Molecular and Histological Evaluation of Sheep Ovarian Tissue Subjected to Lyophilization
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Sample Collection
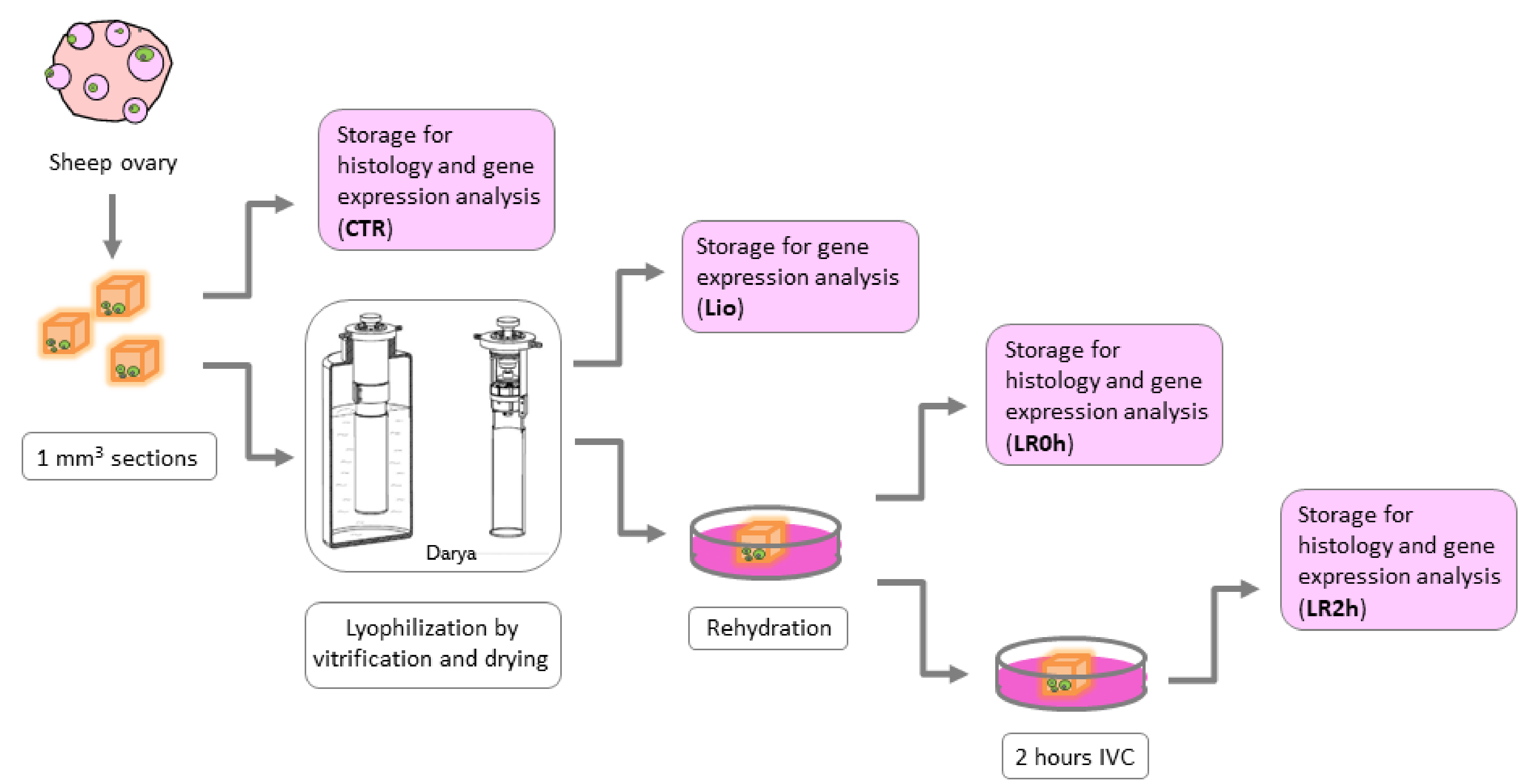
2.3. Experimental Design
- CTR (n = 8): Sections of fresh ovarian tissue stored for further analysis immediately after dissection.
- Lio (n = 8): Sections of ovarian tissue stored for further analysis immediately after lyophilization by vitrification procedure (without warming procedure).
