Simple Summary
Sarcocystidae is a family of apicomplexan protozoa highly prevalent in vertebrates. The definitive hosts of sarcocystids eliminate oocysts or sporocysts that infect intermediate hosts. After infection, mature tissue cysts (sarcocysts) develop in intermediate hosts, mostly in muscle and neurological tissues. Sarcocysts are infectious for definitive hosts, which acquire them through carnivorous or scavenging habits. Intermediate hosts and definitive hosts are the natural hosts of sarcocystids in which infections are usually mildly or not symptomatic. In 2017, muscular and neurological tissues of 22 birds from Magdalena Islands, southern coast of Chile, were screened for the presence of DNA of sarcocystids. DNA of organisms of the genus Sarcocystis was identified in two Chilean skuas (Stercorarius chilensis). The genetic makeup of the parasite detected in skuas was unprecedented and probably represent a new species in the genus. It is well known that Sarcocystis may cause severe infections in aberrant hosts, which are susceptible animals that do not behave as natural hosts for the parasite and have low resistance to the infection, thus more studies are needed to characterize this parasitosis in skuas and other hosts to understand the epidemiology of the infection and its impact on the health of marine fauna.
Abstract
Evidence of sarcocystid infection was investigated in samples of 16 penguins (Spheniscus. magellanicus), four Dominican gulls (Larus dominicanus) and two Chilean skuas (Stercorarius chilensis) found in Madalenas Islands, Chile, in 2017. Samples of skeletal muscle, cardiac muscle and brain from all birds were screened by a pan-sarcocystid nested-PCR targeting a short fragment of the gene encoding the small ribosomal unit (nPCR-18Sa). The only two positive samples by nPCR-18Sa, both from skuas, were tested by a nested-PCR directed to the internal transcribed spacer 1 (nPCR-ITS1), also a pan-sarcocystidae nested-PCR, and to a nested-PCR directed to the B1 gene (nPCR-B1), for the exclusive detection of Toxoplasma gondii. The two nPCR-18Sa-positive samples were nPCR-ITS1-positive and nPCR-B1-negative. The nPCR-ITS1 nucleotide sequences from the two skuas, which were identical to each other, were revealed closely related to homologous sequences of Sarcocystis halieti, species found in seabirds of northern hemisphere. Larger fragments of genes encoding 18S and partial sequences of genes coding for cytochrome oxidase subunit 1 were also analyzed, corroborating ITS1 data. The haplotypes found in the skuas are unprecedent and closely related to species that use birds as the definitive host. Further studies need to be carried out to detect, identify and isolate this parasite to understand the epidemiology of the infection and its impact on the health of marine fauna.
1. Introduction
The phylum Apicomplexa is composed of obligate intracellular parasites that are characterized by having a specialized structure called the apical complex, which is used to invade vertebrate host cells [1] Within this phylum, the Sarcocystidae family comprises more than 196 species of coccidia that form cysts in tissues of intermediate hosts. Although taxonomic controversies still exist, this family has been divided into three subfamilies: Sarcocystinae, represented by the genera Frenkelia and Sarcocystis; Cystoisosporinae, containing the genus Cystoisospora; and Toxoplasmatinae, a subfamily with a few species grouped in the genera Besnoitia, Hammondia, Neospora and Toxoplasma [2,3,4,5].
Toxoplasma gondii is a coccidian parasite with worldwide distribution. It infects virtually all warm-blooded animals, including humans, but only cats (domestic and wild) act as definitive hosts. Toxoplasmosis has been reported in many avian species; however, little information is available in relation to populations of Spheniscus magellanicus, Stercorarius chilensis and Larus dominicanus [6]. Recently, T. gondii antibodies were detected in 57 (43.18%) out of 132 serum samples collected from free-living Magellanic penguins (Spheniscus magellanicus) on Magdalena Island, Chile, with titers that ranged from 20 to 320 [7].
The genus Sarcocystis has an obligate two-host life cycle. Asexual stages develop in intermediate hosts, usually omnivores, through forming cysts in the musculature and central nervous system. Infection of intermediate hosts occurs through their ingestion of food or water contaminated with sporocysts. Sexual stages only develop in the definitive host, which is typically a carnivore or an omnivore, and infection in this case occurs through ingestion of meat contaminated with cysts [8]. Sarcocystids of the genus Sarcocystis may cause severe infections in aberrant hosts, which are susceptible animals that do not behave as natural hosts of the parasite and have low resistance to the infection. Thus, Sarcocystis potentially pose risk to human and animal health, depending on the susceptible host behaving as aberrant host or not [8].
A few studies have documented the presence of Sarcocystis spp. in wild animals in Chile. Presence of cysts of this parasite has been confirmed in muscle tissues of pudu deer (Pudu puda), guanacos (Lama guanicoe) and sea lions (Otaria byronia) [9,10,11]. However, Sarcocystis has not yet been described in Chilean wild birds.
The Chilean skua Stercorarius chilensis is a large predatory seabird that inhabits shore ecosystems along the southern cone of South America from central Peru to northern Argentina, with occasional occurrence on the coasts of Ecuador, Brazil, Uruguay and Antarctica [12]. Skuas belong to the order Charadriiformes and are considered to be opportunist feeders, preying on a wide diversity of animals such as small seabirds, fish, scraps and carrion [13,14]. Populations of skuas may be small, but they do not approach the thresholds for vulnerable classification following a population-size criterion (<10,000 mature individuals) [15].
Considering other coastal birds’ species, the Kelp Gull (Larus dominicanus) is an opportunistic feeder like numerous Laridae and consumes a wide variety of fishes, invertebrates and fisheries waste [16,17]. A high diversity in the use of habitat types has been recorded throughout its distributional range in the Southern Hemisphere, including Argentina, Brazil, Chile, Peru and Uruguay, and the breeding population has been estimated at least 160,000 pairs [17]. In contrast, the Magellanic penguin (S. magellanicus) has approximately 1.1 to 1.6 million breeding pairs that nest along the eastern and western coasts of South America, in Argentina Chile and the Malvinas/Falkland Islands [18]. Spheniscus magellanicus has a primarily piscivorous diet with the presence of some cephalopods and crustaceans [19].
