Genomics Analysis of Bacillus megaterium 1259 as a Probiotic and Its Effects on Performance in Lactating Dairy Cows
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolate
2.2. Genome Sequencing, Assembly, and Annotation
2.3. Multiple Alignments and Phylogenetic Analyses of 16S rRNA Sequences
2.4. Animal Diet, and Experimental Design
2.5. Sampling, Measurement, and Analyses
2.6. Statistical Analysis
3. Results
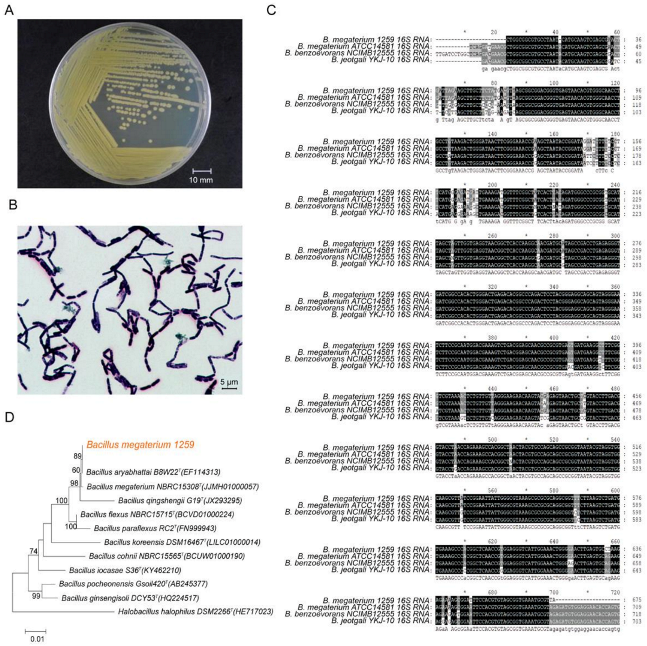
3.1. Identification of BM1259
3.2. Genome Architecture and General Features of BM1259
3.3. Functional Annotation of Predicted Genes from the BM1259 Genome
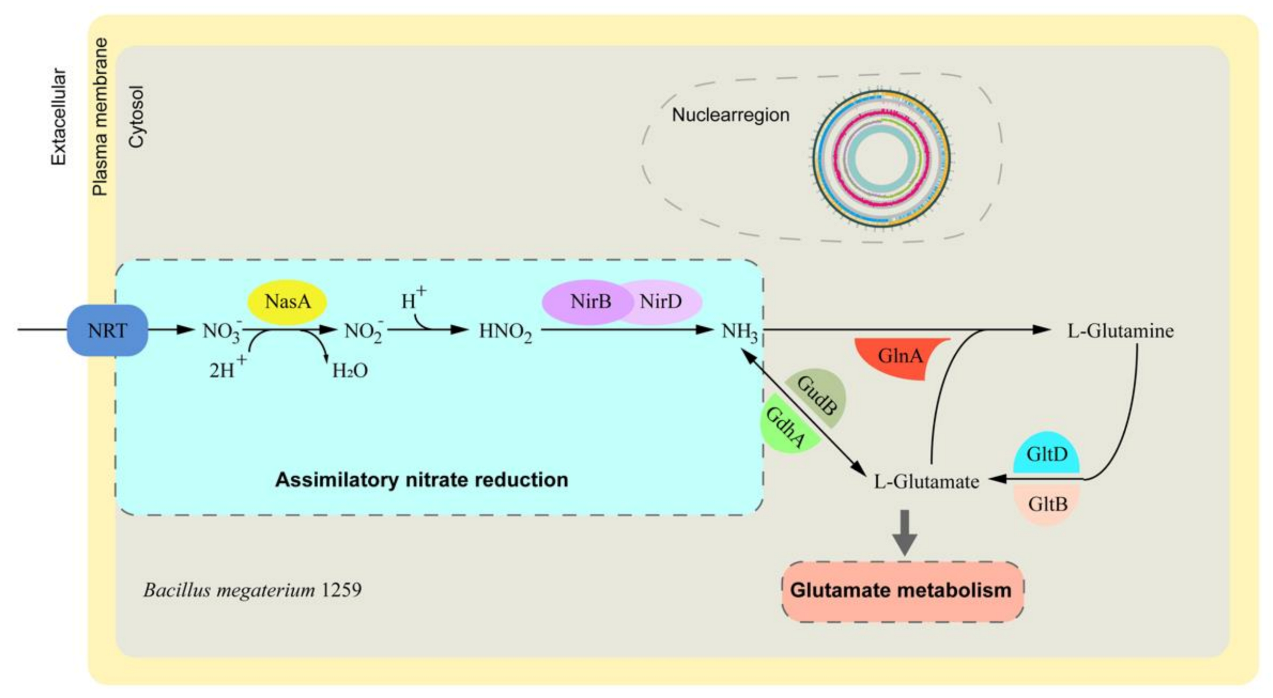
3.4. Nitrate Assimilation-Related Genes and Pathway Identification
3.5. Effects of Dietary BM1259 on Milk Yield and Composition in Lactating Dairy Cows
3.6. Effects of Dietary BM1259 on Ruminal Fermentation and Excrement Nitrogen Biochemical Indicators in Lactating Dairy Cows
3.7. Effects of Dietary BM1259 on Blood Metabolites in Lactating Dairy Cows
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- FAO/WHO. Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. 2001. Available online: http://www.fao.org/tempref/docrep/fao/meeting/009/y6398e.pdf (accessed on 1 October 2001).