- LR0h (n = 8): Sections of ovarian tissue stored for further analysis after lyophilization and rehydration procedures.
- LR2h (n = 8): Sections of ovarian tissue stored for further analysis after lyophilization and rehydration procedures followed by 2 h of in vitro culture.
2.4. Tissue Vitrification and Drying
2.5. Rehydration
2.6. Gene Expression Analysis
2.7. Total RNA Isolation and Reverse Transcription
2.8. Real Time Polymerase Chain Reaction
2.9. Histology
2.10. Statistical Analysis
3. Results
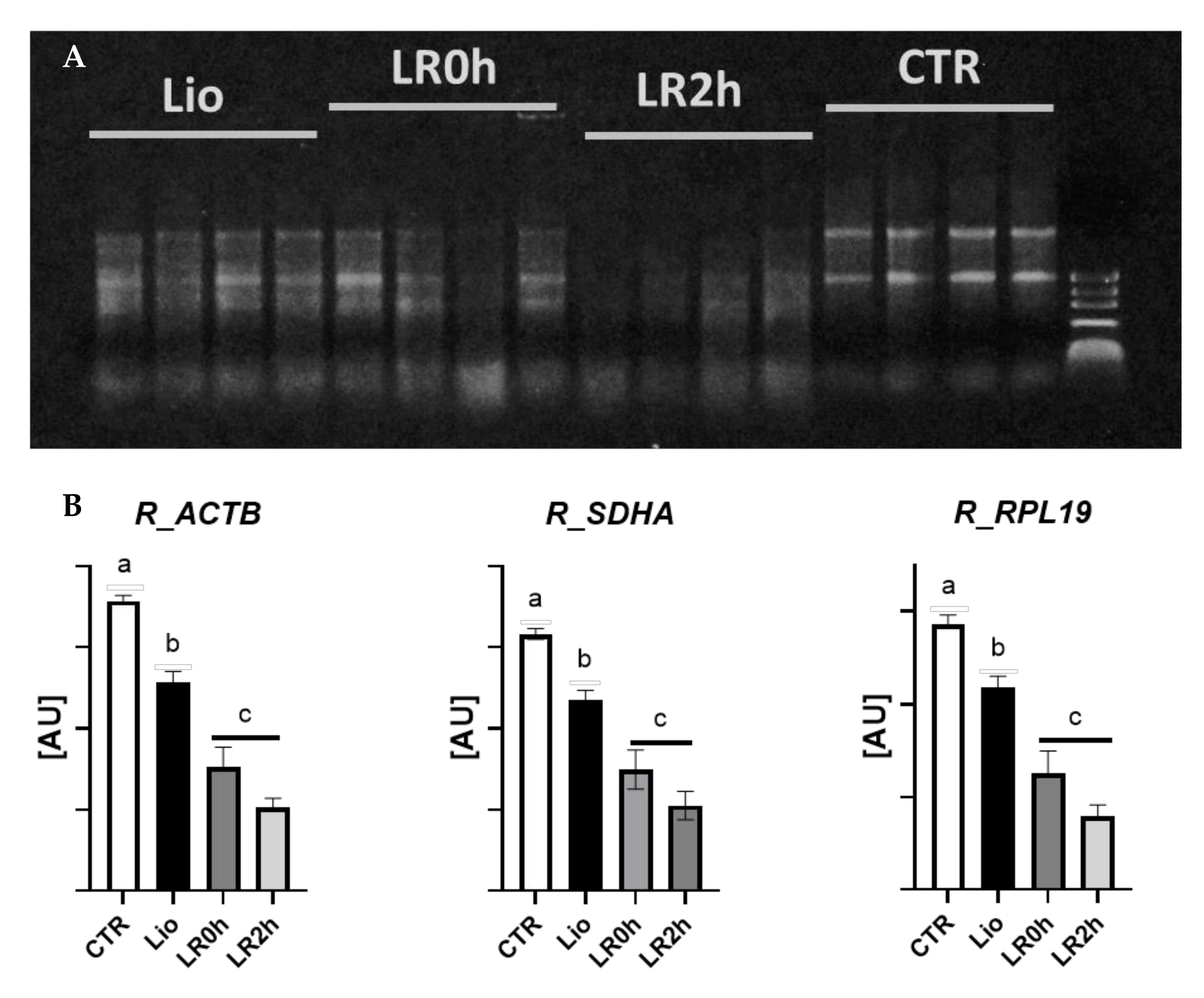
3.1. RNA Integrity
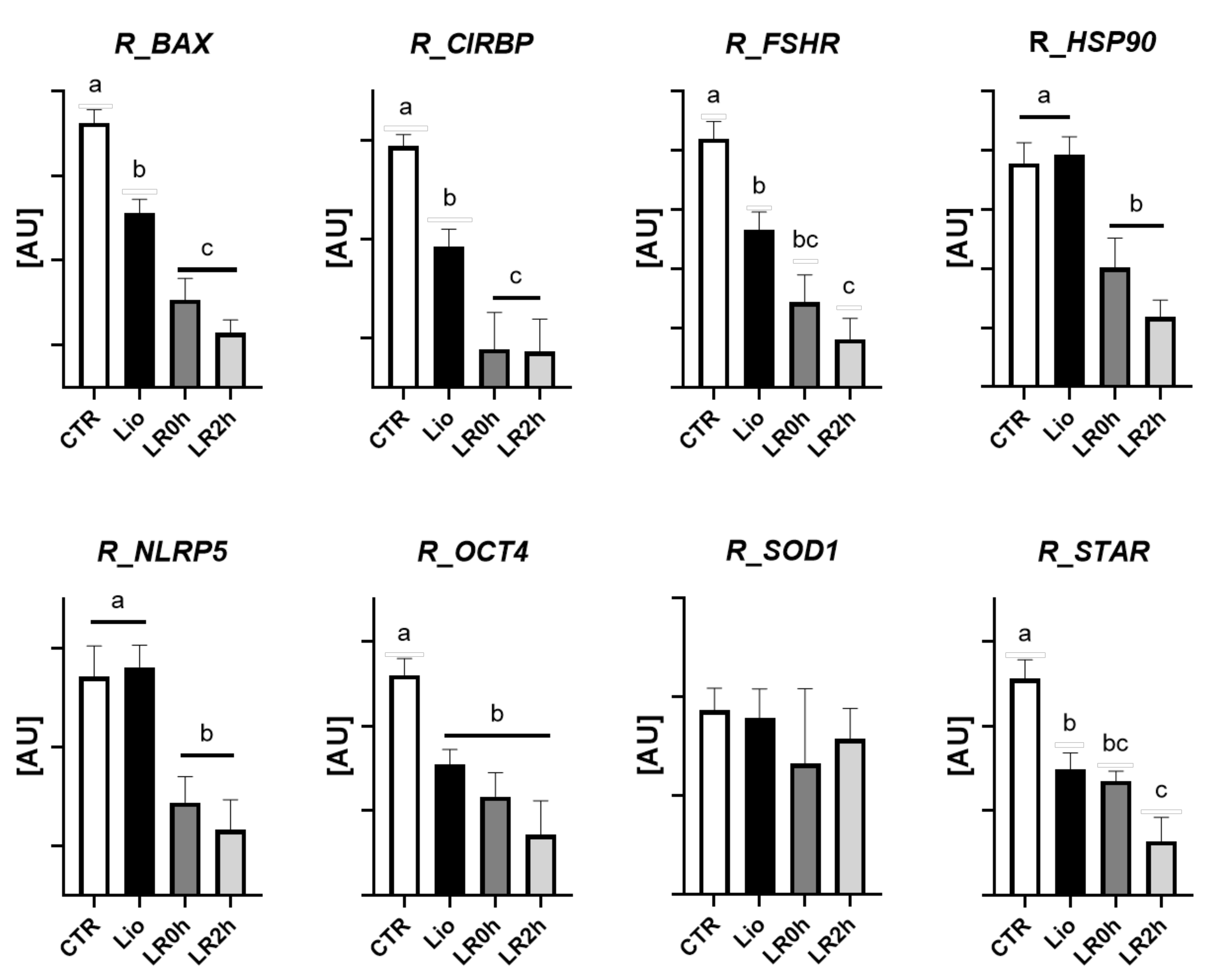
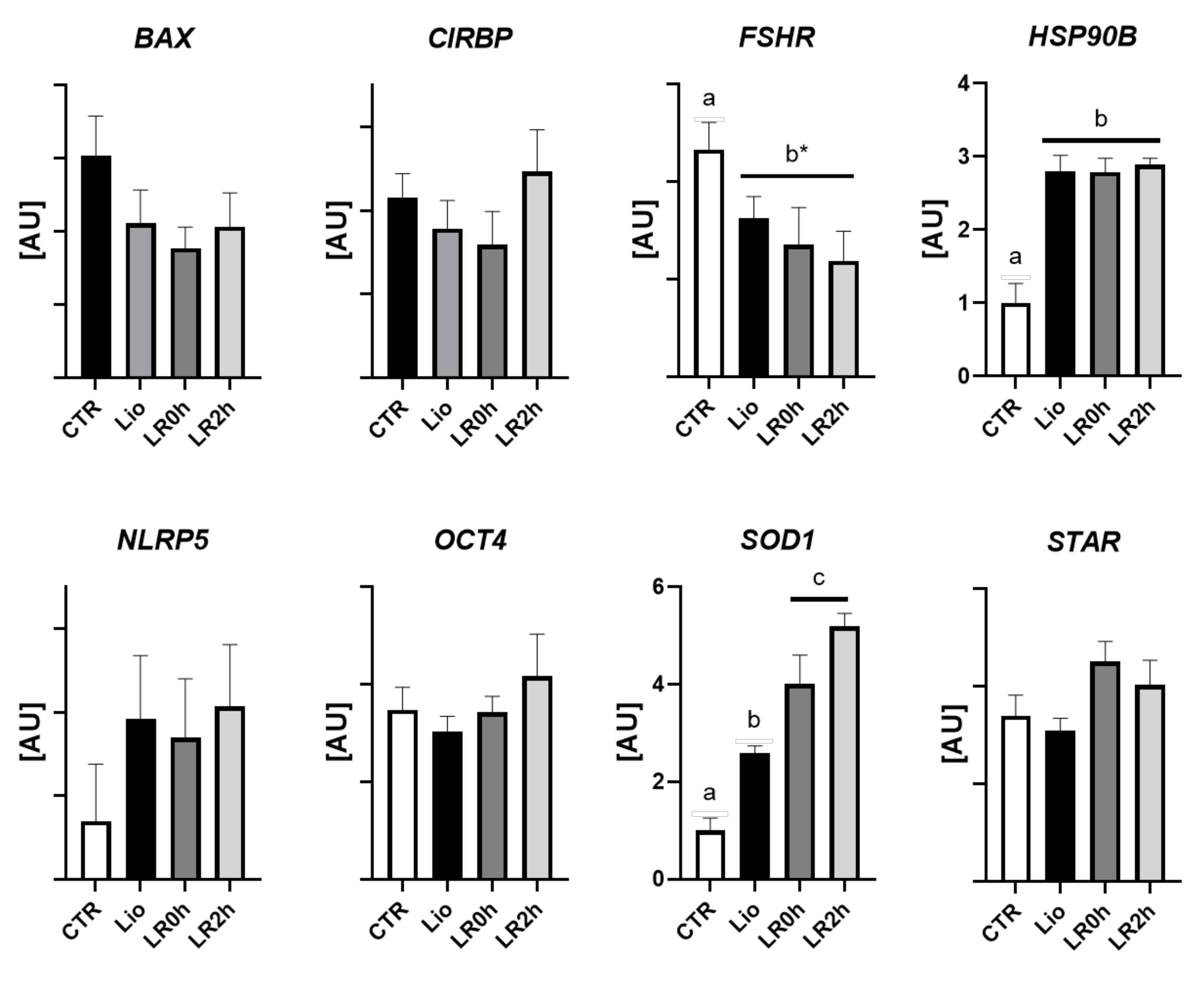
3.2. Gene Expression
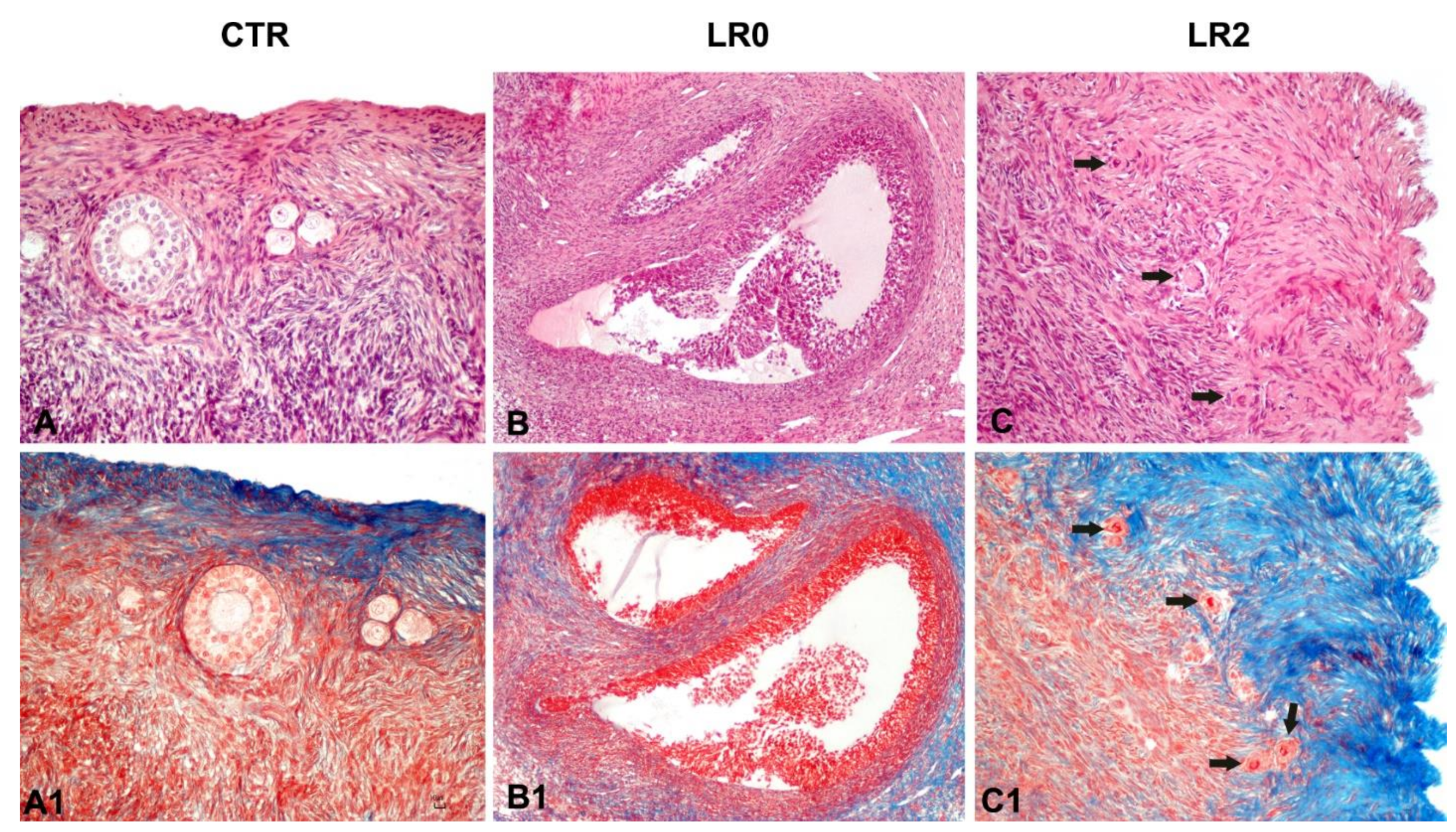
3.3. Histology
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wolkers, W. Freeze-dried cells and tissues. Cryobiology 2013, 67, 398–442. [Google Scholar] [CrossRef]
- Jang, T.H.; Park, S.C.; Yang, J.H.; Kim, J.Y.; Seok, J.H.; Park, U.S.; Choi, C.W.; Lee, S.R.; Han, J. Cryopreservation and its clinical applications. Integr. Med. Res. 2017, 6, 12–18. [Google Scholar] [CrossRef]
- Morgan, C.; Herman, N.; White, P.; Vesey, G. Preservation of micro-organisms by drying: A review. J. Microbiol. Methods 2006, 66, 183–193. [Google Scholar] [CrossRef]
- Arav, A. 082 VitDrying of reproductive cells using the ‘‘Minimum drop size (MDS)” technique. Cryobiology 2013, 67, 398–442. [Google Scholar] [CrossRef]
- Loi, P.; Matsukawa, K.; Ptak, G.; Clinton, M.; Fulka, J., Jr.; Nathan, Y.; Arav, A. Freeze-dried somatic cells direct embryonic development after nuclear transfer. PLoS ONE 2008, 3, e2978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Oldenhof, H.; Sydykov, B.; Bigalk, J.; Sieme, H.; Wolkers, W.F. Freeze-drying of mammalian cells using trehalose: Preservation of DNA integrity. Sci. Rep. 2017, 7, 6198. [Google Scholar] [CrossRef] [PubMed]
