The Koala Immune Response to Chlamydial Infection and Vaccine Development—Advancing Our Immunological Understanding
Simple Summary
Abstract
1. Chlamydia and Koalas
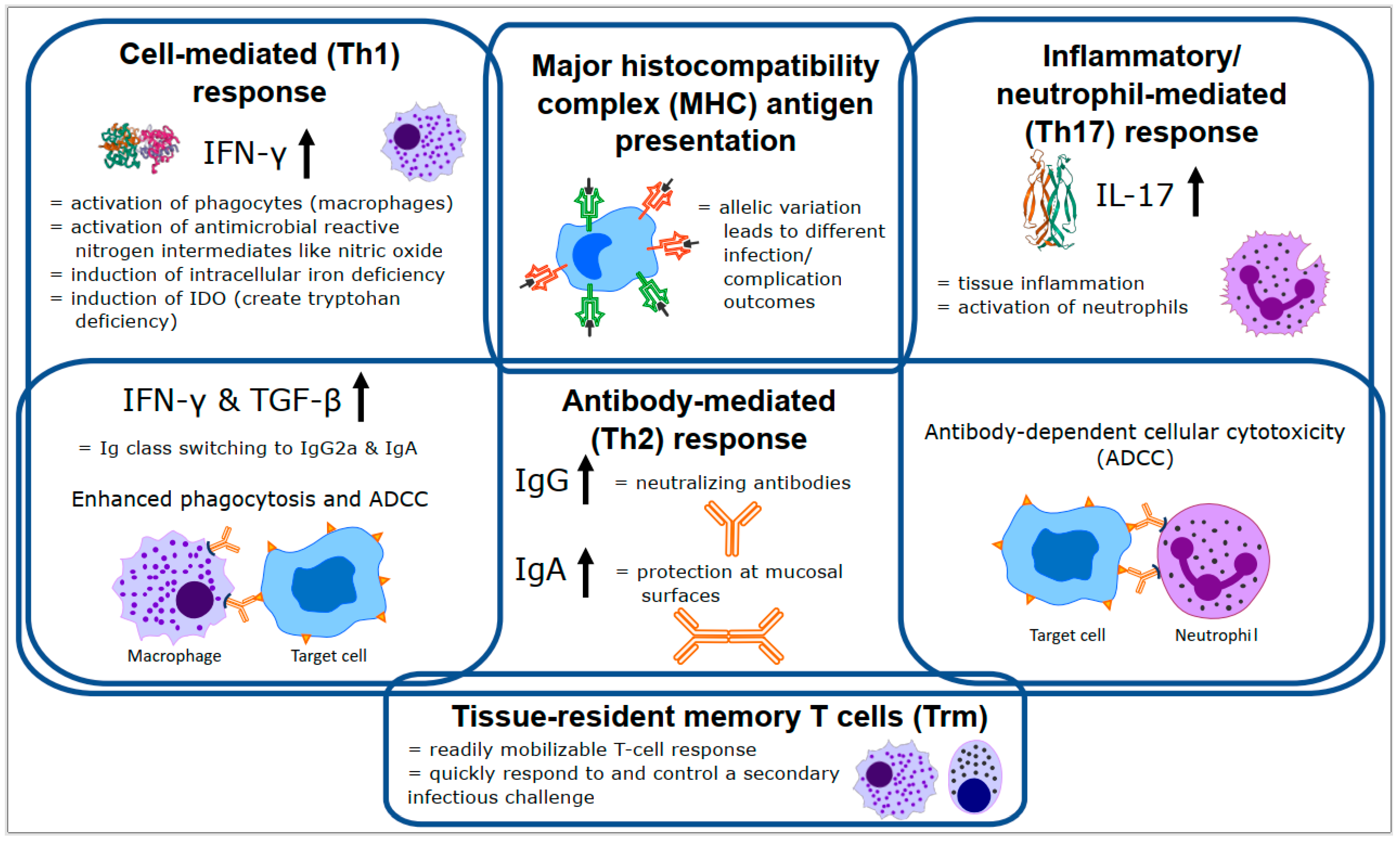
2. Effective Chlamydial Immune Responses
2.1. Immunogenetics
2.2. Cell Mediated (Th1) Responses
2.3. Antibody (Th2) Responses
2.4. Inflammatory/Neutrophil (Th17) Responses
2.5. Coordinated Responses
3. Chlamydial Infection, Disease, and Vaccine Responses in Koalas
3.1. Immunogenetics Related to Chlamydia in Koalas
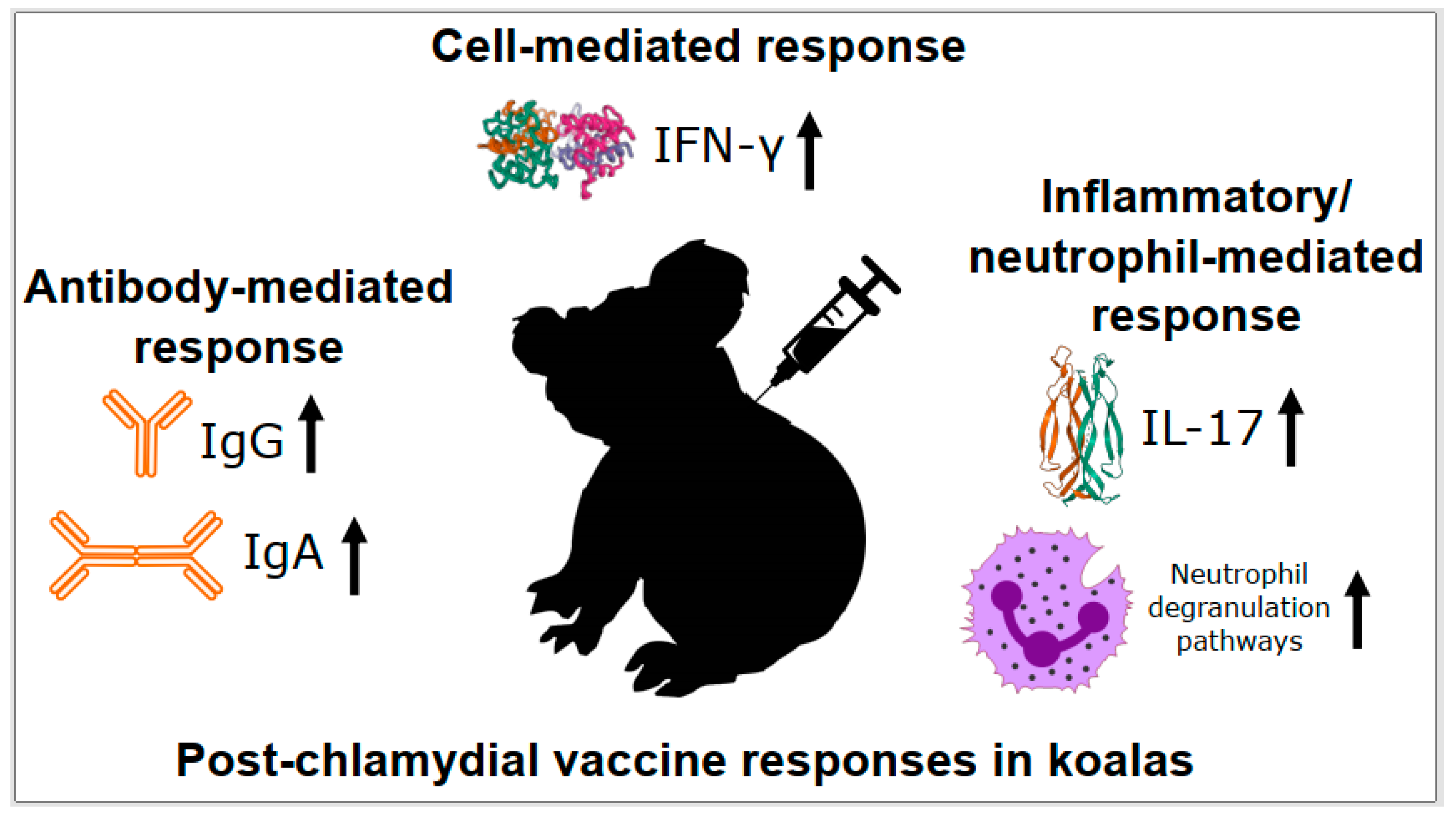
3.2. Cell-Mediated Responses to Chlamydia in Koalas
3.3. Antibody Responses to Chlamydia in Koalas
3.4. Inflammatory/Neutrophil Responses to Chlamydia in Koalas
4. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shewen, P.E. Chlamydial infection in animals: A review. Can. Vet. J. 1980, 21, 2–11. [Google Scholar] [PubMed]
- Collingro, A.; Köstlbacher, S.; Horn, M. Chlamydiae in the Environment. Trends Microbiol. 2020, 28, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Jelocnik, M. Chlamydiae from down under: The curious cases of chlamydial infections in Australia. Microorganisms 2019, 7, 602. [Google Scholar] [CrossRef] [PubMed]
- Quigley, B.L.; Timms, P. Helping koalas battle disease—Recent advances in Chlamydia and koala retrovirus (KoRV) disease understanding and treatment in koalas. FEMS Microbiol. Rev. 2020, 44, 583–605. [Google Scholar] [CrossRef]
- Phillips, S.; Quigley, B.L.; Timms, P. Seventy years of Chlamydia vaccine research—Limitations of the past and directions for the future. Front. Microbiol. 2019, 10, 70. [Google Scholar] [CrossRef]
- Emonds, O.R.B.; Cardillo, M.; Jones, K.E.; MacPhee, R.D.; Beck, R.M.; Grenyer, R.; Price, S.A.; Vos, R.A.; Gittleman, J.L.; Purvis, A. The delayed rise of present-day mammals. Nature 2007, 446, 507–512. [Google Scholar] [CrossRef]
- Jurd, R.D. “Not proper mammals”: Immunity in monotremes and marsupials. Comp. Immunol. Microbiol. Infect. Dis. 1994, 17, 41–52. [Google Scholar] [CrossRef]
- Wilkinson, R.; Kotlarski, I.; Barton, M. Koala lymphoid cells: Analysis of antigen-specific responses. Vet. Immunol. Immunopathol. 1992, 33, 237–247. [Google Scholar] [CrossRef]
- Wilkinson, R.; Kotlarski, I.; Barton, M. Further characterisation of the immune response of the koala. Vet. Immunol. Immunopathol. 1994, 40, 325–339. [Google Scholar] [CrossRef]
