Influence of the Season and Region Factor on Phosphoproteome of Stallion Epididymal Sperm
Abstract
Simple Summary
Abstract
1. Introduction
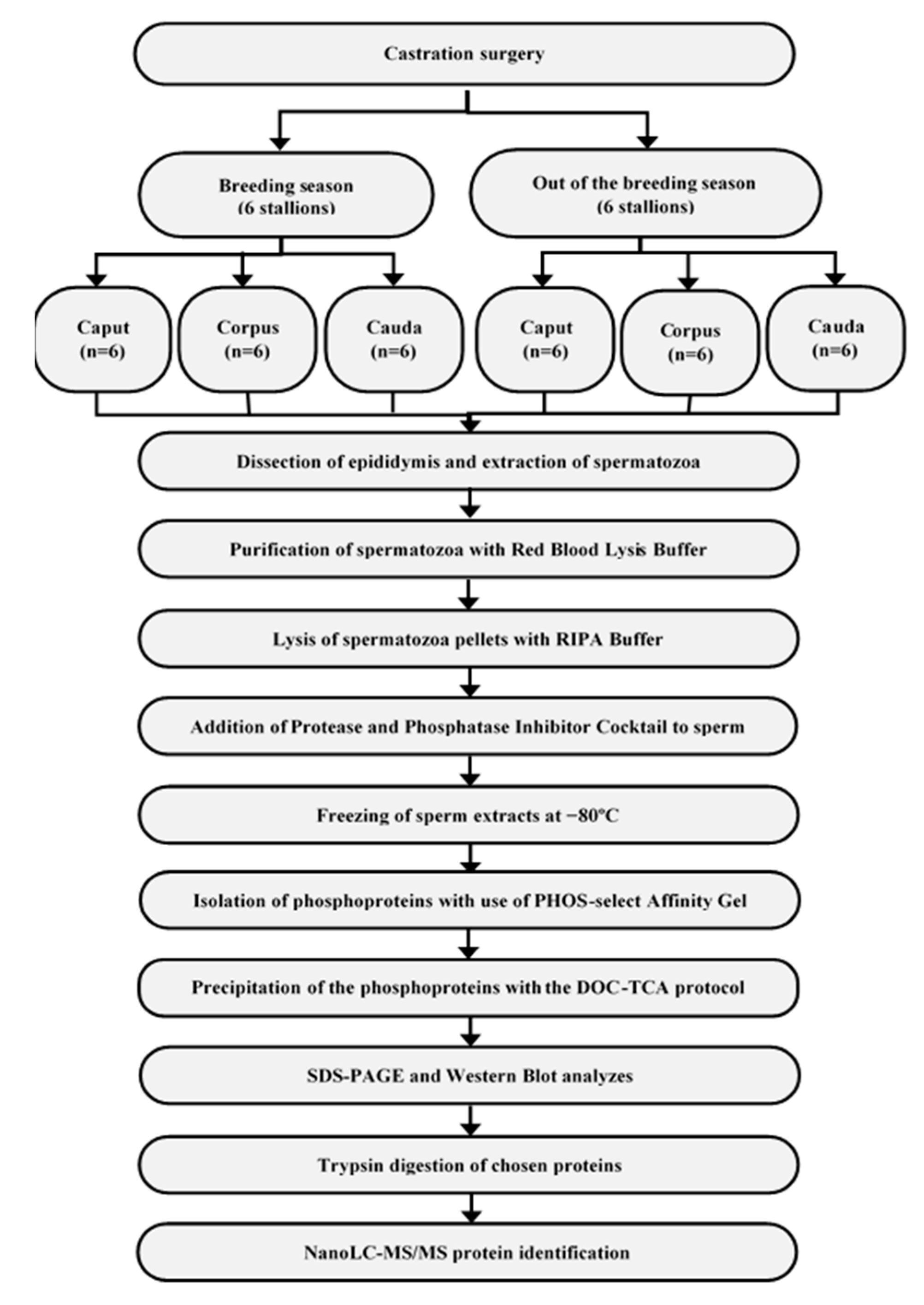
2. Materials and Methods
2.1. Material Collection
2.2. Isolation and Precipitation of Phosphoproteins
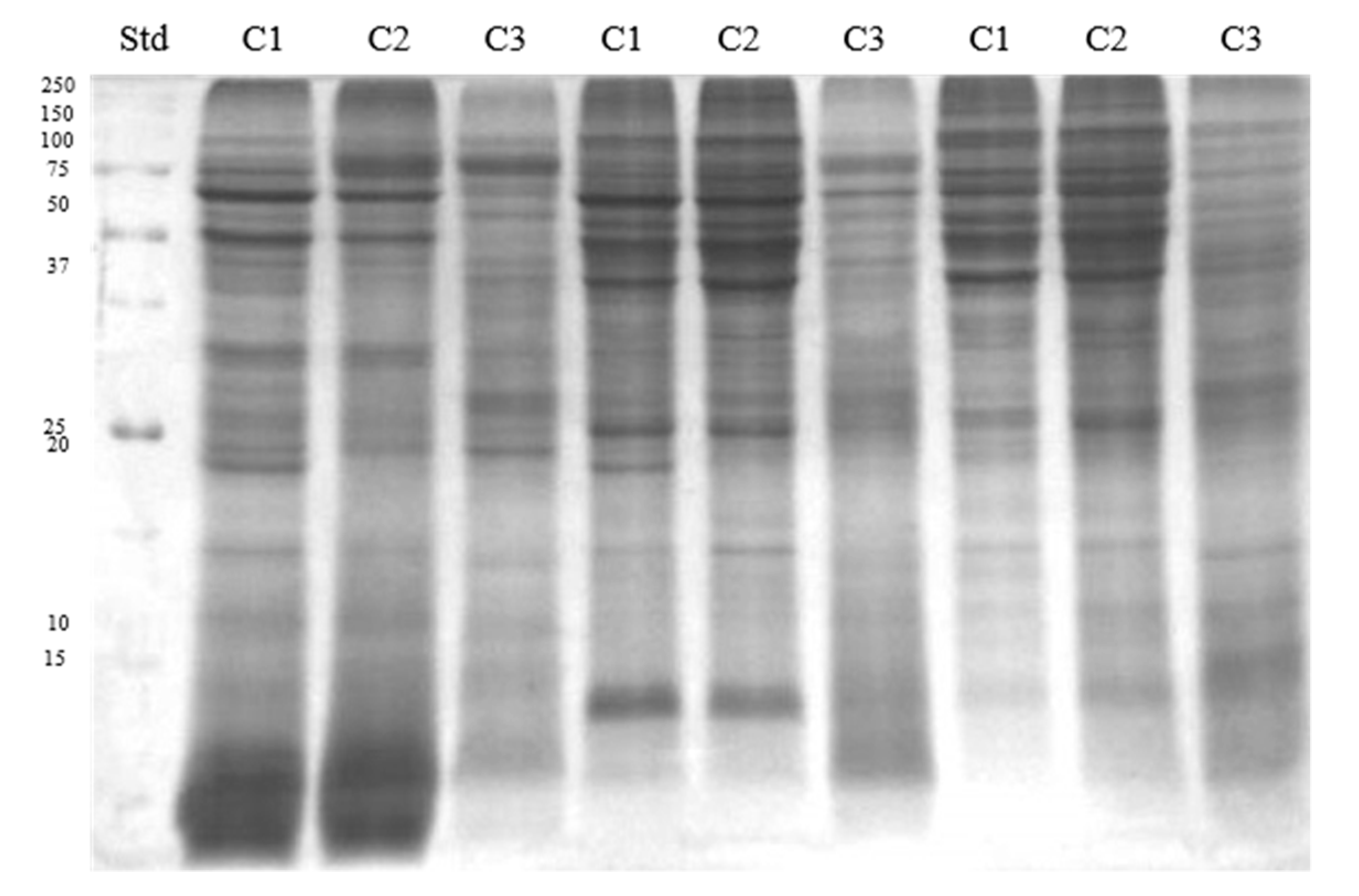
2.3. SDS-PAGE and Western Blotting
2.4. Trypsin Digestion of Chosen Proteins
2.5. NanoLC–MS/MS Protein Identification
2.6. Statistical Analysis
3. Results
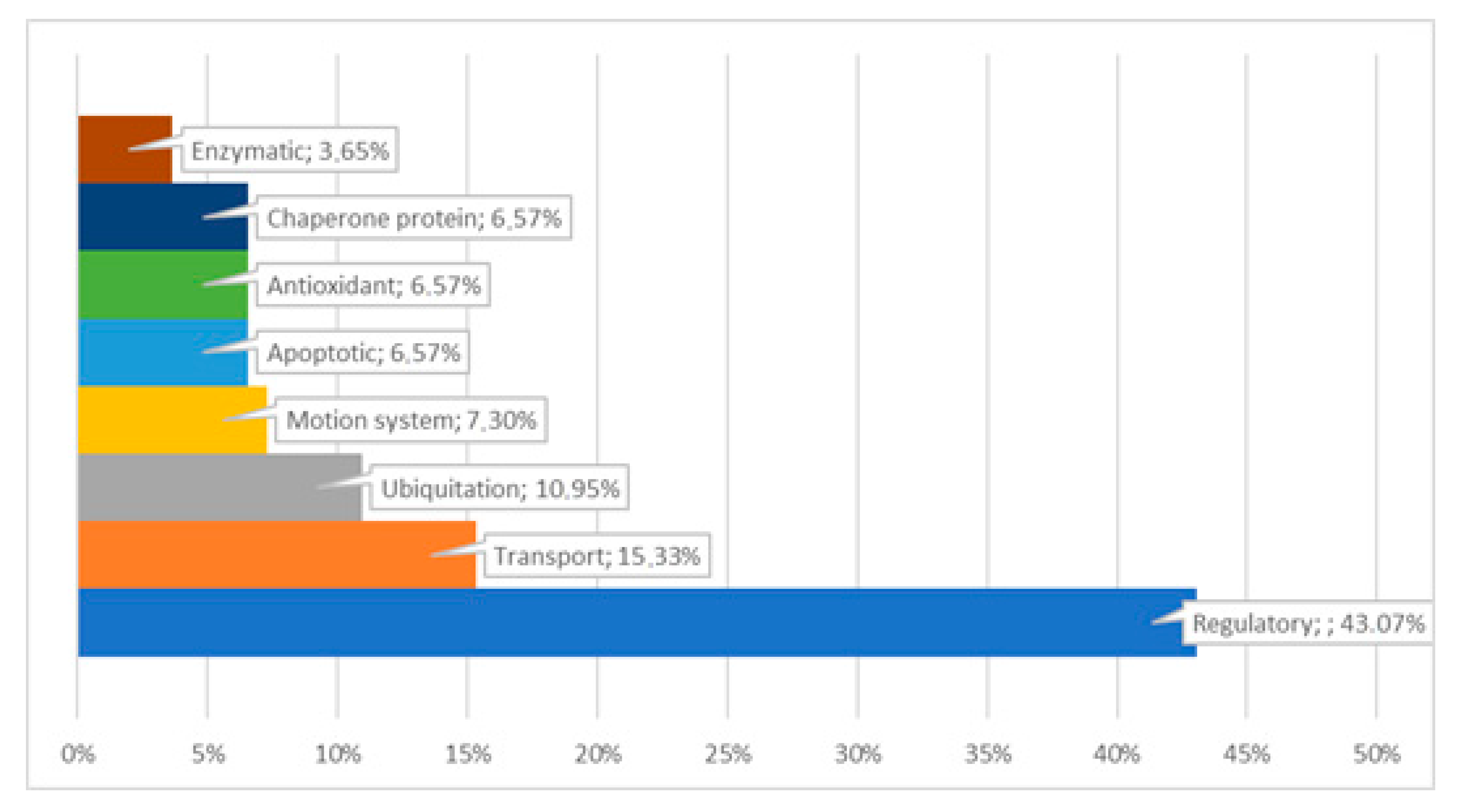
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aitken, R.J.; Nixon, B.; Lin, M.; Koppers, A.J.; Lee, Y.H.; Baker, M.A. Proteomic changes in mammalian spermatozoa during epididymal maturation. Asian J. Androl. 2007, 9, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Labas, V.; Grasseau, I.; Cahier, K.; Gargaros, A.; Harichaux, G.; Teixeira-Gomes, A.P.; Alves, S.; Bourin, M.; Gérard, N.; Blesbois, E. Data for chicken semen proteome and label free quantitative analyses displaying sperm quality biomarkers. Data Brief 2014, 1, 37–41. [Google Scholar] [CrossRef]
- Turner, R.M. Moving to the beat: A review of mammalian sperm motility regulation. Reprod. Fertil. Dev. 2006, 18, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.; Aitken, R.J. Impact of Epididymal Maturation on the Tyrosine Phosphorylation Patterns Exhibited by Rat Spermatozoa. Biol. Reprod. 2001, 64, 1545–1556. [Google Scholar] [CrossRef]
- Urner, F.; Sakkas, D. Protein phosphorylation in mammalian spermatozoa. Reproduction 2003, 125, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Martin-Hidalgo, D.; Serrano, R.; Zaragoza, C.; Garcia-Marin, L.J.; Bragado, M.J. Human sperm phosphoproteome reveals differential phosphoprotein signatures that regulate human sperm motility. J. Proteom. 2020, 215, 103654. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Haines, C.J.; Feng, H.L. Role(s) of the serine/threonine protein phosphatase 1 on mammalian sperm motility. Arch. Androl. 2007, 53, 169–177. [Google Scholar] [CrossRef]