- Iuso, D.; Czernik, M.; Di Egidio, F.; Sampino, S.; Zacchini, F.; Bochenek, M.; Smorag, Z.; Modlinski, J.A.; Ptak, G.; Loi, P. Genomic stability of lyophilized sheep somatic cells before and after nuclear transfer. PLoS ONE 2013, 8, e51317. [Google Scholar] [CrossRef] [PubMed]
- Leboeuf, C.; Ratajczak, P.; Zhao, W.L.; Plassa, L.F.; Court, M.; Pisonero, H.; Murata, H.; Cayuela, J.M.; Ameisen, J.C.; Garin, J.; et al. Long-term preservation at room temperature of freeze-dried human tumor samples dedicated to nucleic acids analyses. Cell Preserv. Technol. 2008, 6, 191–198. [Google Scholar] [CrossRef]
- Wakayama, T.; Yanagimachi, R. Development of normal mice from oocytes injected with freeze-dried spermatozoa. Nat. Biotechnol. 1998, 16, 639–641. [Google Scholar] [CrossRef]
- Ward, M.A.; Kaneko, T.; Kusakabe, H.; Biggers, J.D.; Whittingham, D.G.; Yanagimachi, R. Long-term preservation of mouse spermatozoa after freeze-drying and freezing without cryoprotection. Biol. Reprod. 2003, 69, 2100–2108. [Google Scholar] [CrossRef][Green Version]
- Kusakabe, H.; Szczygiel, M.A.; Whittingham, D.G.; Yanagimachi, R. Maintenance of genetic integrity in frozen and freeze-dried mouse spermatozoa. Proc. Natl. Acad. Sci. USA 2001, 98, 13501–13506. [Google Scholar] [CrossRef] [PubMed]
- Gianaroli, L.; Magli, M.C.; Stanghellini, I.; Crippa, A.; Crivello, A.M.; Pescatori, E.S.; Ferraretti, A.P. DNA integrity is maintained after freeze-drying of human spermatozoa. Fertil. Steril. 2012, 97, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Ozgyin, L.; Horvath, A.; Balint, B.L. Lyophilized human cells stored at room temperature preserve multiple RNA species at excellent quality for RNA sequencing. Oncotarget 2018, 9, 31312. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stebbings, R.; Wang, L.; Sutherland, J.; Kammel, M.; Gaigalas, A.K.; John, M.; Roemer, B.; Kuhne, M.; Schneider, R.J.; Braun, M.; et al. Quantification of cells with specific phenotypes I: Determination of CD4+ cell count per microliter in reconstituted lyophilized human PBMC prelabeled with anti-CD4 FITC antibody. Cytom. Part A 2015, 87, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Stebbings, R.; Gaigalas, A.; Sutherland, J.; Kammel, M.; John, M.; Roemer, B.; Kuhne, M.; Schneider, R.J.; Braun, M.; et al. Quantification of cells with specific phenotypes II: Determination of CD4 expression level on reconstituted lyophilized human PBMC labelled with anti-CD4 FITC antibody. Cytom. Part A 2015, 87, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Keskintepe, L.; Pacholczyk, G.; Machinicka, A.; Norris, K.; Curuk, M.A.; Khan, L.; Brackett, B.G. Bovine blastocyst development from oocytes injected with freeze-dried spermatozoa. Biol. Reprod. 2002, 67, 409–415. [Google Scholar] [CrossRef]
- Liu, J.L.; Kusakabe, H.; Chang, C.C.; Suzuki, H.; Schmidt, D.W.; Julian, M.; Pfeffer, R.; Bormann, C.L.; Tian, X.C.; Yanagimachi, R.; et al. Freeze-dried sperm fertilization leads to full-term development in rabbits. Biol. Reprod. 2004, 70, 1776–1781. [Google Scholar] [CrossRef]
- Hirabayashi, M.; Kato, M.; Ito, J.; Hochi, S. Viable offspring derived from oocytes intracytoplasmically injected with freeze-dried sperm heads. Zygote 2005, 13, 79–85. [Google Scholar] [CrossRef]
- Moisan, A.E.; Leibo, S.P.; Lynn, J.W.; Gomez, M.C.; Pope, C.E.; Dresser, B.L.; Godke, R.A. Embryonic development of felid oocytes injected with freeze-dried or airdried spermatozoa. Cryobiology 2005, 51, 373. [Google Scholar]
