | 3 | - | - |
| Townsville | 80 | 24 | 52 | 13 | 56 |
| Bowen | 130 | 50 | 53 | 15 | 80 |
| Airlie Beach | 1 | 1 | 4 | - | - |
| Gladstone | 54 | 4 | 4 | 4 | 50 |
| Brisbane | 8 | 8 | 15 | 4 | - |
| Total | 275 | 89 | 131 | 36 | 186 |
| Primers and Probes | Sequence | Target Gene | Amplicon Length | Cycling Conditions | Reference | |
|---|---|---|---|---|---|---|
| Cloned plasmids for standard assays | Cm-Dpol-864 | F: 5′-ATG ACG GAC GGA CAA CAG-3′ | ChHV5-DNApol | 864 bp | 2 min at 95 °C; 35 cycles (95 °C for 10 s, 57 °C for 15 s, and 72 °C for 55 s) | The present study |
| R: 5′-GGA GAT GAC GGC TGC TAA-3′ | ||||||
| GAPDH-475 | F: 5′-CCT TTA ATG CGG GTG CTG-3′ | GAPDH | 475 bp | 2 min at 95 °C; 35 cycles (95 °C for 10 s, 57 °C for 15 s, and 72 °C for 30 s) | The present study | |
| R: 5′-CAC GGT TGC TGT ATC CAA-3′ | ||||||
| Cm-Pap-109 | F: 5′-GCC GAT GAT GTC CAC TTA T-3′ | CmPV1-E1 | 109 bp | 2 min at 95 °C; 40 cycles (95 °C for 10 s, 60 °C for 20 s, and 72 °C for 30 s) | Mashkour et al. (2018) | |
| R: 5′-GCT GAA TCC ACA GAG GTA G-3′ | ||||||
| Screening assays | Cm-Pap-109 | F: 5′-GCC GAT GAT GTC CAC TTA T-3′ | CmPV1-E1 | 109 bp | 2 min at 95 °C; 50 cycles (95 °C for 10 s, 60 °C for 10 s) | Mashkour et al. (2018) |
| R: 5′-GCT GAA TCC ACA GAG GTA G-3′ | ||||||
| P: 5′-FAM CGA CCC ATG AAG CCG CTG T BHQ1-3′ | ||||||
| Dpol-ChHV5-82 | F: 5′-CTA CCT TGT CTG GAG GTG GC-3′ | ChHV5-DNApol | 82 bp | 2 min at 95 °C; 50 cycles (95 °C for 10 s, 60 °C for 10 s) | The present study | |
| R: 5′-GGG TGT GAA TAA AAT CCC GCG-3′ | ||||||
| P: 5′-FAM TAG GGC GCG ACA TGC TTC BHQ1-3′ | ||||||
| GAPDH-83 | F: 5′-CTG GTC TCC TGG TAT GGA-3′ | GAPDH | 83 bp | 2 min at 95 °C; 50 cycles (95 °C for 10 s, 60 °C for 10 s) | The present study | |
| R: 5′-CAT GGA CTC CCA ACC TAT C-3′ | ||||||
| P: 5′-FAM AAA CCA CCC TCC AAA TCT GGC BHQ1-3′ | ||||||
| Cc-Pap-99 | F: 5′-AAA GGG CAG TGG GAA ATC TC-3′ | CcPV1-E2 | 99 bp | 2 min at 95 °C; 40 cycles (95 °C for 10 s, 58 °C for 20 s, and 72 °C for 30 s) | The present study | |
| R: 5′-TGT GAT GGC GAC GAT GTG-3′ |
| Viral DNA Detections | ||||
|---|---|---|---|---|
| Sample Type | ChHV5 | CmPV1 | Both ChHV5 and CmPV1 | Neither ChHV5 and CmPV1 |
| Group A | 113/131 (86.3%) | 62/131 (47.3%) | 57/131 (43.5%) | 12/131 (9.2%) |
| Group B | 4/36 (11.1%) | 9/36 (25.0%) | 1/36 (2.8%) | 24/36 (66.7%) |
| Group C | 39/186 (21.0%) | 68/186 (36.6%) | 8/186 (4.3%) | 85/186 (45.7%) |
| Overall | 156/353 (44.2%) | 139/353 (39.4%) | 66/353 (18.7%) | 121/353 (34.3%) |
| ChHV5 (Copy Number Per Cell) | CmPV1 (Copy Number Per Cell) | |
|---|---|---|
| Group A | 15.635 ± 30.152 | 0.0814 ± 0.357 |
| Group B | 0.0204 ± 0.0379 | 0.855 ± 2.351 |
| Group C | 0.100 ± 0.606 | 0.005 ± 0.023 |