To date, more than 25 species of Sarcocystis have been found to use birds as intermediate hosts [8,20]. Sarcocystis falcatula, Sarcocystis calchasi and the recently described unnamed species Sarcocystis sp. Chicken-2016-DF-BR, which can possibly be interpreted as Sarcocystis wenzeli [21] are species that may be pathogenic for intermediate hosts [22,23,24].
Focusing on T. gondi and the genus Sarcocystis, the aim of this study was to screen for the evidence of new species or species genotypes of Sarcocystidae in seabird carcasses from southern Chile, a region with scarce data on the occurrence of this group of parasites. Molecular evidence of a unique haplotype of genus Sarcocystis was found in two Chilean skuas (S. chilensis).
2. Materials and Methods
2.1. Ethical Considerations
Sample collections on Magdalena Island were conducted under license no. 039/2016 issued by the National Forestry Corporation (Corporación Nacional Forestal; CONAF), and permit no. 2799 issued by the National Fisheries Service (Servicio Nacional de Pesca; SERNAPESCA), Chile. This study was approved by the Ethics Committee on Animal Use (CEUA-no. 9701041113) of the School of Veterinary Medicine, University of São Paulo (FMVZ-USP).
2.2. Collection of Samples
In January 2017, fragments from the pectoral muscle, heart and brain, comprising approximately 5–10 g each, were collected from fresh seabird carcasses on Magdalena Island. This island is located in the Strait of Magellan, near the city of Punta Arenas, in southern Chile (52°55′10.0″ S; 70°34′37.7″ W), and constitutes a natural reserve named “Monumento Natural Los Pinguinos”. Necropsies were performed in situ and samples were stored in sterile microtubes at −20 °C until the time of analysis. Samples were collected from 22 birds: 16 penguins (S. magellanicus), four Dominican gulls (L. dominicanus) and two Chilean skuas (S. chilensis), totaling 66 samples (22 from pectoral muscles, 22 from hearts and 22 from brains).
2.3. Molecular Identification
Tissue samples of 25–50 mg were subjected to DNA extraction using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) following the manufacturer’s recommendations, with the exception of final elution of the product into 50 µL of elution buffer from Qiagen Kit. As internal control for the evaluation of the successfulness of the DNA extraction, DNA samples were tested by conventional PCR targeting mitochondrial hypervariable region in the penguins derived samples and by PCR directed to mitochondrial 16S rRNA gene in the tissues from the other seabirds [25,26,27].
Initial screening targeting the Sarcocystidae family was performed using a pan-sarcocystid nested PCR based on primers [28] directed to a short fragment of 18S rDNA gene (nPCR-18Sa). The nPCR-18Sa positive samples were further investigated for the presence of DNA of T. gondii to amplify partial fragments of gene B1 (nPCR-B1) using the primers described by [29]. The nPCR-18Sa positive samples were also tested by a second pan-sarcocystid nested PCR directed to internal transcribed spacer 1 (nPCR-ITS1) [30,31] The nPCR-ITS1 were used in order to obtain genetic sequences capable of differentiating the species of the Sarcocystid screened with nPCR-18Sa. The nPCR-ITS1 positive samples were further tested with a third pan-sarcocystid nested PCR, now targeting a larger fragment of 18S rDNA gene (nPCR-18Sb) using primers described by [32], as well with a Sarcocystis specific nested PCR [30] directed to cytochrome oxidase subunit I (nPCR-CO1). The primers are depicted in Table 1.

Table 1.
Primers for the detection of Sarcocystidae in tissues of seabirds from Magdalena Island, Chile.
The first round of nPCR-18Sa were performed with 3.0 µL of extracted DNA, 1.8 µL of 10× PCR Buffer (KCL 50 mM; Tris HCl 10 mM; pH 9.0) (Life Technologies Corporation, Carlsbad, CA 92008 USA), 0.7 µL of MgCL2 (50 mM), 1.4 µL of mixed dNTPs (10 mM), 0.1 µL of each primer (25 µM), 0.14 µL of PlatinumTM Taq DNA Polymerase (5 U/µL) (Life Technologies Corporation, Carlsbad, CA 92008 USA) and ultrapure autoclaved water to a volume of 18 µL per reaction. The PCR thermal cycling consisted of an initial incubation at 94 °C for 30 sec, followed by 30 cycles (94 °C for 25 sec, 55 °C for 1 min, 72 °C for 1.5 min) and a final extension at 72 °C for 10 min. For the second rounds: 1 µL of template derived from the first reactions, 2.5 µL of 10× PCR Buffer (KCL 50 mM; Tris HCl 10 mM; pH 9.0) (Life Technologies Corporation, Carlsbad, CA 92008 USA), 2.5 µL of MgCL2 (50 mM), 4.0 µL of mixed dNTPs (10 mM), 1.25 µL of each primer (10µM), 0.15 µL of PlatinumTM Taq DNA Polymerase (5 U/µL) (Life Technologies Corporation, Carlsbad, CA 92008 USA)and ultrapure autoclaved water to a volume of 25 µL per reaction. The PCR thermal cycling consisted of an initial incubation at 94 °C for 4 min, followed by 30 cycles (94 °C for 30 s, 55 °C for 1 min, 72 °C for 2.0 min) and a final extension at 72 °C for 10 min. For the second rounds the same quantities of the reagent mixture with primers at 50 μM, using 2 μL of the product of the PCR diluted in ultra-pure water (1:2). The nPCR thermal cycling consisted of an initial incubation at 94 °C for 4 min, followed by 35 cycles (94 °C for 30 s, 55 °C for 1 min, 72 °C for 2.0 min) and a final extension at 72 °C for 10 min.