- Jha, K.N.; Salicioni, A.M.; Arcelay, E.; Chertihin, O.; Kumari, S.; Herr, J.C.; Visconti, P.E. Evidence for the involvement of proline-directed serine/threonine phosphorylation in sperm capacitation. Mol. Hum. Reprod. 2006, 12, 781–789. [Google Scholar] [CrossRef]
- Pawson, T.; Scott, J.D. Signaling through scaffold, anchoring, and adaptor proteins. Science 1997, 278, 2075–2080. [Google Scholar] [CrossRef]
- Dou, Y.; Yao, B.; Zhang, C. PhosphoSVM: Prediction of phosphorylation sites by integrating various protein sequence attributes with a support vector machine. Amino Acids 2014, 46, 1459–1469. [Google Scholar] [CrossRef]
- Morais, D.B.; de Paula, T.A.R.; Barros, M.S.; Balarini, M.K.; de Freitas, M.B.D.; da Matta, S.L.P. Stages and duration of the seminiferous epithelium cycle in the bat Sturnira lilium. J. Anat. 2013, 222, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.M.; Irvine, C.H.G.; Evans, M.J. Seasonal changes in serum levels of FSH, LH and testosterone and in semen parameters in stallions. Theriogenology 1983, 19, 311–322. [Google Scholar] [CrossRef]
- Schön, J.; Blottner, S. Seasonal variations in the epididymis of the roe deer (Capreolus capreolus). Anim. Reprod. Sci. 2009, 111, 344–352. [Google Scholar] [CrossRef]
- Belleannee, C.; Belghazi, M.; Labas, V.; Teixeira-Gomes, A.P.; Gatti, J.L.; Dacheux, J.L.; Dacheux, F. Purification and identification of sperm surface proteins and changes during epididymal maturation. Proteomics 2011, 11, 1952–1964. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Bensadoun, A.; Weinstein, D. Assay of proteins in the presence of interfering materials. Anal. Biochem. 1976, 70, 241–250. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Tyanova, S.; Cox, J.; Olsen, J.; Mann, M.; Frishman, D. Phosphorylation variation during the cell cycle scales with structural propensities of proteins. PLoS Comput. Biol. 2013, 9, e1002842. [Google Scholar] [CrossRef]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef]
- Nishi, H.; Fong, J.H.; Chang, C.; Teichmann, S.A.; Panchenko, A.R. Regulation of protein–protein binding by coupling between phosphorylation and intrinsic disorder: Analysis of human protein complexes. Mol. BioSyst. 2013, 9, 1620–1626. [Google Scholar] [CrossRef]