- Martins, C.F.; Bao, S.N.; Dodea, M.N.; Correa, G.A.; Rumpf, R. Effects of freeze-drying on cytology; ultrastructure; DNA fragmentation; and fertilizing ability of bovine sperm. Theriogenology 2007, 67, 1307–1315. [Google Scholar] [CrossRef][Green Version]
- Hochi, S.; Watanabe, K.; Kato, M.; Hirabayashi, M. Live rats resulting from injection of oocytes with spermatozoa freeze—Dried and stored for one year. Mol. Reprod. Dev. 2008, 70, 1776–1781. [Google Scholar] [CrossRef]
- Kusakabe, H.; Yanagimachi, R.; Kamiguchi, Y. Mouse and human spermatozoa can be freeze-dried without damaging their chromosomes. Hum. Reprod. 2008, 23, 233–239. [Google Scholar] [CrossRef]
- Watanabe, H.; Asano, T.; Abe, Y.; Fukui, Y.; Suzuki, H. Pronuclear formation of freeze-dried canine spermatozoa microinjected into mouse oocytes. J. Assist. Reprod. Genet. 2009, 26, 531–536. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choi, Y.H.; Varner, D.; Love, C.C.; Hartman, D.L.; Hinrichs, K. Production of live foals via intracytoplasmic injection of lyophilized sperm and sperm extract in the horse. Reproduction 2011, 142, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Abdalla, H.; Morita, H.; Kuwayana, M.; Hirabayasi, M.; Hochi, S. Procedure for bovine ICSI; not sperm freeze-drying; impairs the function of the microtubule-organizing center. J. Reprod. Dev. 2011, 57, 428–432. [Google Scholar] [CrossRef]
- Men, N.T.; Kikuchi, K.; Nakai, N.; Fukuda, A.; Tanihara, F.; Noguchi, J.; Kaneko, H.; Linh, N.V.; Nguyen, B.S.; Nagai, T.; et al. Effect of trehalose on DNA integrity of freeze-dried boar sperm; fertilization; and embryo development after intracytoplasmic sperm injection. Theriogenology 2013, 80, 1033–1044. [Google Scholar] [CrossRef]