- Belov, K.; Miller, R.D.; Old, J.M.; Young, L.J. Marsupial immunology bounding ahead. Aust. J. Zool. 2013, 61, 21–40. [Google Scholar] [CrossRef]
- Madden, D.; Whaite, A.; Jones, E.; Belov, K.; Timms, P.; Polkinghorne, A. Koala immunology and infectious diseases: How much can the koala bear? Dev. Comp. Immunol. 2018, 82, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.N.; O’Meally, D.; Chen, Z.; Etherington, G.J.; Ho, S.Y.W.; Nash, W.J.; Grueber, C.E.; Cheng, Y.; Whittington, C.M.; Dennison, S.; et al. Adaptation and conservation insights from the koala genome. Nat. Genet. 2018, 50, 1102–1111. [Google Scholar] [CrossRef] [PubMed]
- Desclozeaux, M.; Robbins, A.; Jelocnik, M.; Khan, S.A.; Hanger, J.; Gerdts, V.; Potter, A.; Polkinghorne, A.; Timms, P. Immunization of a wild koala population with a recombinant Chlamydia pecorum Major Outer Membrane Protein (MOMP) or Polymorphic Membrane Protein (PMP) based vaccine: New insights into immune response, protection and clearance. PLoS ONE 2017, 12, e0178786. [Google Scholar] [CrossRef] [PubMed]
- Nyari, S.; Booth, R.; Quigley, B.L.; Waugh, C.A.; Timms, P. Therapeutic effect of a Chlamydia pecorum recombinant major outer membrane protein vaccine on ocular disease in koalas (Phascolarctos cinereus). PLoS ONE 2019, 14, e0210245. [Google Scholar] [CrossRef]
- Phillips, S.; Quigley, B.L.; Olagoke, O.; Booth, R.; Pyne, M.; Timms, P. Vaccination of koalas during antibiotic treatment for Chlamydia-induced cystitis induces an improved antibody response to Chlamydia pecorum. Sci. Rep. 2020, 10, 10152. [Google Scholar] [CrossRef]
- Rank, R.G.; Whittum-Hudson, J.A. Animal models for ocular infections. Methods Enzymol. 1994, 235, 69–83. [Google Scholar] [CrossRef]
- Vasilevsky, S.; Greub, G.; Haefliger, D.N.; Baud, D. Genital Chlamydia trachomatis: Understanding the roles of innate and adaptive immunity in vaccine research. Clin. Microbiol. Rev. 2014, 27, 346–370. [Google Scholar] [CrossRef]
- Hafner, L.; Beagley, K.; Timms, P. Chlamydia trachomatis infection: Host immune responses and potential vaccines. Mucosal Immunol. 2008, 1, 116–130. [Google Scholar] [CrossRef]
- Redgrove, K.A.; McLaughlin, E.A. The role of the immune response in Chlamydia trachomatis infection of the male genital tract: A double-edged sword. Front. Immunol. 2014, 5, 534. [Google Scholar] [CrossRef]