- Vanbelle, M.; Teller, E.; Focant, M. Probiotics in animal nutrition: A review. Arch. Anim. Nutr. 1990, 40, 543–567. [Google Scholar] [CrossRef] [PubMed]
- Gaggìa, F.; Mattarelli, P.; Biavati, B. Probiotics and prebiotics in animal feeding for safe food production. Int. J. Food Microbiol. 2010, 141, S15–S28. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhou, C.; Liu, G.; Jiang, H.; Lu, Q.; Tan, Z.; Wu, X.; Fang, J. Application of lactic acid bacteria, yeast and bacillus as feed additive in dairy cattle. J. Food Agric. Environ. 2013, 11, 626–629. [Google Scholar]
- Song, D.J.; Kang, H.Y.; Wang, J.Q.; Peng, H.; Bu, D.P. Effect of feeding bacillus subtilis natto on hindgut fermentation and microbiota of Holstein dairy cows. Asian Australas. J. Anim. Sci. 2014, 27, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrzak-Piekarczyk, T.; Puzia, W.; Żylińska, J.; Cieśla, J.; Gulewicz, K.A.; Bardowski, J.K.; Górecki, R.K. Potential of Lactobacillus plantarum IBB 3036 and Lactobacillus salivarius IBB 3154 to persistence in chicken after in ovo delivery. Microbiology 2019, 8, e00620. [Google Scholar] [CrossRef]
- Sun, P.; Wang, J.Q.; Deng, L.F. Effects of Bacillus subtilis natto on milk production, rumen fermentation and ruminal microbiome of dairy cows. Animal 2013, 7, 216–222. [Google Scholar] [CrossRef]
- Qiao, G.H.; Shan, A.S.; Ma, N.; Ma, Q.Q.; Sun, Z.W. Effect of supplemental Bacillus cultures on rumen fermentation and milk yield in Chinese Holstein cows. J. Anim. Physiol. Anim. Nutr. 2010, 94, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Thorsen, L.; Kpikpi, E.N.; Stuer-Lauridsen, B.; Cantor, M.D.; Nielsen, B.; Brockmann, E.; Derkx, P.M.; Jespersen, L. Characterization of Bacillus spp. strains for use as probiotic additives in pig feed. Appl. Microbiol. Biotechnol. 2014, 98, 1105–1118. [Google Scholar] [CrossRef]
- Kritas, S.K.; Govaris, A.; Christodoulopoulos, G.; Burriel, A.R. Effect of Bacillus licheniformis and Bacillus subtilis supplementation of ewe’s feed on sheep milk production and young lamb mortality. J. Vet. Med. A Physiol. Pathol. Clin. Med. 2006, 53, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Vary, P.S.; Biedendieck, R.; Fuerch, T.; Meinhardt, F.; Rohde, M.; Deckwer, W.D.; Jahn, D. Bacillus megaterium—from simple soil bacterium to industrial protein production host. Appl. Microbiol. Biotechnol. 2007, 76, 957–967. [Google Scholar] [CrossRef]
- Vos, P.; Garrity, G.; Jones, D.; Krieg, N.R.; Ludwig, W.; Rainey, F.A.; Schleifer, K.H.; Whitman, W.B. (Eds.) Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA, 2009; Volume 3. [Google Scholar]
- Kildea, S.; Ransbotyn, V.; Khan, M.R.; Fagan, B.; Leonard, G.; Mullins, E.; Doohan, F.M. Bacillus megaterium shows potential for the biocontrol of Septoria tritici blotch of wheat. Biol. Control 2008, 47, 37–45. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Ranamukhaarachchi, S.L.; Hannaway, D.B. Efficacy of antagonist strains of Bacillus megaterium, Enterobacter cloacae, Pichia guilliermondii and Candida ethanolica against bacterial wilt disease of tomato. J. Phytol. 2011, 3, 1–10. [Google Scholar]