- Nishi, H.; Shaytan, A.; Panchenko, A.R. Physicochemical mechanism of protein regulation by phosphorylation. Front. Genet. 2014, 270, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Buffone, M.G.; Doncel, G.F.; Marin Briggiler, C.I.; Vazquez-Levin, M.H.; Calamera, J.C. Human sperm subpopulations: Relationship between functional quality and protein tyrosine phosphorylation. Hum. Reprod. 2004, 19, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Jankovicova, J.; Michalkova, K.; Secova, P.; Horovská, L.; Manaskova-Postlerova, P.; Antalíkova, J. Evaluation of protein phosphorylation in bull sperm during their maturation in the epididymis. Cell Tissue Res. 2018, 371, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Visconti, P.E.; Kopf, G.S. Regulation of Protein Phosphorylation during Sperm Capacitation. Biol. Reprod. 1998, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kruger, M.; Linke, W.A. The Giant Protein Titin: A regulatory node that integrates myocyte signaling pathways. J. Biol. Chem. 2011, 286, 9905–9912. [Google Scholar] [CrossRef]
- Milardi, D.; Grande, G.; Vincenzoni, F.; Messana, I.; Pontecorvi, A.; De Marinis, L.; Castagnola, M.; Marana, R. Proteomic approach in the identification of fertility pattern in seminal plasma of fertile men. Fertil. Steril. 2012, 97, 67–73. [Google Scholar] [CrossRef]
- Pobre, K.F.R.; Poet, G.J.; Hendershot, L.M. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J. Biol. Chem. 2019, 294, 2098–2108. [Google Scholar] [CrossRef]
- Suozzi, K.C.; Wu, X.; Fuchs, E. Spectraplakins: Master orchestrators of cytoskeletal dynamics. J. Cell Biol. 2012, 197, 465. [Google Scholar] [CrossRef]
- Aslam, M.K.M.; Sharma, V.K.; Pandey, S.; Kumaresan, A.; Srinivasan, A.; Datta, T.K.; Mohanty, T.K.; Yadav, S. Identification of biomarker candidates for fertility in spermatozoa of crossbred bulls through comparative proteomics. Theriogenology 2018, 119, 43–51. [Google Scholar] [CrossRef]