- Punt, J.; Stranford, S.A.; Jones, P.P.; Owen, J.A. Kuby Immunology; Mcmillian Education: New York, NY, USA, 2018. [Google Scholar]
- Morré, S.A.; Karimi, O.; Ouburg, S. Chlamydia trachomatis: Identification of susceptibility markers for ocular and sexually transmitted infection by immunogenetics. FEMS Immunol. Med. Microbiol. 2009, 55, 140–153. [Google Scholar] [CrossRef]
- Kimani, J.; Maclean, I.W.; Bwayo, J.J.; MacDonald, K.; Oyugi, J.; Maitha, G.M.; Peeling, R.W.; Cheang, M.; Nagelkerke, N.J.; Plummer, F.A.; et al. Risk factors for Chlamydia trachomatis pelvic inflammatory disease among sex workers in Nairobi, Kenya. J. Infect. Dis. 1996, 173, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.M.; Tang, J.; Brown, L.; Press, C.G.; Geisler, W.M. HLA-DQB1 * 06 is a risk marker for chlamydia reinfection in African American women. Genes Immun. 2019, 20, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Brunham, R.C.; Rey-Ladino, J. Immunology of Chlamydia infection: Implications for a Chlamydia trachomatis vaccine. Nat. Rev. Immunol. 2005, 5, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.M.; de la Maza, L.M. Activation of mouse peritoneal macrophages in vitro or in vivo by recombinant murine gamma interferon inhibits the growth of Chlamydia trachomatis serovar L1. Infect. Immun. 1988, 56, 3322–3325. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Stout, R.; Campbell, W.F. Nitric oxide production: A mechanism of Chlamydia trachomatis inhibition in interferon-gamma-treated RAW264.7 cells. FEMS Immunol. Med. Microbiol. 1996, 14, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.Y.; Jeong, K.S.; Kang, B.N.; Park, S.J.; Yoon, W.K.; Kim, S.H.; Kim, T.H. Modulation of transferrin synthesis, transferrin receptor expression, iNOS expression and NO production in mouse macrophages by cytokines, either alone or in combination. Anticancer Res. 2000, 20, 3331–3338. [Google Scholar] [PubMed]
- Akers, J.C.; Tan, M. Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis. J. Bacteriol. 2006, 188, 4236–4243. [Google Scholar] [CrossRef]
- Kaiko, G.E.; Horvat, J.C.; Beagley, K.W.; Hansbro, P.M. Immunological decision-making: How does the immune system decide to mount a helper T-cell response? Immunology 2008, 123, 326–338. [Google Scholar] [CrossRef]