The first round of nPCR-B1 were performed with 1.0 µL of extracted DNA, 2.5 µL of 10× PCR Buffer (KCL 50 mM; Tris HCl 10 mM; pH 9.0), (Life Technologies Corporation, Carlsbad, CA 92008 USA), 0.75 µL of MgCL2 (50 mM), 4.0 µL of mixed dNTPs (10 mM), 1.25 µL of each primer (10 µM), 0.15 µL of PlatinumTM Taq DNA Polymerase (5 U/µL) (Life Technologies Corporation, Carlsbad, CA 92008 USA)and ultrapure autoclaved water to a volume of 25 µL per reaction. The PCR thermal cycling consisted of an initial incubation at 94 °C for 3 min, followed by 25 cycles (94 °C for 25 s, 55 °C for 1 min, 72 °C for 1.5 min) and a final extension at 72 °C for 10 min. For the second rounds: 1 µL of template derived from the first reactions, 2.5 µL of 10× PCR Buffer (KCL 50 mM; Tris HCl 10 mM; pH 9.0) (Life Technologies Corporation, Carlsbad, CA 92008 USA), 2.5 µL of MgCL2 (50 mM), 4.0 µL of mixed dNTPs (10 mM), 1.25 µL of each primer (10 µM), 0.15 µL of PlatinumTM Taq DNA Polymerase (Life Technologies Corporation, Carlsbad, CA 92008 USA) and ultrapure autoclaved water to a volume of 25 µL per reaction. The nPCR thermal cycling consisted of an initial incubation at 94 °C for 3 min, followed by 35 cycles (94 °C for 25 s, 55 °C for 1 min, 72 °C for 1.5 min) and a final extension at 72 °C for 10 min.
The first round of nPCR-18Sb, nPCR-ITS1 and nPCR-CO1 were performed with 4 µL of extracted DNA, 2.5 µL of 10× PCR Buffer (KCL 50 mM; Tris HCl 10 mM; pH 9.0) (Life Technologies Corporation, Carlsbad, CA 92008 USA), 1.0 µL of MgCL2 (50 mM), 0.5 µL of mixed dNTPs (10 mM), 1.0 µL of each primer (10 µM), 0.2 µL of PlatinumTM Taq DNA Polymerase (Life Technologies Corporation, Carlsbad, CA 92008 USA) (5 U/µL) (Termofischer Scientific) and ultrapure autoclaved water to a volume of 25 µL per reaction. The PCR thermal cycling consisted of an initial incubation at 94 °C for 3 min, followed by 35 cycles (94 °C for 30 s, 56 °C for 30 s, 72 °C for 50 s) and a final extension at 72 °C for 5 min. For the second rounds: 2 µL of template derived from the first reactions, 2.5 µL of 10× PCR Buffer (KCL 50 mM; Tris HCl 10 mM; pH 9.0) (Life Technologies Corporation, Carlsbad, CA 92008 USA), 1.0 µL of MgCL2 (50 mM), 0.5 µL of mixed dNTPs (10 mM), 2.5 µL of each primer (10 µM), 0.2 µL of PlatinumTM Taq DNA ) (Life Technologies Corporation, Carlsbad, CA 92008 USA) (5 U/µL) and ultrapure autoclaved water to a volume of 25 µL per reaction. The thermal cycling was the same used in the first round.
DNA of Sarcocystis neurona, Neospora caninum and Hammondia hammondi was used as positive controls and ultrapure DNAse-free water as the negative control for all reactions.
PCR products were resolved on 2.0% agarose gels and viewed through UV transillumination. Amplicons of the expected sizes were treated with ExoSAP-IT (Affimetryx/Thermo Fisher Scientific, Santa Clara, CA, USA), prepared for sequencing using the Big Dye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) and sequenced in an ABI automated sequencer (ABI 3500 Genetic Analyzer, Applied Biosystems). Sequencing was performed using the same primers as in the nPCR consensus. Sequence edition and contig assemblies were done by using the software Codoncode aligner, Codoncode Corporation. Final sequences were compared with homologous available in GenBank, using the BLASTn algorithm (Table S1) (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
For the phylogenies, sequences were aligned using the Clustal W program, as implemented in the BioEdit Sequence Alignment Editor [34]. The phylogenetic tree based on ITS1 was inferred using MEGA X [35], through the maximum likelihood method and T92 model of evolutionary distances. Branch supports were tested through 1000 bootstrap replications.
3. Results
3.1. Molecular Identification
Sixty-six tissue samples from 22 seabirds were screened by nPCR-18Sa and only two samples of pectoral muscle from two Chilean skuas were positive. None of these two samples were positive for the T. gondii-specific nested-PCR (nPCR-B1). The two nPCR-18Sa-positive samples were also positive by nPCR-ITS1, nPCR-18Sb and nPCR-CO1 (Figure 1, left panel). After sequencing nPCR-ITS1, nPCR-18Sb and nPCR-CO1 amplicons and removal of primer-derived sequences, 861, 783 and 547 base pairs were obtained, respectively. Fragments of the sequences obtained are shown in Figure 1, right panel. The homologous sequences of the two samples were identical to each other, thus only one set were submitted to the GenBank, under the accession numbers MW160469, MW161469, MW157378. Through BLAST searches, ITS1, CO1 and 18S genetic sequences were compared with sequences producing the most significant alignments, with query coverage ≥ 99% and percentage similarities ≥ 99.00% in the cases of CO1 and 18S. All ITS1 sequences with query cover ≥ 96% were used for analyses of the genetic sequence of the skuas.