| Group A | Group B | Group C | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ChHV5 | CmPV1 | p-Value | ChHV5 | CmPV1 | p-Value | ChHV5 | CmPV1 | p-Value | ||
| Cairns | No. (%) positive | 3/3 (100.0%) | 3/3 (100.0%) | 1.000 | - | - | - | - | - | - |
| Average copy number/cell | 39.300 ± 56.788 | 0.002 ± 0.002 | * | - | - | - | - | - | * | |
| Townsville | No. (%) positive | 51/52 (98.1%) | 26/52 (50.0%) | <0.00001 | 2/13 (15.4%) | 2/13 (15.4%) | 1.000 | 5/56 (8.9%) | 11/56 (19.6%) | 0.105 |
| Average copy number/cell | 11.364 ± 16.120 | 0.010 ± 0.030 | * | 0.039 ± 0.053 | 0.023 ± 0.033 | 0.758 ± 1.695 | 0.009 ± 0.024 | * | ||
| Bowen | No. (%) positive | 39/53 (73.6%) | 26/53 (49.1%) | 0.0095 | 1/15 (6.7%) | 6/15 (40.0%) | 0.0309 | 17/80 (21.25%) | 52/80 (65%) | <0.00001 |
| Average copy number/cell | 16.456 ± 28.911 | 0.088 ± 0.257 | - | - | 0.231 ± 0.918 | 0.005 ± 0.024 | * | |||
| Airlie Beach | No. (%) positive | 4/4 (100%) | 2/4 (50.0%) | 0.429 | - | - | - | - | - | - |
| Average copy number/cell | 3.554 ± 0.835 | 0.001 ± 0.0004 | * | - | - | - | - | - | * | |
| Gladstone | No. (%) positive | 2/4 (50.0%) | 2/4 (50.0%) | 1.000 | 0/4 (0.0%) | 1/4 (25.0%) | 1.000 | 22/50 (44.0%) | 6/50 (12.0%) | 0.0003 |
| Average copy number/cell | 2.061 ± 2.900 | 0.003 ± 0.003 | * | - | - | - | 0.000004 ± 0.00001 | 0.0003 ± 0.0008 | * | |
| Brisbane | No. (%) positive | 14/15 (93.3%) | 4/15 (26.6%) | <0.001 | 1/4 (25.0%) | 0/4 (0.0%) | 1.000 | - | - | - |
| Average copy number/cell | 29.220 ± 58.466 | 0.634 ± 1.267 | * | - | - | - | - | - | * | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mashkour, N.; Jones, K.; Wirth, W.; Burgess, G.; Ariel, E. The Concurrent Detection of Chelonid Alphaherpesvirus 5 and Chelonia mydas Papillomavirus 1 in Tumoured and Non-Tumoured Green Turtles. Animals 2021, 11, 697. https://doi.org/10.3390/ani11030697
Mashkour N, Jones K, Wirth W, Burgess G, Ariel E. The Concurrent Detection of Chelonid Alphaherpesvirus 5 and Chelonia mydas Papillomavirus 1 in Tumoured and Non-Tumoured Green Turtles. Animals. 2021; 11(3):697. https://doi.org/10.3390/ani11030697
Chicago/Turabian StyleMashkour, Narges, Karina Jones, Wytamma Wirth, Graham Burgess, and Ellen Ariel. 2021. "The Concurrent Detection of Chelonid Alphaherpesvirus 5 and Chelonia mydas Papillomavirus 1 in Tumoured and Non-Tumoured Green Turtles" Animals 11, no. 3: 697. https://doi.org/10.3390/ani11030697
APA StyleMashkour, N., Jones, K., Wirth, W., Burgess, G., & Ariel, E. (2021). The Concurrent Detection of Chelonid Alphaherpesvirus 5 and Chelonia mydas Papillomavirus 1 in Tumoured and Non-Tumoured Green Turtles. Animals, 11(3), 697. https://doi.org/10.3390/ani11030697