- Nakamura, N. Ubiquitination regulates the morphogenesis and function of sperm organelles. Cells 2013, 2, 732–750. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.J.; Manandhar, G.; Sutovsky, M.; Li, R.; Jonáková, V.; Oko, R.; Park, C.S.; Prather, R.S.; Sutovsky, P. Ubiquitin C-terminal hydrolase-activity is involved in sperm acrosomal function and anti-polyspermy defense during porcine fertilization. Biol. Reprod. 2007, 77, 780–793. [Google Scholar] [CrossRef] [PubMed]
- Gob, E.; Schmitt, J.; Benavente, R.; Alsheimer, M. Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS ONE 2010, 10, e12072. [Google Scholar]
- Zhao, X.J.; Tang, R.Z.; Wang, M.L.; Guo, W.L.; Liu, J.; Li, L.; Xing, W.J. Distribution of PDIA3 transcript and protein in rat testis and sperm cells. Reprod. Domest. Anim. 2013, 48, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Turano, C.; Coppari, S.; Altieri, F.; Ferraro, A. Proteins of the PDI family: Unpredicted nonER locations and functions. J. Cell. Physiol. 2002, 193, 154–163. [Google Scholar] [CrossRef]
- Ellerman, D.A.; Myles, D.G.; Primakoff, P. A role for sperm surface protein disulfide isomerase activity in gamete fusion: Evidence for the participation of ERp57. Dev. Cell 2006, 10, 831–837. [Google Scholar] [CrossRef]
- Di Marzo, N.; Chisci, E.; Giovannoni, R. The role of hydrogen peroxide in redox-dependent signaling homeostatic and pathological response inmammalian cells. Cells 2018, 7, 156. [Google Scholar] [CrossRef]
- Ryu, D.Y.; Kim, K.U.; Kwon, W.S.; Rahman, M.S.; Khatun, A.; Pang, M.G. Peroxiredoxin activity is a major landmark of male fertility. Sci. Rep. 2017, 7, 17174. [Google Scholar] [CrossRef]
- Nagdas, S.K.; Buchanan, T.; Raychoudhury, S. Identification of peroxiredoxin-5 in bovine cauda epididymal sperm. Mol. Cell. Biochem. 2014, 387, 113–121. [Google Scholar] [CrossRef][Green Version]
- Matanis, T.; Akhmanova, A.; Wulf, P.; Del Nery, E.; Weide, T.; Stepanova, T.; Galjart, N.; Grosveld, F.; Goud, B.; De Zeeuw, C.; et al. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 2002, 4, 986–992. [Google Scholar] [CrossRef]