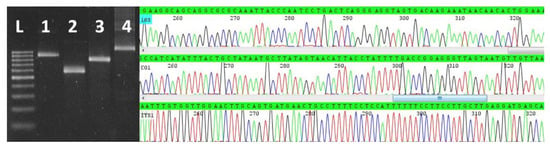
Figure 1.
Left panel: Agarose gel electrophoresis of nPCR-ITS1 (1), nPCR-CO1 (2), and nPCR-18Sb (3) amplicons from Sarcocystis sp. ex Stercorarius chilensis, nPCR-ITS1 amplicons (4) from Sarcocystis neurona and Ladder Scada 100 bp, Sinapse, Inc. (L). Right panel: segments of the electropherogram obtained after sequencing nPCR-18Sb (top), nPCR-CO1 (middle), and nPCR-ITS1 (bottom) amplicons from Sarcocystis sp. ex Stercorarius chilensis.
The ITS1 fragment from the skuas showed 96.14–96.28% identity to sequences of Sarcocystis halieti from herring gulls (Larus argentatus) (MN450340–MN450356), great cormorants (Phalacrocorax carbo) (MH130209, JQ733513) and white-tailed sea-eagles (Haliaeetus albicilla) (MF946589–MF946596). Sarcocystis sp. from Cooper’s hawk (Accipiter cooperii) (KY348755), Sarcocystis columbae from common woodpigeons (Columba palumbus) (GU253885, HM125052) and herring gulls (Larus argentatus) (MN450338–MN450339) and Sarcocystis corvusi from Eurasian jackdaws (Corvus monedula) (JN256119) showed less than 94% sequence identity with homologous sequences from the Chilean skuas at ITS1 locus.
In contrast to ITS1, much less genetic variability was observed within the CO1 and 18S coding genes. The haplotype obtained from the skuas was named Sarcocystis sp. ex Stercorarius chilensis.
The CO1 sequences from Sarcocystis sp. ex Stercorarius chilensis were 100% identical with homologous sequences of S. corvusi (MH138314), S. columbae (MH138312) and S. halieti (MH138308-09; MF946583). Sarcocystis fulicae (MH138316), Sarcocystis wobeseri (MH138315), Sarcocystis cornixi (MH138313) and Sarcocystis sp. ex Accipiter cooperii (KY348756) differed by only one nucleotide substitution from Sarcocystis sp. ex Stercorarius chilensis at this locus. Regarding the 18S rRNA gene, the maximum percentage identity was found between Sarcocystis sp. ex Stercorarius chilensis and S. halieti (99.74%).
3.2. Phylogeny
The ITS1-based phylogeny demonstrated that the species that shares the most recent ancestral commonality with Sarcocystis sp. ex Stercorarius chilensis was S. halieti (Figure 2).
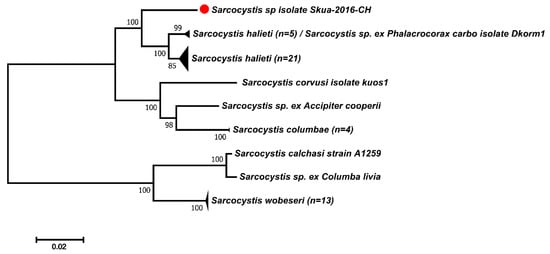
Figure 2.
Phylogenetic tree of Sarcocystis species based on ITS1 sequences. The tree was constructed using the maximum likelihood method and Tamura 3 parameters nucleotide substitution model. The final alignment contained 49 sequences and 814 aligned nucleotide positions. All positions containing gaps and missing data were eliminated (complete deletion option). Numbers in branches represent bootstrap values after 1000 replicates. The red dot identifies the sequence of Sarcocystis sp. ex Stercorarius chilensis.
These sequences were separated with high support from a major clade comprising the species Sarcocystis sp. ex Columba livia (FJ232948), Sarcocystis calchasi (KC733715–KC733718) and Sarcocystis wobeseri (MN450365–MN450373, HM159421, JN256121), which exploit Anseriformes, Charadriiformes, Columbiformes, Psittaciformes and other birds as intermediate hosts.
Altogether, the Sarcocystis species that were most similar to Sarcocystis sp. ex Stercorarius chilensis used birds as intermediate hosts.
4. Discussion
Toxoplasma gondii has high genotypic diversity and several new genotypes have been described in wildlife [36,37,38], which has aided to understand the shape of molecular evolution and the epidemiology of the infection [38]. Similarly, several species descriptions have been made for the genus Sarcocystis, most of them with the aid of molecular methods [39,40,41,42,43]. This study presents the results of a molecular screening of Sarcocystidae focusing on animal species and geographical areas where these parasites have rarely or not yet been identified. DNA of organisms of the genus Sarcocystis was identified in two Chilean skuas, whereas DNA of T. gondii were not found in any sample. The Sarcocystis haplotype detected in skuas was named Sarcocystis sp. ex Stercorarius chilensis.
Although all the samples tested negative for the presence of T. gondii, antibodies against this parasite were detected previously in 57 (43.18%) of the 132 serum samples from free-living Magellanic penguins from the same region, with titers that ranged from 20 to 320 [7]. Herein, infected animals were not encountered possibly because the sampling was insufficient to find at least one positive animal, as the prevalence of the infection in seabirds in the sampled area are not known and the sampling might have not been representative of the population surveyed. In addition, the mass of tissue that was tested might have been insufficient because of the very sparse and focal distribution of T. gondii cysts in the tissues of the HI, thus, digestion of samples previously to the DNA extraction and subsequent DNA detection would be more appropriate than direct DNA extraction, as used in this study [44].