- Igietseme, J.U.; Eko, F.O.; He, Q.; Black, C.M. Antibody regulation of T-cell immunity: Implications for vaccine strategies against intracellular pathogens. Expert Rev. Vaccines 2004, 3, 23–34. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L.A. A reappraisal of humoral immunity based on mechanisms of antibody-mediated protection against intracellular pathogens. Adv. Immunol. 2006, 91, 1–44. [Google Scholar] [CrossRef]
- Brunham, R.C.; Kuo, C.C.; Cles, L.; Holmes, K.K. Correlation of host immune response with quantitative recovery of Chlamydia trachomatis from the human endocervix. Infect. Immun. 1983, 39, 1491–1494. [Google Scholar] [CrossRef] [PubMed]
- Eko, F.O.; Ekong, E.; He, Q.; Black, C.M.; Igietseme, J.U. Induction of immune memory by a multisubunit chlamydial vaccine. Vaccine 2011, 29, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Morrison, S.G.; Morrison, R.P. A predominant role for antibody in acquired immunity to chlamydial genital tract reinfection. J. Immunol. 2005, 175, 7536–7542. [Google Scholar] [CrossRef] [PubMed]
- Bartolini, E.; Ianni, E.; Frigimelica, E.; Petracca, R.; Galli, G.; Berlanda Scorza, F.; Norais, N.; Laera, D.; Giusti, F.; Pierleoni, A.; et al. Recombinant outer membrane vesicles carrying Chlamydia muridarum HtrA induce antibodies that neutralize chlamydial infection in vitro. J. Extracell. Vesicles 2013, 2, 20181. [Google Scholar] [CrossRef] [PubMed]
- Moore, T.; Ananaba, G.A.; Bolier, J.; Bowers, S.; Belay, T.; Eko, F.O.; Igietseme, J.U. Fc receptor regulation of protective immunity against Chlamydia trachomatis. Immunology 2002, 105, 213–221. [Google Scholar] [CrossRef]
- Barteneva, N.; Theodor, I.; Peterson, E.M.; de la Maza, L.M. Role of neutrophils in controlling early stages of a Chlamydia trachomatis infection. Infect. Immun. 1996, 64, 4830–4833. [Google Scholar] [CrossRef]
- Deenick, E.K.; Hasbold, J.; Hodgkin, P.D. Decision criteria for resolving isotype switching conflicts by B cells. Eur. J. Immunol. 2005, 35, 2949–2955. [Google Scholar] [CrossRef]
- Naglak, E.K.; Morrison, S.G.; Morrison, R.P. Neutrophils are central to antibody-mediated protection against genital Chlamydia. Infect. Immun. 2017, 85, e00409-17. [Google Scholar] [CrossRef]
- Morrison, S.G.; Morrison, R.P. In situ analysis of the evolution of the primary immune response in murine Chlamydia trachomatis genital tract infection. Infect. Immun. 2000, 68, 2870–2879. [Google Scholar] [CrossRef]