- Campos, A.G.; Gil, L.; Malo, C.; Martnınez, F.; de Blas, I. Effect of different mono-disaccharides on the freeze-dried boar spermatozoa: A preliminary study. CryoLetters 2014, 35, 277–285. [Google Scholar]
- Kaneko, T. Preservation of mammalian sperm by freeze-drying. Cryobiology 2014, 69, 510. [Google Scholar] [CrossRef]
- Olaciregui, M.; Luño, V.; Gonzalez, N.; de Blas, I.; Gil, L. Freeze-dried dog sperm: Dynamics of DNA integrity. Cryobiology 2015, 71, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Polge, C.; Smith, A.; Parkes, A. Revival of spermatozoa after vitrification and dehydration at low temperatures. Nature 1949, 164, 666. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.C.; Yen, J.M.; Kuo, T.C.; Gong, K.B.; Hsu, K.H.; Hsu, T.T. Comparison of the developmental potential of 2-week-old preantral follicles derived from vitrified ovarian tissue slices; vitrified whole ovaries and vitrified/transplanted newborn mouse ovaries using the metal surface method. BMC Biotechnol. 2008, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.A.; Kim, H.Y.; Kim, J.W.; Lee, G.; Lee, E.; Ahn, J.Y.; Park, J.H.; Lim, J.M. Effectiveness of slow freezing and vitrification for long-term preservation of mouse ovarian tissue. Theriogenology 2011, 75, 1045–1051. [Google Scholar] [CrossRef]
- Mouttham, L.; Comizzoli, P. The preservation of vital functions in cat ovarian tissues during vitrification depends more on the temperature of the cryoprotectant exposure than on the sucrose supplementation. Cryobiology 2016, 73, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Tanpradit, N.; Chatdarong, K.; Comizzoli, P. Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) pre-exposure ensures follicle integrity during In Vitro culture of ovarian tissue but not during cryopreservation in the domestic cat model. J. Assist. Reprod. Genet. 2016, 33, 1621–1631. [Google Scholar] [CrossRef]