- Oates, E.C.; Rossor, A.M.; Hafezparast, M.; Gonzalez, M.; Speziani, F.; MacArthur, D.G.; Lek, M.; Cottenie, E.; Scoto, M.; Foley, A.R.; et al. Mutations in BICD2 cause dominant congenital spinal muscular atrophy and hereditary spastic paraplegia. Am. J. Hum. Genet. 2013, 92, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Yardimci, H.; van Duffelen, M.; Mao, Y.; Rosenfeld, S.S.; Selvin, P.R. The mitotic kinesin CENP-E is a processive transport motor. Proc. Natl. Acad. Sci. USA 2008, 105, 6016–6021. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.L.; Uvesky, V.N. Intrinsic Disorder and Posttranslational Modifications: The Darker Side of the Biological Dark Matter. Front. Genet. 2018, 9, 158. [Google Scholar] [CrossRef]
- Medini, R.; Bhagya, M.; Samson, S. Seasonal changes in the protein profile and enzyme activity of the epididymal luminal fluid in the lizard, Eutropis carinata (Schneider, 1801). Anim. Biol. 2018, 68, 387–404. [Google Scholar] [CrossRef]
- Dasheux, J.L.; Belleannée, C.; Guyonnet, B.; Labas, V.; Teixeira-Gomes, A.P.; Ecroyd, H.; Druart, X.; Gatti, J.L.; Dacheux, F. The contribution of proteomics to understanding epididymal maturation of mammalian spermatozoa. Syst. Biol. Reprod. Med. 2012, 58, 197–210. [Google Scholar]
- Cheema, R.S.; Bansal, A.; Bilaspuri, G.S.; Gandotra, V. Correlation between the proteins and protein profile(s) of different regions of epididymis and their contents in goat buck. Anim. Sci. Pap. 2011, 29, 75–84. [Google Scholar]
- Dias, G.M.; López, M.L.; Ferreira, A.T.S.; Chapeaurouge, D.A.; Rodrigues, A.; Perales, J.; Retamal, C.A. Thiol-disulfide proteins of stallion epididymal spermatozoa. Anim. Rep. Sci. 2014, 145, 29–39. [Google Scholar] [CrossRef] [PubMed]

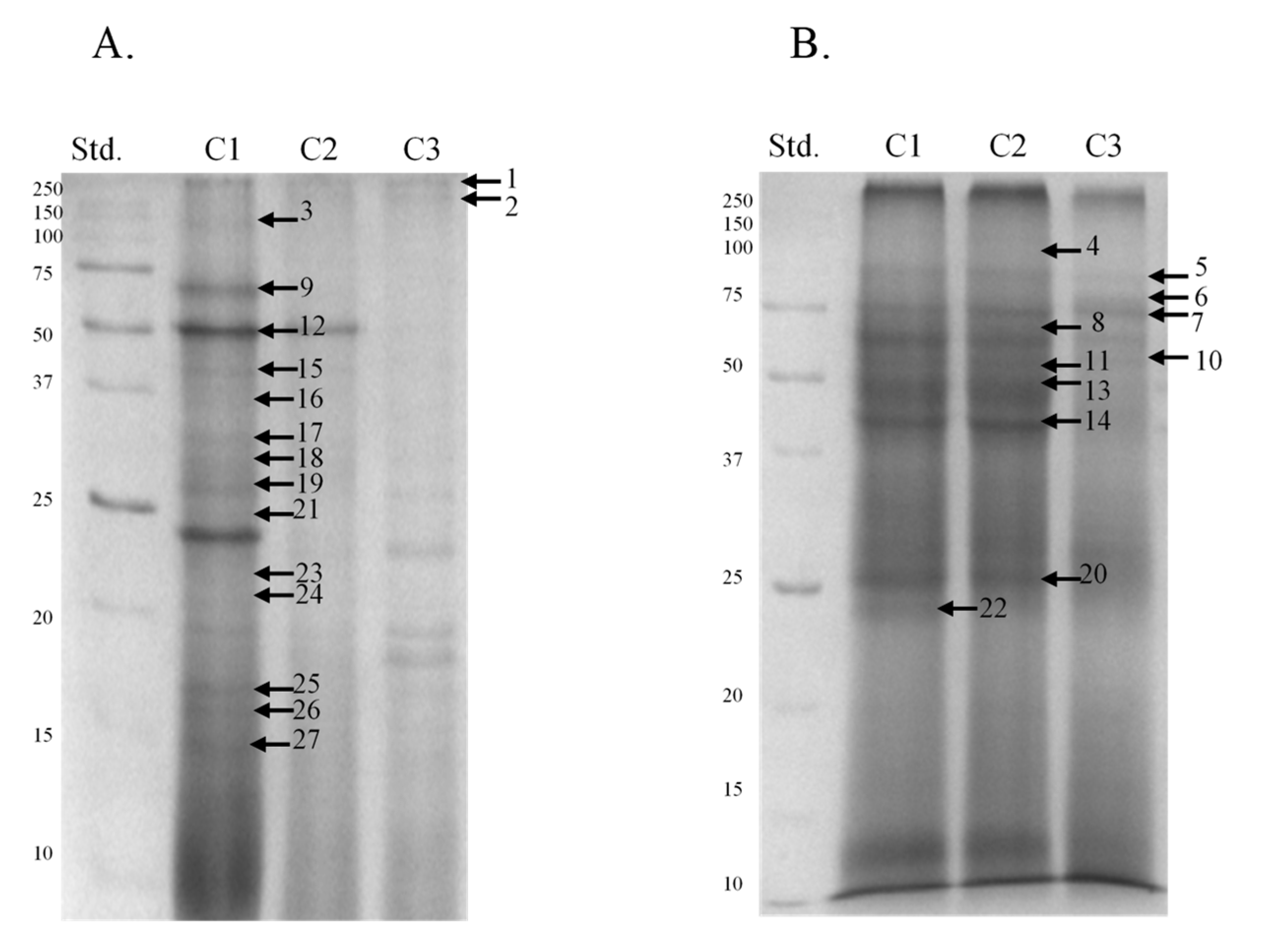
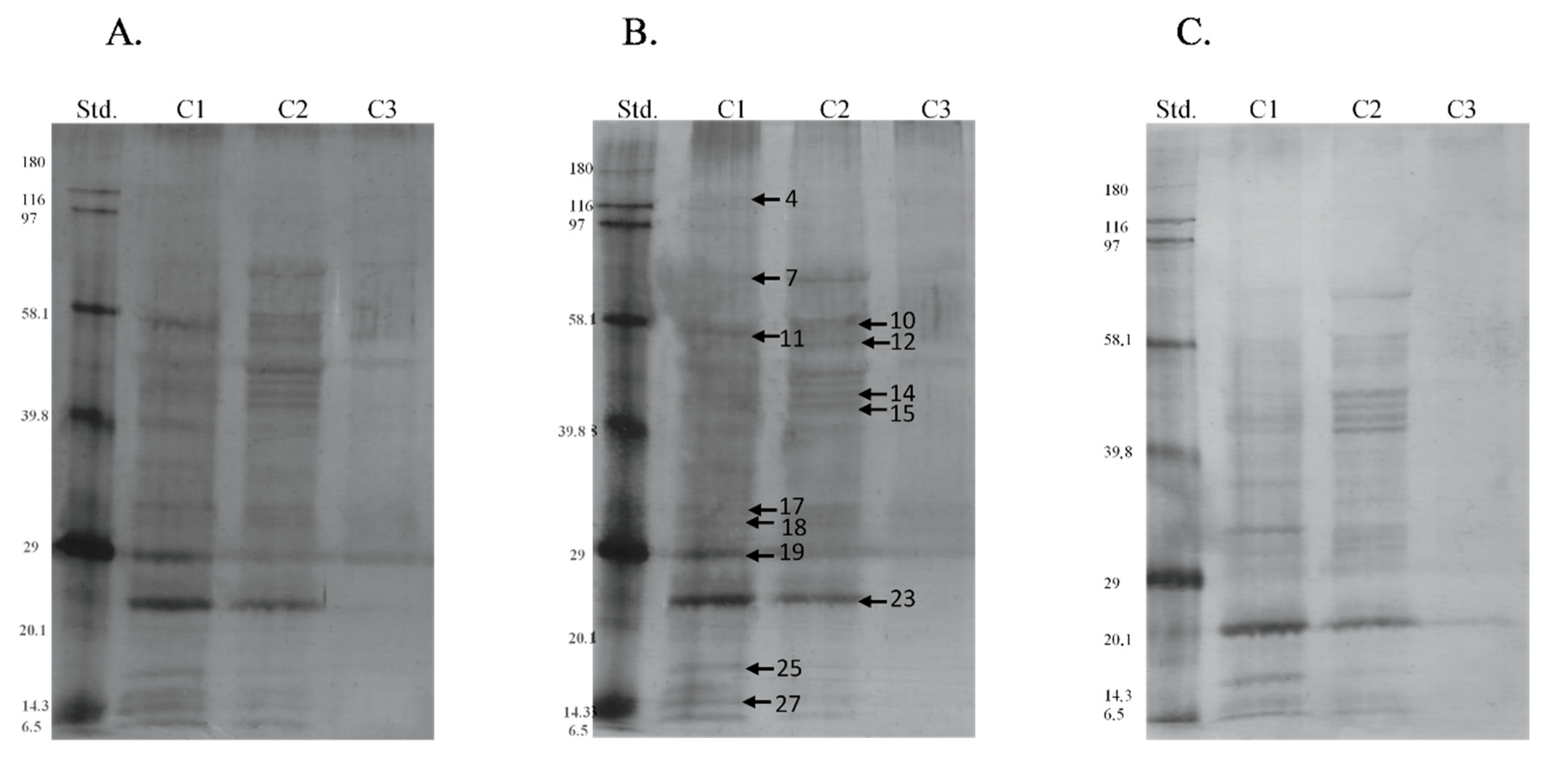
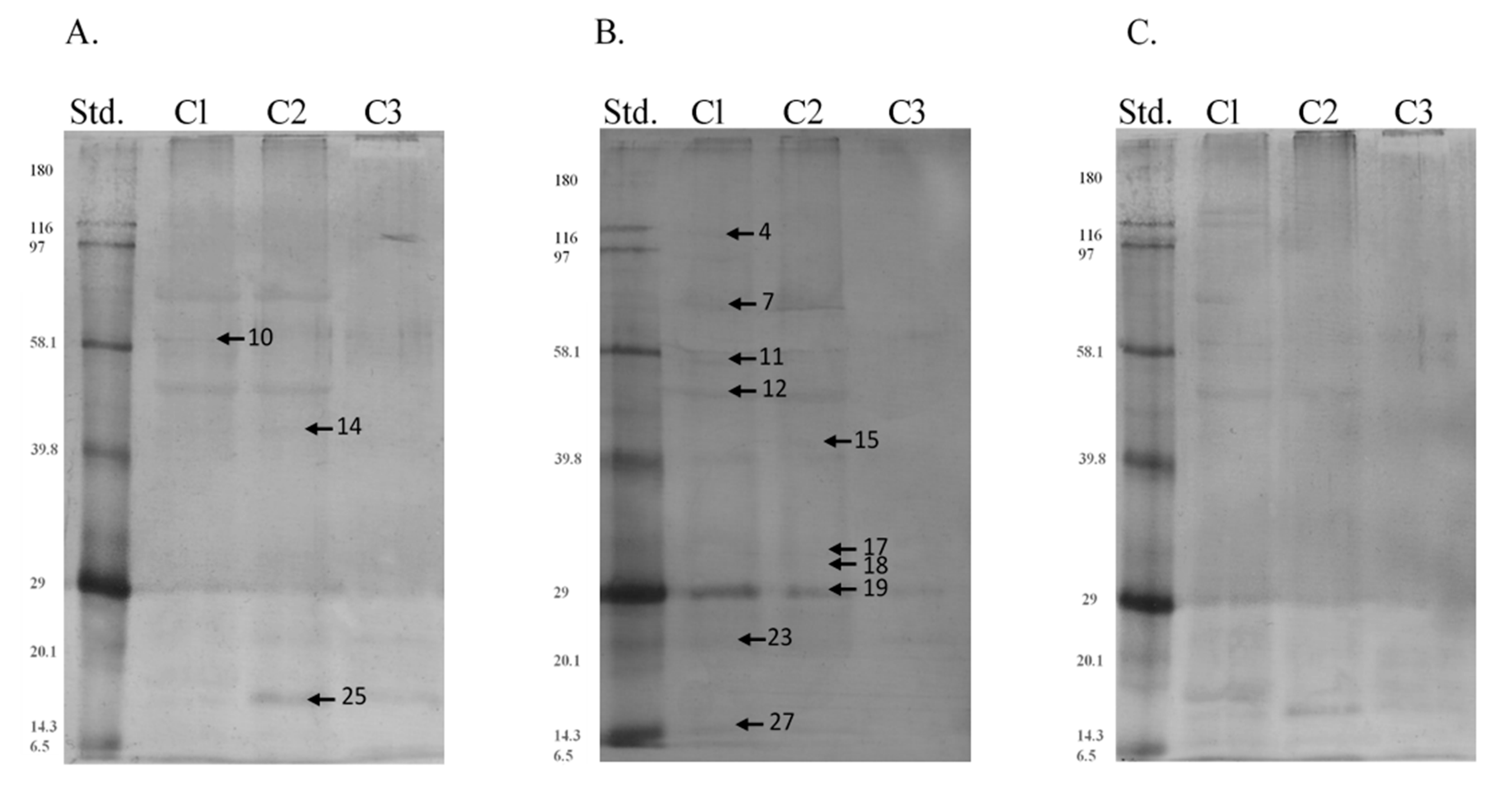
| Band | Identified Protein | M. W. (kDa) | pI | Score | Sequence Cov. % | Peptide Matches | |
|---|---|---|---|---|---|---|---|
| MultiAnalyst | Mascot | ||||||
| 1 | Elongation factor 1-alpha | 280 | 50.1 | 9.7 | 83.9 | 4.1 | 2 |
| Titin | 3904.1 | 5.8 | 63.2 | 0.2 | 6 | ||
| Phosphate carrier protein, mitochondrial | 39.4 | 10.1 | 57.0 | 3.4 | 1 | ||