Oocysts of T. gondii can sporulate and survive in seawater for months [45,46]. Marine mammals in different groups (cetaceans, pinnipeds and sirenians) and seabirds might become infected through consumption of water containing the oocysts. Thus, T. gondii oocysts from felidae feces might enter the marine environment and contaminate both the water and several invertebrate species, which could act as vectors of infection for mammals and seabirds [47]. Mice can be experimentally infected when fed with T. gondii-contaminated oysters (Crassostrea virginica) [45] proving that T. gondii was able to survive for several months in these mollusks [48]. Anchovies and Pacific sardines can be experimentally contaminated with T. gondii oocysts, which indicates that migratory filter feeders may serve as biotic vectors for this parasite [49]. Another study proved that freshwater crustaceans were able to bioaccumulate T. gondii oocysts. It should be noted that crustaceans are part of penguins’ and many seabirds’ food chain [50,51]. Thus, although the birds screened here were found not infected by T. gondii, marine fauna are at risk of acquiring the infection, by ingesting oocysts carried by transport hosts (oysters, fish and other) or through predation of intermediate hosts in the marine or in the coastal environment.
Based on molecular data, Sarcocystis sp. ex Stercorarius chilensis is an undescribed Sarcocystis species, closely related to S. halieti. The molecular identification based on ITS1, CO1 and 18S rRNA gene sequences showed a closed relationship between Sarcocystis sp. from Chilean skuas and other Sarcocystis spp. that use birds as intermediate hosts and predatory birds as definitive hosts. As expected, the most variable locus was ITS1, and phylogenies based on 18S rRNA and CO1 genes showed insufficient discrimination power to differentiate between species within the genus [39,41].
The most similar sequences to ITS1 of Sarcocystis sp. ex Stercorarius chilensis are those from Sarcocystis spp. that use hawks as definitive hosts. Sarcocystis sp. ex Stercorarius chilensis grouped together with S. halieti, a species that uses white-tailed sea-eagles (Haliaeetus albicilla) and Eurasian sparrowhawks (Accipiter nisus) as definitive hosts [39]. Other taxa found through ITS1-based BLAST searches encompass Sarcocystis spp. that also use hawks as definitive hosts (Accipiter cooperii, Accipiter nisus), except for S. corvusi, for which this information remains unknown [52,53]. Accipiter hawks (Accipiter gentilis, Accipiter nisus) are definitive hosts for S. calchasi [54,55,56].
Several studies have expanded the knowledge on the host specificity of Sarcocystis, as unequivocal identification of the parasite can be achieved after identifying sarcocysts and oocysts to species level using molecular methods. Sarcocystis halieti and Sarcocystis lari were found to have formed oocysts in the intestine of white-tailed sea eagle (Haliaeetus albicilla), showing for the first time the potential role of sea eagle as definitive host of those species of Sarcocystis [53]. Likewise, european seabirds were found to harbor several species of Sarcocystis after DNA of Sarcocystis lari, S. wobeseri, S. columbae and S. halieti were detected in sarcocysts infecting muscle of herring gulls (Larus argentatus), great black-backed gulls (Larus marinus) and great cormorants (Phalacrocorax carbo) in Lithuania [40,41,42].
The four morphologically indistinguishable Sarcocystis species, Sarcocystis lari, S. wobeseri, S. columbae and S. halieti, could only be differentiated in L. argentatus by means of ITS1 sequence analysis [42]. Likewise, only ITS1 clearly discriminated Sarcocystis sp. ex Stercorarius chilensis from S. halieti, which reinforces that molecular characterization using this marker is of paramount importance to distinguish closed related species within the genus.
It is well known that a single animal can host more than one Sarcocystis species [40]. Here, sarcocysts were not individually excised and subjected to molecular examination, notwithstanding, the possibility of mixed infected samples of skuas was discarded because single peaks and clean sequence throughout the chromatograms were obtained for each sequence. Thus, a haplotype could be confidently assigned to the samples.
Although screening Sarcocystis by using molecular methods without morphological characterization of parasitic structures is obviously not enough to name a new species, this procedure may provide subsides to future studies on the epidemiology of the infection and its impact on the health of marine fauna. To our knowledge, Sarcocystis in south American seabirds were identified only once [43], which suggests that a wide field of research on diversity of sarcocystidae can be explored on this continent.
5. Conclusions
Although few animals have been screened in this study and morphological characterization of the parasites was not carried out, evidence of an unprecedented haplotype of Sarcocystis was found in skuas from Chile, which demonstrate that molecular screening of Sarcocystis can be a valuable tool to prospect for new species, contributing to knowledge on the epidemiology of sarcocystosis and life cycle of Sarcocystis. Sporocysts shed with feces, sarcocysts in tissues or rapid dividing structures in acute sarcocystosis (schyzonts and merozoites) can be more easily and accurately identified as data on Sarcocystis genetic sequences increases. Nevertheless, a complete study encompassing aspects of life cycle and morphological data is necessary to fully describe Sarcocystis sp. ex Stercorarius chilensis and additional studies are needed to better understand the epidemiology of the infection and its impact on the health of marine fauna.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/11/2/245/s1, Table S1. Results from BLAST search using sequences of ITS1, COX1 and 18S of Sarcocystis sp. ex Stercorarius chilensis.
Author Contributions
Conceptualization, I.C.L.A., S.M.G. and R.M.S. writing—original draft preparation, I.C.L.A. and R.M.S.; review and editing, I.C.L.A., S.M.G., H.A.B.L., S.M.-L. and R.M.S.; visualization, I.C.L.A., S.M.G., H.A.B.L., S.M.-L. and R.M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo), Brazil and by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil. R.M.S. and S.M.G. received a fellowship from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico).