- Yuniarti, A.; Guntoro, D.A.; Hariati, A.M. Response of indigenous Bacillus megaterium supplementation on the growth of Litopenaeus vannamei (Boone), a new target species for shrimp culture in East Java of Indonesia. J. Basic Appl. Sci. Res. 2013, 3, 747–754. [Google Scholar]
- Yao, J.; Wang, L.; Zhang, W.; Liu, M.; Niu, J. Effects of Bacillus megaterium on growth performance, serum biochemical parameters, antioxidant capacity, and immune function in suckling calves. Open Life Sci. 2020, 15, 1033–1041. [Google Scholar] [CrossRef]
- Weili, Z.; Lihuai, Y.; Tianrong, X.; Zhengxu, L.; Denghui, X.; Zhibing, L.; Yanyun, Z. Comparison of BM1259 and yucca extract on ammonia nitrogen emission of manureduring storage periodin laying hens. Feed Ind. 2014, 2014, 13–16. (In Chinese) [Google Scholar]
- Senol Cali, D.; Kim, J.S.; Ghose, S.; Alkan, C.; Mutlu, O. Nanopore sequencing technology and tools for genome assembly: Computational analysis of the current state, bottlenecks and future directions. Brief. Bioinform. 2019, 20, 1542–1559. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Li, Y.; Hu, J.; Xiang, Y.; Zhang, Y.; Chen, D.; Liu, F. Identification and comparative expression profiles of chemosensory genes in major chemoreception organs of a notorious pests, Laodelphax striatellus. Comp. Biochem Phys Genom. Proteom. 2020, 33, 100646. [Google Scholar] [CrossRef]
- Wang, D.M.; Wang, C.; Liu, H.Y.; Liu, J.X.; Ferguson, J.D. Effects of rumen-protected γ-aminobutyric acid on feed intake, lactation performance, and antioxidative status in early lactating dairy cows. J. Dairy Sci. 2013, 96, 3222–3227. [Google Scholar] [CrossRef] [PubMed]
- Davidson, S.; Hopkins, B.A.; Odle, J.; Brownie, C.; Fellner, V.; Whitlow, L.W. Supplementing limited methionine diets with rumen-protected methionine, betaine, and choline in early lactation Holstein cows. J. Dairy Sci. 2008, 91, 1552–1559. [Google Scholar] [CrossRef] [PubMed]
- Bary, A. Vergleichende Morphologie und Biologie der Pilze, Mycetozoen und Bacterien; Wilhelm Engelmann: Leipzig, Germany, 1884. [Google Scholar]
- Foerster, H.F.; Foster, J.W. Response of Bacillus spores to combinations of germinative compounds. J. Bacteriol. 1966, 91, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Panbangred, W.; Weeradechapon, K.; Udomvaraphant, S.; Fujiyama, K.; Meevootisom, V. High expression of the penicillin G acylase gene (pac) from Bacillus megaterium UN1 in its own pac minus mutant. J. Appl. Microbiol. 2000, 89, 152–157. [Google Scholar] [CrossRef]
- Lwoff, A.; Siminovitch, L.; Kjeldgaard, N. Induction of the production of bacteriophages in lysogenic bacteria. Ann. Inst. Pasteur. 1950, 79, 815–859. (In French) [Google Scholar]