| 2 | Titin | 160 | 3813.7 | 6.0 | 81.3 | 0.2 | 7 |
| 3 | Titin | 150 | 3813.7 | 6.0 | 72.4 | 0.3 | 8 |
| Endoplasmic reticulum chaperone BiP | 72.3 | 4.9 | 59.9 | 3.8 | 2 | ||
| 4 | Endoplasmin | 110 | 92.4 | 4.6 | 89.6 | 4.6 | 4 |
| Titin | 3813.7 | 6.0 | 81.4 | 0.4 | 10 | ||
| Leucine-rich repeat ser/thr-protein kinase 2 | 285.9 | 6.4 | 59.3 | 1.5 | 2 | ||
| 5 | Titin | 100 | 3904.1 | 5.8 | 57.4 | 0.2 | 6 |
| Spectrin beta chain, non-erythrocytic 1 | 274.1 | 5.3 | 56.1 | 0.8 | 2 | ||
| 6 | Endoplasmic reticulum chaperone BiP | 83 | 72.3 | 4.9 | 107.1 | 3.2 | 4 |
| Titin | 3813.7 | 6.0 | 81.4 | 0.4 | 7 | ||
| 7 | Endoplasmic reticulum chaperone BiP | 75 | 72.3 | 4.9 | 63.5 | 22.5 | 2 |
| 8 | Endoplasmic reticulum chaperone BiP | 73 | 72.3 | 4.9 | 174.0 | 11.0 | 6 |
| Serum albumin | 68.5 | 5.9 | 77.2 | 3.6 | 2 | ||
| 9 | Heat shock-related 70 kDa protein 2 | 70 | 69.6 | 5.4 | 141.3 | 9.8 | 6 |
| Heat shock 70 kDa protein 1-like | 70.3 | 6.0 | 137.8 | 1.9 | 1 | ||
| Titin | 3813.7 | 6.0 | 107.5 | 0.2 | 8 | ||
| Serum albumin | 68.5 | 5.9 | 98.6 | 6.9 | 4 | ||
| Endoplasmic reticulum chaperone BiP | 72.3 | 4.9 | 54.9 | 4.1 | 2 | ||
| Fibrous sheath-interacting protein 2 | 780.1 | 6.3 | 52.0 | 0.5 | 3 | ||
| 10 | Serum albumin | 61 | 68.6 | 5.9 | 362.0 | 13.8 | 10 |
| Endoplasmic reticulum chaperone BiP | 72.3 | 4.9 | 156.8 | 9.0 | 4 | ||
| Titin | 3813.7 | 6.0 | 69.7 | 0.3 | 7 | ||
| Centromere-associated protein E | 286.3 | 5.1 | 54.7 | 1.4 | 4 | ||
| 11 | Protein disulfide-isomerase A3 (Fragments) | 57 | 23.4 | 4.7 | 54.5 | 11.1 | 2 |
| 12 | Endoplasmic reticulum chaperone BiP | 50 | 72.3 | 4.9 | 162.6 | 7.8 | 5 |
| Protein disulfide-isomerase A3 | 56.9 | 6.2 | 136.3 | 10.9 | 5 | ||
| Titin | 3813.7 | 6.0 | 82.5 | 0.3 | 8 | ||
| Protein disulfide-isomerase | 56.9 | 4.7 | 73.7 | 8.1 | 4 | ||
| BCL-6 corepressor-like protein 1 | 190.4 | 9.0 | 64.0 | 3.4 | 5 | ||
| 13 | Serum albumin | 48 | 68.5 | 5.9 | 69.4 | 4.0 | 2 |
| Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 | 837.8 | 5.3 | 57.1 | 0.6 | 4 | ||
| 14 | Endoplasmic reticulum chaperone BiP | 44 | 72.3 | 4.9 | 163.1 | 12.8 | 7 |