Institutional Review Board Statement
This study was approved by the Ethics Committee on Animal Use (CEUA-no. 9701041113 - 2014) of the School of Veterinary Medicine, University of São Paulo (FMVZ-USP). Sample collections on Magdalena Island were conducted under license no. 039/2016 issued by the National Forestry Corporation (Corporación Nacional Forestal; CONAF), and permit no. 2799 issued by the National Fisheries Service (Servicio Nacional de Pesca; SERNAPESCA), Chile
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
To Roberto Fernández and Ricardo Cid (CONAF) for their valuable assistance during the sample collections on Magdalena Island.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sam-Yellowe, T.Y. Rhoptry organelles of the apicomplexa: Their role in host cell invasion and intracellular survival. Parasitol. Today 1996, 12, 308–316. [Google Scholar] [CrossRef]
- Morrison, D.A.; Bornstein, S.; Thebo, P.; Wernery, U.; Kinne, J.; Mattsson, J.G. The current status of the small subunit rRNA phylogeny of the coccidia (Sporozoa). Int. J. Parasitol. 2004, 34, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Mugridge, N.B.; Morrison, D.A.; Johnson, A.M.; Luton, K.; Dubey, J.P.; Votýpka, J.; Tenter, A.M. Phylogenetic relationships of the genus Frenkelia: A review of its history and new knowledge gained from comparison of large subunit ribosomal ribonucleic acid gene sequences. Int. J. Parasitol. 1999, 29, 957–972. [Google Scholar] [CrossRef]
- Samarasinghe, B.; Johnson, J.; Ryan, U. Phylogenetic analysis of Cystoisospora species at the rRNA ITS1 locus and development of a PCR-RFLP assay. Exp. Parasitol. 2008, 118, 592–595. [Google Scholar] [CrossRef] [PubMed]
- Tenter, A.M.; Barta, J.R.; Beveridge, I.; Duszynski, D.W.; Mehlhorn, H.; Morrison, D.A.; Andrew Thompson, R.C.; Conrad, P.A. The conceptual basis for a new classification of the coccidian. Int. J. Parasitol. 2002, 32, 595–616. [Google Scholar] [CrossRef]
- Dubey, J.P. Toxoplasmosis of Animals and Humans, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Acosta, I.C.L.; Souza-Filho, A.F.; Muñoz-Leal, S.; Soares, H.S.; Heinemann, M.B.; Moreno, L.; González-Acuña, D.; Gennari, S.M. Evaluation of antibodies against Toxoplasma gondii and Leptospira spp. in Magellanic penguins (Spheniscus magellanicus) on Magdalena Island, Chile. Vet. Parasitol. Reg. Stud. Rep. 2019, 16, 100282. [Google Scholar] [CrossRef]
- Dubey, J.P.J.; Calero-Bernal, R.; Rosenthal, B.M.B.; Speer, C.A.C.; Fayer, R. Sarcocystosis of Animals and Humans; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Rioseco, H.; Cubillos, V.; González, H.; Díaz, L. Sarcosporidiosis en pudues (Pudu pudu, Molina, 1782) Primera comunicación en Chile. Arch. Med. Vet. 1976, 8, 122–123. [Google Scholar]
- Gorman, T.R.; Alcaíno, H.A.; Muñuz, H.; Cunazza, C. Sarcocystis sp. in guanaco (Lama guanicoe) and effect of temperature on its viability. Vet. Parasitol. 1984, 15, 95–101. [Google Scholar] [CrossRef]
- Sepúlveda, M.A.; Seguel, M.; Alvarado-Rybak, M.; Verdugo, C.; Muñoz-Zanzi, C.; Tamayo, R. Postmortem findings in four South American sea lions (Otaria byronia) from an urban colony in Valdivia, Chile. J. Wildl. Dis. 2015, 51, 279–282. [Google Scholar] [CrossRef]
- Tavares, D.C.; Moura, J.F.; de Amorim, C.E.; Boldrini, M.A.; Siciliano, S. Aves, Stercorariidae, Chilean Skua Stercorarius chilensis Bonaparte, 1857: First documented record for the state of Espírito Santo, southeastern Brazil. Check List 2012, 8, 560. [Google Scholar] [CrossRef][Green Version]
- Furness, R.W. The Skuas; Poyser, Poyser Monographs; Bloomsbury Collections: London, UK, 1987. [Google Scholar]
- Robinson, I. Seabirds. In Handbook of Avian Medicine; Elsevier Ltd.: Amsterdam, The Netherlands, 2009; pp. 377–403. [Google Scholar]
- BirdLife International. Species Factsheet: Catharacta Chilensis. Available online: http://www.birdlife.org (accessed on 16 July 2020).