- Oskam, I.C.; Asadi, B.A.; Santos, R.R. Histologic and ultrastructural features of cryopreserved ovine ovarian tissue: Deleterious effect of 1;2-propanediol applying different thawing protocols. Fertil. Steril. 2010, 93, 2764–2766. [Google Scholar] [CrossRef]
- Gandolfi, F.; Paffoni, A.; Brambilla, E.P.; Bonetti, S.; Brevini, T.A.; Ragni, G. Efficiency of equilibrium cooling and vitrification procedures for the cryopreservation of ovarian tissue: Comparative analysis between human and animal models. Fertil. Steril. 2006, 85, 1150–1156. [Google Scholar] [CrossRef]
- Mouttham, L.; Fortune, J.E.; Comizzoli, P. Damage to fetal bovine ovarian tissue caused by cryoprotectant exposure and vitrification is mitigated during tissue culture. J. Assist. Reprod. Genet. 2015, 32, 1239–1250. [Google Scholar] [CrossRef]
- Kardak, A.; Leibo, S.P.; Devireddy, R. Membrane transport properties of equine and macaque ovarian tissues frozen in mixtures of dimethylsulfoxide and ethylene glycol. J. Biomech. Eng. 2007, 129, 688–694. [Google Scholar] [CrossRef] [PubMed]
- ASRM Practice Committee. Fertility preservation in patients undergoing gonadotoxic therapy or gonadectomy: A committee opinion. Fertil. Steril. 2019, 112, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.C.; Adams, D.M.; Amelkina, O.; White, K.K.; Amoretti, L.A.; Whitaker, M.G.; Comizzoli, P. Influence of microwave-assisted dehydration on morphological integrity and viability of cat ovarian tissues: First steps toward long-term preservation of complex biomaterials at supra-zero temperatures. PLoS ONE 2019, 14, e0225440. [Google Scholar] [CrossRef]