- Johnson, R.M.; Brunham, R.C. Tissue-Resident T cells as the central paradigm of Chlamydia immunity. Infect. Immun. 2016, 84, 868–873. [Google Scholar] [CrossRef]
- Cheng, Y.; Polkinghorne, A.; Gillett, A.; Jones, E.A.; O’Meally, D.; Timms, P.; Belov, K. Characterisation of MHC class I genes in the koala. Immunogenetics 2018, 70, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Quigley, B.L.; Tzipori, G.; Nilsson, K.; Timms, P. High throughput immunogenetic typing of koalas suggests possible link between MHC alleles and cancer. Immunogenetics 2020, 72, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Punt, J.; Stranford, S.; Jones, P.; Owen, J. Kuby Immunology, 8th ed.; Macmillan Science and Education USA: New York, NY, USA, 2019; Volume 8, p. 944. [Google Scholar]
- Lau, Q.; Jobbins, S.E.; Belov, K.; Higgins, D.P. Characterisation of four major histocompatibility complex class II genes of the koala (Phascolarctos cinereus). Immunogenetics 2013, 65, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Abts, K.C.; Ivy, J.A.; de Woody, J.A. Demographic, environmental and genetic determinants of mating success in captive koalas (Phascolarctos cinereus). Zoo Biol. 2018, 37, 416–433. [Google Scholar] [CrossRef]
- Lau, Q.; Griffith, J.E.; Higgins, D.P. Identification of MHCII variants associated with chlamydial disease in the koala (Phascolarctos cinereus). PeerJ 2014, 2, e443. [Google Scholar] [CrossRef]
- Robbins, A.; Hanger, J.; Jelocnik, M.; Quigley, B.L.; Timms, P. Koala immunogenetics and chlamydial strain type are more directly involved in chlamydial disease progression in koalas from two south east Queensland koala populations than koala retrovirus subtypes. Sci. Rep. 2020, 10, 15013. [Google Scholar] [CrossRef]
- Quigley, B.L.; Carver, S.; Hanger, J.; Vidgen, M.E.; Timms, P. The relative contribution of causal factors in the transition from infection to clinical chlamydial disease. Sci. Rep. 2018, 8, 8893. [Google Scholar] [CrossRef]
- Mathew, M.; Pavasovic, A.; Prentis, P.J.; Beagley, K.W.; Timms, P.; Polkinghorne, A. Molecular characterisation and expression analysis of interferon gamma in response to natural Chlamydia infection in the koala, Phascolarctos cinereus. Gene 2013, 527, 570–577. [Google Scholar] [CrossRef]