- Ludynia, K.; Garthe, S.; Luna-Jorquera, G. Seasonal and regional variation in the diet of the Kelp Gull in northern Chile. Waterbirds 2005, 28, 359–365. [Google Scholar] [CrossRef]
- Yorio, P.; Branco, J.O.; Lenzi, J.; Luna-Jorquera, G.; Zavalaga, C. Distribution and Trends in Kelp Gull (Larus dominicanus) Coastal Breeding Populations in South America. Waterbirds 2016, 39, 114–135. [Google Scholar] [CrossRef]
- Boersma, P.D.; Frere, E.; Kane, O.; Pozzi, L.M.; Pütz, K.; Rey, A.R. Magellanic penguins. In Penguins: Natural History and Conservation; Borboroglu, P.G., Boersma, P.D., Eds.; University of Washington Press: Seattle, WA, USA, 2013; pp. 233–263. [Google Scholar]
- Frere, E.; Gandini, P.; Lichtschein, V. Variacion latitudinal en la dieta del pinguino de Magallanes (Spheniscus magellanicus) en la Costa Patagonica, Argentina. Ornitol. Neotrop. 1996, 7, 35–41. [Google Scholar]
- Odening, K. The present state of species-systematics in Sarcocystis Lankester, 1882 (Protista, Sporozoa, Coccidia). Syst. Parasitol. 1998, 41, 209–233. [Google Scholar] [CrossRef]
- Pan, J.; Ma, C.; Huang, Z.; Ye, Y.; Zeng, H.; Deng, S.; Hu, J.; Tao, J. Morphological and molecular characterization of Sarcocystis wenzeli in chickens (Gallus gallus) in China. Res. Sq. 2020, 13, 1–7. [Google Scholar] [CrossRef]
- Konradt, G.; Bianchi, M.V.; Leite-Filho, R.V.; da Silva, B.Z.; Soares, R.M.; Pavarini, S.P.; Driemeier, D. Necrotizing meningoencephalitis caused by Sarcocystis falcatula in bare-faced ibis (Phimosus infuscatus). Parasitol. Res. 2017, 116, 809–812. [Google Scholar] [CrossRef]
- Olias, P.; Maier, K.; Wuenschmann, A.; Reed, L.; Armién, A.G.; Shaw, D.P.; Gruber, A.D.; Lierz, M. Sarcocystis calchasi has an expanded host range and induces neurological disease in cockatiels (Nymphicus hollandicus) and North American rock pigeons (Columbia livia f. dom.). Vet. Parasitol. 2014, 200, 59–65. [Google Scholar] [CrossRef]
- Wilson, T.M.; Sousa, S.K.H.; Paludo, G.R.; de Melo, C.B.; Llano, H.A.B.; Soares, R.M.; Castro, M.B. An undescribed species of Sarcocystis associated with necrotizing meningoencephalitis in naturally infected backyard chickens in the Midwest of Brazil. Parasitol. Int. 2020, 76, 102098. [Google Scholar] [CrossRef]
- Roeder, A.D.; Ritchie, P.A.; Lambert, D.M. New DNA markers for penguins. Conserv. Genet. 2002, 3, 341–344. [Google Scholar] [CrossRef]
- Pons, J.M.; Hassanin, A.; Crochet, P.A. Phylogenetic relationships within the Laridae (Charadriiformes, Aves) inferred from mitochondrial markers. Mol. Phylogenet. Evol. 2005, 37, 686–699. [Google Scholar] [CrossRef]
- Han, Y.D.; Baek, Y.S.; Kim, J.H.; Choi, H.G.; Kim, S. Complete mitochondrial genome of the South Polar Skua Stercorarius maccormicki (Charadriiformes, Stercorariidae) in Antarctica. Mitochondrial DNA 2016, 27, 1783–1784. [Google Scholar]
- Su, C.; Shwab, E.K.; Zhou, P.; Zhu, X.Q.; Dubey, J.P. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology 2010, 137, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yai, L.E.O.; Cañon-Franco, W.A.; Geraldi, V.C.; Summa, M.E.L.; Camargo, M.C.G.O.; Dubey, J.P.; Gennari, S.M. Seroprevalence of Neospora caninum and Toxoplasma gondii antibodies in the South American opossum (Didelphis marsupialis) from the city of São Paulo, Brazil. J. Parasitol. 2003, 89, 870–871. [Google Scholar] [CrossRef] [PubMed]
- Soares, R.M.; Lopes, E.G.; Keid, L.B.; Sercundes, M.K.; Martins, J.; Richtzenhain, L.J. Identification of Hammondia heydorni oocysts by a heminested-PCR (hnPCR-AP10) based on the H. heydorni RAPD fragment AP10. Vet. Parasitol. 2011, 175, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Gondim, L.F.P.; Soares, R.M.; Tavares, A.S.; Silva, W.B.; de Jesus, R.F.; Llano, H.A.B.; Gondim, L.Q. Sarcocystis falcatula-like derived from opossum in Northeastern Brazil: In vitro propagation in avian cells, molecular characterization and bioassay in birds. Int. J. Parasitol. Parasites Wildl. 2019, 10, 132–137. [Google Scholar] [CrossRef]
- Li, Z.Q.; Yang, Y.X.; Zuo, S.W.; Attwood, X.W.; Chen, Y.P.; Zhang, A. PCR-based RFLP analysis of Sarcocystis cruzi (Protozoa: Sarcocystidae) in Yunnan Province, PR China, reveals the water buffalo (Bubalus bubalis) as a natural intermediate host. J. Parasitol. 2002, 88, 1259–1261. [Google Scholar] [CrossRef]
- Šlapeta, J.R.; Koudela, B.; Votýpka, J.; Modrý, D.; Hořejš, R.; Lukeš, J. Coprodiagnosis of Hammondia heydorni in dogs by PCR based amplification of ITS 1 rRNA: Differentiation from morphologically indistinguishable oocysts of Neospora caninum. Vet. J. 2002, 163, 147–154. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Sibley, L.D.; Khan, A.; Ajioka, J.W.; Rosenthal, B.M. Genetic diversity of Toxoplasma gondii in animals and humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 2749–2761. [Google Scholar] [CrossRef] [PubMed]