- Bunk, B.; Biedendieck, R.; Jahn, D.; Vary, P.S. Encyclopedia of Industrial Biotechnology. In Chapter: Bacillus Megaterium and other Bacilli: Industrial Applications; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Vary, P.S. Prime time for Bacillus megaterium. Microbiology 1994, 140, 1001–1013. [Google Scholar] [CrossRef]
- Eppinger, M.; Bunk, B.; Johns, M.A.; Edirisinghe, J.N.; Kutumbaka, K.K.; Koenig, S.S.; Creasy, H.H.; Rosovitz, M.J.; Riley, D.R.; Daugherty, S.; et al. Genome sequences of the biotechnologically important Bacillus megaterium strains QM B1551 and DSM319. J. Bacteriol. 2011, 193, 4199–4213. [Google Scholar] [CrossRef]
- Lin, J.T.; Stewart, V. Nitrate assimilation by bacteria. Adv. Microb. Physiol. 1998, 39, 1–30. [Google Scholar]
- Shi, W.; Lu, W.; Liu, Q.; Zhi, Y.; Zhou, P. The identification of the nitrate assimilation related genes in the novel Bacillus megaterium NCT-2 accounts for its ability to use nitrate as its only source of nitrogen. Funct. Integr. Genom. 2014, 14, 219–227. [Google Scholar] [CrossRef]
- Chu, S.; Zhang, D.; Zhi, Y.; Wang, B.; Chi, C.P.; Zhang, D.; Liu, Y.; Zhou, P. Enhanced removal of nitrate in the maize rhizosphere by plant growth-promoting Bacillus megaterium NCT-2, and its colonization pattern in response to nitrate. Chemosphere 2018, 208, 316–324. [Google Scholar] [CrossRef]
- Fuller, R. Probiotics in man and animals. J. Bacteriol. 1989, 66, 365–378. [Google Scholar]
- Hosoi, T.; Ametani, A.; Kiuchi, K.; Kaminogawa, S. Improved growth and viability of lactobacilli in the presence of Bacillus subtilis (natto), catalase, or subtilisin. Can. J. Microbiol. 2000, 46, 892–897. [Google Scholar] [CrossRef]
- Hong, H.A.; Duc, L.H.; Cutting, S.M. The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. 2005, 29, 813–835. [Google Scholar] [CrossRef] [PubMed]
- Frizzo, L.S.; Soto, L.P.; Bertozzi, E.; Zbrun, M.V.; Sequeira, G.J.; Dalla Santina, R.; Rodriguez Armesto, R.; Rosmini, M.R. The effect of supplementation with three lactic acid bacteria from bovine origin on growth performance and health status of young calves. J. Anim. Vet. Adv. 2008, 7, 400–408. [Google Scholar]
- Fritts, C.A.; Kersey, J.H.; Motl, M.A.; Kroger, E.C.; Yan, F.; Si, J.; Jiang, Q.; Campos, M.M.; Waldroup, A.L.; Waldroup, P.W. Bacillus subtilis C-3102 (Calsporin) improves live performance and microbiological status of broiler chickens. J. Appl. Poult. Res. 2000, 9, 149–155. [Google Scholar] [CrossRef]
- Teo, A.Y.; Tan, H.M. Inhibition of Clostridium perfringens by a novel strain of Bacillus subtilis isolated from the gastrointestinal tracts of healthy chickens. Appl. Environ. Microbiol. 2005, 71, 4185–4190. [Google Scholar] [CrossRef]
- Samanya, M.; Yamauchi, K.E. Histological alterations of intestinal villi in chickens fed dried Bacillus subtilis var. natto. Comp. Biochem. Phys. 2002, 133, 95–104. [Google Scholar] [CrossRef]