- Mathew, M.; Beagley, K.W.; Timms, P.; Polkinghorne, A. Preliminary characterisation of tumor necrosis factor alpha and interleukin-10 responses to Chlamydia pecorum infection in the koala (Phascolarctos cinereus). PLoS ONE 2013, 8, e59958. [Google Scholar] [CrossRef]
- Mathew, M.; Waugh, C.; Beagley, K.W.; Timms, P.; Polkinghorne, A. Interleukin 17A is an immune marker for chlamydial disease severity and pathogenesis in the koala (Phascolarctos cinereus). Dev. Comp. Immunol. 2014, 46, 423–429. [Google Scholar] [CrossRef]
- Waugh, C.; Khan, S.A.; Carver, S.; Hanger, J.; Loader, J.; Polkinghorne, A.; Beagley, K.; Timms, P. A prototype recombinant-protein based Chlamydia pecorum vaccine results in reduced chlamydial burden and less clinical disease in free-ranging koalas (Phascolarctos cinereus). PLoS ONE 2016, 11, e0146934. [Google Scholar] [CrossRef] [PubMed]
- Lizárraga, D.; Timms, P.; Quigley, B.L.; Hanger, J.; Carver, S. Capturing complex vaccine-immune-disease relationships for free-ranging koalas: Higher chlamydial loads are associated with less IL17 expression and more chlamydial disease. Front. Vet. Sci. 2020, 7, 530686. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Jelocnik, M.; Huston, W.M.; Timms, P.; Polkinghorne, A. Characterization of the in vitro Chlamydia pecorum response to gamma interferon. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, N.L.; Fraser, T.A.; Bertelli, C.; Jelocnik, M.; Gillett, A.; Funnell, O.; Flanagan, C.; Myers, G.S.; Timms, P.; Polkinghorne, A. Comparative genomics of koala, cattle and sheep strains of Chlamydia pecorum. BMC Genom. 2014, 15, 667. [Google Scholar] [CrossRef] [PubMed]
- Mojica, S.; Huot Creasy, H.; Daugherty, S.; Read, T.D.; Kim, T.; Kaltenboeck, B.; Bavoil, P.; Myers, G. Gemone sequence of the obligate intracellular animal pathogen Chlamydia pecorum E58. J. Bacteriol. 2011, 193, 3690. [Google Scholar] [CrossRef]
- Kollipara, A.; Wan, C.; Rawlinson, G.; Brumm, J.; Nilsson, K.; Polkinghorne, A.; Beagley, K.; Timms, P. Antigenic specificity of a monovalent versus polyvalent MOMP based Chlamydia pecorum vaccine in koalas (Phascolarctos cinereus). Vaccine 2013, 31, 1217–1223. [Google Scholar] [CrossRef][Green Version]
- Kollipara, A.; Polkinghorne, A.; Beagley, K.W.; Timms, P. Vaccination of koalas with a recombinant Chlamydia pecorum major outer membrane protein induces antibodies of different specificity compared to those following a natural live infection. PLoS ONE 2013, 8, e74808. [Google Scholar] [CrossRef]