| Titin | 3813.7 | 6.0 | 85.5 | 0.2 | 8 | ||
| Protein disulfide-isomerase | 56.9 | 4.7 | 60.9 | 3.3 | 2 | ||
| 15 | Endoplasmic reticulum chaperone BiP | 41 | 72.3 | 4.9 | 234.6 | 11.3 | 5 |
| Titin | 3813.7 | 6.0 | 81.7 | 0.2 | 7 | ||
| Protein disulfide-isomerase A6 | 48.1 | 4.9 | 80.5 | 3.4 | 1 | ||
| Protein TALPID3 | 169.2 | 5.4 | 51.0 | 1.8 | 2 | ||
| E3 ubiquitin-protein ligase HECTD1 | 289.2 | 5.2 | 50.8 | 1.8 | 4 | ||
| 16 | Endoplasmic reticulum chaperone BiP | 35 | 72.3 | 4.9 | 109.6 | 5.0 | 3 |
| Protein disulfide-isomerase A6 | 48.1 | 4.9 | 60.5 | 3.4 | 1 | ||
| Titin | 3813.7 | 6.0 | 54.7 | 0.2 | 5 | ||
| 17 | Endoplasmic reticulum chaperone BiP | 32 | 28.9 | 4.6 | 77.4 | 7.3 | 2 |
| 18 | Endoplasmic reticulum chaperone BiP | 31 | 72.3 | 4.9 | 158.0 | 8.6 | 4 |
| Dystonin | 860.1 | 5.1 | 75.3 | 0.5 | 3 | ||
| 19 | Endoplasmic reticulum chaperone BiP | 29 | 72.3 | 4.9 | 112.4 | 6.0 | 4 |
| E3 ubiquitin-protein ligase LRSAM1 | 83.9 | 5.8 | 50.3 | 5.4 | 3 | ||
| 20 | Serum albumin | 26 | 68.5 | 5.9 | 86.9 | 4.0 | 2 |
| Titin | 3813.7 | 6.0 | 78.0 | 0.2 | 7 | ||
| Heat shock protein beta-1 | 22.4 | 6.0 | 50.4 | 5.0 | 1 | ||
| 21 | Heat shock protein beta-1 | 25 | 22.4 | 6.0 | 89.6 | 8.5 | 2 |
| 22 | Titin | 24 | 3813.7 | 6.0 | 95.0 | 0.3 | 9 |
| Heat shock protein beta-1 | 22.4 | 6.0 | 55.3 | 5.0 | 1 | ||
| Nesprin-1 | 1010.5 | 5.4 | 51.1 | 0.4 | 4 | ||
| 23 | Nesprin-1 | 23 | 1010.5 | 5.4 | 50.6 | 0.4 | 4 |
| 24 | Titin | 22 | 3813.7 | 6.0 | 54.6 | 0.2 | 6 |
| Dystonin | 860.1 | 5.1 | 51.6 | 0.7 | 3 | ||
| 25 | Titin | 17 | 3813.7 | 6.0 | 73.3 | 0.2 | 6 |
| Peroxiredoxin-5, mitochondrial | 22.2 | 10.2 | 50.1 | 10.7 | 2 | ||
| 26 | Nesprin-1 | 15 | 1010.5 | 5.4 | 54.9 | 0.7 | 5 |
| Protein bicaudal D homolog 2 | 93.5 | 5.2 | 50.2 | 3.6 | 3 | ||
| 27 | Canalicular multispecific organic anion transporter 1 | 12 | 175.4 | 9.5 | 50.2 | 1.9 | 3 |
| Band | Protein | Season | Type of P-Residues | Median | SD |
|---|---|---|---|---|---|
| 7 | 75 kDa Endoplasmic reticulum chaperone BiP | s | thr | 6.515 a | 5.751 |
| os | 0.000 b | 3.670 | |||