- Sun, P.; Wang, J.Q.; Zhang, H.T. Effects of Bacillus subtilis natto on performance and immune function of preweaning calves. J. Dairy Sci. 2010, 93, 5851–5855. [Google Scholar] [CrossRef] [PubMed]
- McDonald, P.; Edwards, R.A.; Greenhalgh, J.F.; Morgan, C.A.; Sinclair, L.A. Animal Nutrition, 5th ed.; John Wiley and Sons - Scientific Publishers: New York, NY, USA, 1995; pp. 49–127. [Google Scholar]
- Leng, R.A.; Leonard, G.J. Measurement of the rates of production of acetic, propionic and butyric acids in the rumen of sheep. Br. J. Nutr. 1965, 19, 469–484. [Google Scholar] [CrossRef]
- Khan, M.A.; Lee, H.J.; Lee, W.S.; Kim, H.S.; Kim, S.B.; Park, S.B.; Baek, K.S.; Ha, J.K.; Choi, Y.J. Starch source evaluation in calf starter: II. Ruminal parameters, rumen development, nutrient digestibilities, and nitrogen utilization in Holstein calves. J. Dairy Sci. 2008, 91, 1140–1149. [Google Scholar] [CrossRef]
- Bach, A.; Calsamiglia, S.; Stern, M.D. Nitrogen metabolism in the rumen. J. Dairy Sci. 2005, 88, E9–E21. [Google Scholar] [CrossRef]
- De Boer, I.J.; Smits, M.C.; Mollenhorst, H.; Van Duinkerken, G.; Monteny, G.J. Prediction of ammonia emission from dairy barns using feed characteristics part I: Relation between feed characteristics and urinary urea concentration. J. Dairy Sci. 2002, 85, 3382–3388. [Google Scholar] [CrossRef]
- Santoso, U.; Ohtani, S.; Tanaka, K.; Sakaida, M. Dried Bacillus subtilis culture reduced ammonia gas release in poultry house. Asian Australas. J. Anim. Sci. 1999, 12, 806–809. [Google Scholar] [CrossRef]
- Santoso, U.; Tanaka, K.; Ohtani, S. Effect of dried Bacillus subtilis culture on growth, body composition and hepatic lipogenic enzyme activity in female broiler chicks. Br. J. Nutr. 1995, 74, 523–529. [Google Scholar] [CrossRef] [PubMed]

| Item | Value |
|---|---|
| Ingredients | |
| Ground corn | 15.83 |
| Flaked corn | 5.72 |
| Soybean meal | 3.52 |
| Cottonseed meal | 2.86 |
| Corn germ meal | 2.5 |
| Beet pulp | 4 |
| DDGS | 5.68 |
| Brewer grains | 2.46 |
| Soybean hull | 5.8 |
| Cottonseed | 3 |
| CaCO3 | 0.35 |
| CaHPO4 | 0.44 |
| NaCl | 0.22 |
| Sodium bicarbonate | 0.33 |
| Yeast culture | 0.11 |
| Alfalfa | 4 |
| Corn silage | 43 |
| Premix 1 | 0.18 |
| Total | 100 |
| Nutrient levels | |
| CP | 18.37 |
| NDF | 37.06 |
| ADF | 16.21 |
| Ca | 0.71 |
| P | 0.43 |
| NEL/(MJ·Kg) | 6.91 |
| Items | C | T1 | T2 | T3 |
|---|---|---|---|---|
| Yield, kg/d | ||||
| Milk | 30.21 ± 2.40 | 32.42 ± 3.02 | 29.27 ± 4.31 | 30.49 ± 5.01 |
| 4% FCM | 35.19 ± 3.84 a | 35.84 ± 2.07 a | 37.38 ± 0.56 ab | 39.25 ± 1.46 b |
| Milk composition, % | ||||
| Protein | 2.94 ± 0.26 | 2.99 ± 0.20 | 3.15 ± 0.24 | 3.11 ± 0.27 |
| Fat | 2.89 ± 0.26 | 3.12 ± 0.35 | 3.06 ± 0.27 | 3.09 ± 0.18 |
| Lactose | 5.06 ± 0.16 | 4.90 ± 0.04 | 4.54 ± 0.17 | 4.91 ± 0.11 |
| TS | 11.53 ± 2.05 | 11.63 ± 1.28 | 11.42 ± 0.95 | 11.74 ± 1.44 |
| MUN | 7.50 ± 0.42 a | 10.43 ± 0.36 b | 8.91 ± 0.22 ab | 10.24 ± 0.32 b |
| Feed efficiency | 1.34 ± 0.17 | 1.51 ± 0.43 | 1.81 ± 0.58 | 1.43 ± 0.34 |