- Khan, S.A.; Polkinghorne, A.; Waugh, C.; Hanger, J.; Loader, J.; Beagley, K.; Timms, P. Humoral immune responses in koalas (Phascolarctos cinereus) either naturally infected with Chlamydia pecorum or following administration of a recombinant chlamydial major outer membrane protein vaccine. Vaccine 2016, 34, 775–782. [Google Scholar] [CrossRef]
- Nyari, S.; Khan, S.A.; Rawlinson, G.; Waugh, C.A.; Potter, A.; Gerdts, V.; Timms, P. Vaccination of koalas (Phascolarctos cinereus) against Chlamydia pecorum using synthetic peptides derived from the major outer membrane protein. PLoS ONE 2018, 13, e0200112. [Google Scholar] [CrossRef]

| A | Class | MHC Loci | Number of Alleles | B | Class | HLA Loci | Number of Alleles |
|---|---|---|---|---|---|---|---|
| Class I | UA | 21 | Class I | A | 2480 | ||
| UB | 5 | B | 3221 | ||||
| UC | 10 | C | 2196 | ||||
| Class II | DAα | 3 | E | 8 | |||
| DAβ | 44 | F | 4 | ||||
| DBα | 3 | G | 18 | ||||
| DBβ | 26 | Class II | DMα | 4 | |||
| DCα | No work yet done | DMβ | 7 | ||||
| DCβ | 3 | DOα | 3 | ||||
| DMα | No work yet done | DOβ | 5 | ||||
| DMβ | 4 | DPα1 | 22 | ||||
| DPβ1 | 591 | ||||||
| DQα1 | 34 | ||||||
| DQβ1 | 678 | ||||||
| DRα | 2 | ||||||
| DRβ1 | 1440 | ||||||
| DRβ3 | 106 | ||||||
| DRβ4 | 42 | ||||||
| DRβ5 | 39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quigley, B.L.; Timms, P. The Koala Immune Response to Chlamydial Infection and Vaccine Development—Advancing Our Immunological Understanding. Animals 2021, 11, 380. https://doi.org/10.3390/ani11020380
Quigley BL, Timms P. The Koala Immune Response to Chlamydial Infection and Vaccine Development—Advancing Our Immunological Understanding. Animals. 2021; 11(2):380. https://doi.org/10.3390/ani11020380
Chicago/Turabian StyleQuigley, Bonnie L, and Peter Timms. 2021. "The Koala Immune Response to Chlamydial Infection and Vaccine Development—Advancing Our Immunological Understanding" Animals 11, no. 2: 380. https://doi.org/10.3390/ani11020380
APA StyleQuigley, B. L., & Timms, P. (2021). The Koala Immune Response to Chlamydial Infection and Vaccine Development—Advancing Our Immunological Understanding. Animals, 11(2), 380. https://doi.org/10.3390/ani11020380