| 12 | 50 kDa Protein disulfide-isomerase A3 | s | ser | 7.125 a | 5.509 |
| os | 3.105 b | 3.224 | |||
| 17 | 32 kDa Endoplasmic reticulum chaperone BiP | s | tyr | 2.985 a | 2.605 |
| os | 0.000 b | 2.008 | |||
| s | thr | 3.220 a | 2.431 | ||
| os | 0.000 b | 2.202 | |||
| 23 | 23 kDa Nesprin-1 | s | ser | 17.555 a | 15.126 |
| os | 2.910 b | 5.688 | |||
| s | tyr | 21.055 a | 14.799 | ||
| os | 8.080 b | 12.271 | |||
| 25 | 17 Peroxiredoxin-5, mitochondrial | s | thr | 7.860 a | 7.746 |
| os | 0.000 b | 6.504 | |||
| 26 | 15 Protein bicaudal D homolog 2 | s | thr | 4.175 a | 3.882 |
| os | 0.000 b | 2.699 |
| Band | Protein | Region | Type of P-Residues | Median | SD |
|---|---|---|---|---|---|
| 10 | 61 kDa Endoplasmic reticulum chaperone BiP, albumin | c1 | tyr | 1.755 a | 2.689 |
| c2 | 1.600 a | 4.884 | |||
| c3 | 9.345 b | 5.459 | |||
| 12 | 50 kDa Protein disulfide-isomerase A3 | c1 | thr | 11.700 a | 10.589 |
| c2 | 11.330 a | 10.278 | |||
| c3 | 0.000 b | 7.530 | |||
| 27 | 15 kDa Protein bicaudal D homolog 2 | c1 | ser | 4.375 a | 4.345 |
| c2 | 3.430 a | 5.780 | |||
| c3 | 0.000 b | 2.546 | |||
| c1 | tyr | 8.430 a | 5.903 | ||
| c2 | 3.215 b | 7.186 | |||
| c3 | 0.000 c | 6.327 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dyrda, K.; Orzołek, A.; Ner-Kluza, J.; Wysocki, P. Influence of the Season and Region Factor on Phosphoproteome of Stallion Epididymal Sperm. Animals 2021, 11, 3487. https://doi.org/10.3390/ani11123487
Dyrda K, Orzołek A, Ner-Kluza J, Wysocki P. Influence of the Season and Region Factor on Phosphoproteome of Stallion Epididymal Sperm. Animals. 2021; 11(12):3487. https://doi.org/10.3390/ani11123487
Chicago/Turabian StyleDyrda, Katarzyna, Aleksandra Orzołek, Joanna Ner-Kluza, and Paweł Wysocki. 2021. "Influence of the Season and Region Factor on Phosphoproteome of Stallion Epididymal Sperm" Animals 11, no. 12: 3487. https://doi.org/10.3390/ani11123487
APA StyleDyrda, K., Orzołek, A., Ner-Kluza, J., & Wysocki, P. (2021). Influence of the Season and Region Factor on Phosphoproteome of Stallion Epididymal Sperm. Animals, 11(12), 3487. https://doi.org/10.3390/ani11123487