| Items | C | T1 | T2 | T3 |
|---|---|---|---|---|
| Ruminal pH | 6.42 ± 0.25 | 6.41 ± 0.25 | 6.30 ± 0.24 | 6.46 ± 0.25 |
| NH3-N (mg N/dL) | 17.40 ± 5.40 | 21.42 ± 2.26 | 20.95 ± 2.07 | 20.42 ± 5.19 |
| Total VFA (mmol/L) | 84.68 ± 6.06 | 92.52 ± 7.45 | 94.54 ± 8.40 | 93.02 ± 27.81 |
| Acetate (C2) (%) | 65.40 ± 0.01 a | 64.60 ± 0.02 a | 61.25 ± 0.04 b | 66.20 ± 0.01 a |
| Propionate (C3) (%) | 17.50 ± 0.01 a | 17.80 ± 0.01 a | 21.50 ± 0.05 b | 17.40 ± 0.01 a |
| Butyrate (C4) (%) | 13.40 ± 0.01 | 14.00 ± 0.01 | 12.50 ± 0.02 | 13.00 ± 0.01 |
| C2:C3 | 3.73 ± 0.17 ab | 3.64 ± 0.27 ab | 3.35 ± 0.51 a | 3.84 ± 0.27 b |
| Items | C | T1 | T2 | T3 |
|---|---|---|---|---|
| Feces | ||||
| BUN (mg/g) | 4.36 ± 0.21 a | 3.98 ± 0.37 ab | 4.23 ± 0.26 ab | 4.01 ± 0.31 b |
| UA (mg//g) | 0.97 ± 0.03 | 1.07 ± 0.13 | 0.63 ± 0.09 | 0.72 ± 0.43 |
| NH3-N (umol/g) | 603.39 ± 91.22 a | 611.81 ± 87.36 ab | 573.53 ± 72.99 b | 581.00 ± 100.03 ab |
| Urase (umol/g.24 h) | 4.27 ± 0.76 | 4.36 ± 0.43 | 4.11 ± 0.58 | 4.56 ± 0.91 |
| Uricase (%) | 0.44 ± 0.01 | 0.31 ± 0.08 | 0.33 ± 0.07 | 0.49 ± 0.03 |
| Urine | ||||
| BUN (mmol/L) | 397.03 ± 50.91 a | 376.33 ± 71.23 ab | 346.33 ± 107.06 ab | 372.11 ± 61.23 b |
| UA (mmol/L) | 7.36 ± 0.91 | 8.22 ± 1.02 | 6.27 ± 2.11 | 6.11 ± 3.87 |
| Items | C | T1 | T2 | T3 |
|---|---|---|---|---|
| TP (g/L) | 77.18 ± 5.57 a | 85.56 ± 6.09 b | 88.28 ± 5.96 b | 83.19 ± 2.94 ab |
| ALB (g/L) | 29.77 ± 2.38 | 28.93 ± 1.77 | 27.49 ± 1.07 | 28.57 ± 1.01 |
| GLOB (g/L) | 45.04 ± 4.52 a | 60.15 ± 7.11 b | 60.09 ± 5.57 b | 55.70 ± 3.00 b |
| AST (U/L) | 68.27 ± 1.89 | 77.88 ± 4.45 | 82.56 ± 1.76 | 81.31 ± 1.25 |
| ALT (U/L) | 28.99 ± 12.37 a | 29.17 ± 8.67 a | 28.65 ± 9.40 a | 34.94 ± 3.60 b |
| BUN (mmol/L) | 4.30 ± 0.31 a | 4.80 ± 0.23 b | 4.60 ± 0.22 ab | 4.93 ± 0.35 b |
| UA (µmol/L) | 76.04 ± 7.50 | 80.33 ± 8.55 | 75.08 ± 2.33 | 77.55 ± 4.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, B.; Wang, L.; Ma, Q.; Yu, T.; Liu, D.; Dai, Y.; Zhao, G. Genomics Analysis of Bacillus megaterium 1259 as a Probiotic and Its Effects on Performance in Lactating Dairy Cows. Animals 2021, 11, 397. https://doi.org/10.3390/ani11020397
Deng B, Wang L, Ma Q, Yu T, Liu D, Dai Y, Zhao G. Genomics Analysis of Bacillus megaterium 1259 as a Probiotic and Its Effects on Performance in Lactating Dairy Cows. Animals. 2021; 11(2):397. https://doi.org/10.3390/ani11020397
Chicago/Turabian StyleDeng, Bobo, Lin Wang, Qianbo Ma, Tongshui Yu, Dalin Liu, Yi Dai, and Guoqi Zhao. 2021. "Genomics Analysis of Bacillus megaterium 1259 as a Probiotic and Its Effects on Performance in Lactating Dairy Cows" Animals 11, no. 2: 397. https://doi.org/10.3390/ani11020397
APA StyleDeng, B., Wang, L., Ma, Q., Yu, T., Liu, D., Dai, Y., & Zhao, G. (2021). Genomics Analysis of Bacillus megaterium 1259 as a Probiotic and Its Effects on Performance in Lactating Dairy Cows. Animals, 11(2), 397. https://doi.org/10.3390/ani11020397