- Vitaliano, S.N.; Soares, H.S.; Minervino, A.H.H.; Santos, A.L.Q.; Werther, K.; Marvulo, M.F.V.; Siqueira, D.B.; Pena, H.F.J.; Soares, R.M.; Su, C.; et al. Genetic characterization of Toxoplasma gondii from Brazilian wildlife revealed abundant new genotypes. Int. J. Parasitol. Parasites Wildl. 2014, 3, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.A.; Longcore, T.; Barbieri, M.; Dabritz, H.; Hill, D.; Klein, P.N.; Lepczyk, C.; Lilly, E.L.; Milcarsky, R.M.J.; Su, C.; et al. The one health approach to toxoplasmosis: Epidemiology, control, and prevention strategies. EcoHealth 2019, 16, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Gjerde, B.; Vikøren, T.; Hamnes, I.S. Molecular identification of Sarcocystis halieti n. sp., Sarcocystis lari and Sarcocystis truncata in the intestine of a white-tailed sea eagle (Haliaeetus albicilla) in Norway. Int. J. Parasitol. Parasites Wildl. 2018, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Prakas, P.; Butkauskas, D.; Juozaitytė-Ngugu, E. Molecular identification of four Sarcocystis species in the herring gull, Larus argentatus, from Lithuania. Parasit Vectors 2020, 13, 1–6. [Google Scholar] [CrossRef]
- Prakas, P.; Butkauskas, D.; Švažas, S.; Stanevičius, V. Morphological and genetic characterisation of Sarcocystis halieti from the great cormorant (Phalacrocorax carbo). Parasitol. Res. 2018, 117, 3663–3667. [Google Scholar] [CrossRef]
- Prakas, P.; Kutkiene, L.; Butkauskas, D.; Sruoga, A.; Zalakevicius, M. Description of Sarcocystis lari sp. n. (Apicomplexa: Sarcocystidae) from the great black-backed gull, Larus marinus (Charadriiformes: Laridae), on the basis of cyst morphology and molecular data. Folia Parasitol. 2014, 61, 11–17. [Google Scholar] [CrossRef]
- Acosta, I.C.L.; Soares, R.M.; Mayorga, L.F.S.P.; Alves, B.F.; Soares, H.S.; Gennari, S.M. Occurrence of tissue cyst forming coccidia in Magellanic penguins (Spheniscus magellanicus) rescued on the coast of Brazil. PLoS ONE 2018, 13, e0212467. [Google Scholar] [CrossRef]
- Dubey, J.P. Long-term persistence of Toxoplasma gondii in tissues of pigs inoculated with T. gondii oocysts and effect of freezing on viability of tissue cysts in pork. Am. J. Vet. Res. 1988, 49, 910–913. [Google Scholar]
- Lindsay, D.S.; Collins, M.V.; Mitchell, S.M.; Cole, R.A.; Flick, G.J.; Wetch, C.N.; Lindquist, A.; Dubey, J.P. Sporulation and survival of Toxoplasma gondii oocysts in seawater. J. Eukaryot. Microbiol. 2003, 50, 687–688. [Google Scholar] [CrossRef]
- Fayer, R.; Dubey, J.P.; Lindsay, D.S. Zoonotic protozoa: From land to sea. Trends Parasitol. 2004, 20, 531–536. [Google Scholar] [CrossRef]
- Cole, R.A.; Lindsay, D.S.; Howe, D.K.; Roderick, C.L.; Dubey, J.P.; Thomas, N.J.; Baeten, L.A. Biological and molecular characterization of Toxoplasma gondii strains obtained from southern sea otters (Enhydra lutris nereis). J. Parasitol. 2000, 86, 526–530. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Phelps, K.K.; Smith, S.A.; Flick, G.; Sumner, S.S.; Dubey, J.P. Removal of Toxoplasma gondii oocysts from sea water by eastern oysters (Crassostrea virginica). J. Eukaryot. Microbiol. 2001, 48, 197S–198S. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, D.S.; Collins, M.V.; Mitchell, S.M.; Wetch, C.N.; Rosypal, A.C.; Flick, G.J.; Zajac, A.M.; Lindquist, A.; Dubey, J.P. Survival of Toxoplasma gondii oocysts in Eastern oysters (Crassostrea virginica). J. Parasitol. 2004, 90, 1054–1057. [Google Scholar] [CrossRef] [PubMed]
- Massie, G.N.; Ware, M.W.; Villegas, E.N.; Black, M.W. Uptake and transmission of Toxoplasma gondii oocysts by migratory, filter-feeding fish. Vet. Parasitol. 2010, 169, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Bigot-Clivot, A.; Ladeiro, M.P.; Lepoutre, A.; Bastien, F.; Bonnard, I.; Dubey, J.P.; Villena, I.; Aubert, D.; Geffard, O.; François, A.; et al. Bioaccumulation of Toxoplasma and Cryptosporidium by the freshwater crustacean Gammarus fossarum: Involvement in biomonitoring surveys and trophic transfer. Ecotoxicol. Environ. Saf. 2016, 133, 188–194. [Google Scholar] [CrossRef]
- Mayr, S.L.; Maier, K.; Müller, J.; Enderlein, D.; Gruber, A.D.; Lierz, M. Accipiter hawks (Accipitridae) confirmed as definitive hosts of Sarcocystis turdusi, Sarcocystis cornixi and Sarcocystis sp. ex Phalacrocorax carbo. Parasitol. Res. 2016, 115, 3041–3047. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Verma, S.K.; Scott, D.; Dubey, J.P.; von Dohlen, A.R. Isolation, molecular characterization, and in vitro schizogonic development of Sarcocystis sp. ex Accipiter cooperii from a naturally infected Cooper’s hawk (Accipiter cooperii). Parasitol. Int. 2017, 66, 106–111. [Google Scholar] [CrossRef]
- Olias, P.; Olias, L.; Krücken, J.; Lierz, M.; Gruber, A.D. High prevalence of Sarcocystis calchasi sporocysts in European Accipiter hawks. Vet. Parasitol. 2011, 175, 230–236. [Google Scholar] [CrossRef]
- Olias, P.; Gruber, A.D.; Hafez, H.M.; Heydorn, A.O.; Mehlhorn, H.; Lierz, M. Sarcocystis calchasi sp. nov. of the domestic pigeon (Columba livia f. domestica) and the Northern goshawk (Accipiter gentilis): Light and electron microscopical characteristics. Parasitol. Res. 2010, 106, 577–585. [Google Scholar] [CrossRef]
- Olias, P.; Olias, L.; Lierz, M.; Mehlhorn, H.; Gruber, A.D. Sarcocystis calchasi is distinct to Sarcocystis columbae sp. nov. from the wood pigeon (Columba palumbus) and Sarcocystis sp. from the sparrow hawk (Accipiter nisus). Vet. Parasitol. 2010, 171, 7–14. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).