- Arav, A.; Idda, A.; Nieddu, S.M.; Natan, Y.; Ledda, S. High post-thaw survival of ram sperm after partial freeze-drying. J. Assist. Reprod. Genet. 2018, 35, 1149–1155. [Google Scholar] [CrossRef]
- Bebbere, D.; Pinna, S.; Nieddu, S.; Natan, D.; Arav, A.; Ledda, S. Gene expression analysis of ovine prepubertal testicular tissue vitrified with a novel cryodevice (E.Vit). J. Assist. Reprod. Genet. 2019, 36, 214646-2154. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The ultimate qPCR experiment: Producing publication quality; reproducible data the first time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Vorachek, W.R.; Hugejiletu; Bobe, G.; Hall, J.A. Reference gene selection for quantitative PCR studies in sheep neutrophils. Int. J. Mol. Sci. 2013, 14, 11484–11495. [Google Scholar] [CrossRef]
- Röhn, G.; Koch, A.; Krischek, B.; Stavrinou, P.; Goldbrunner, R.; Timmer, M. ACTB and SDHA Are Suitable Endogenous Reference Genes for Gene Expression Studies in Human Astrocytomas Using Quantitative RT-PCR. Technol. Cancer Res. Treat. 2018, 17, 1533033818802318. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Tagiri, M.; Hwang, I.S.; Takahashi, M.; Hirabayasi, M. Adverse effect of cake collapse on the functional integrity of freeze-dried bull spermatozoa. Cryobiology 2014, 68, 354–360. [Google Scholar] [CrossRef]
- Watson, P.F.; Duncan, A.E. Effect of salt concentration and unfrozen water fraction on the viability of slowly frozen ram spermatozoa. Cryobiology 1988, 25, 131–142. [Google Scholar] [CrossRef]
- Schöler, H.R.; Ruppert, S.; Suzuki, N.; Chowdhury, K.; Gruss, P. New type of POU domain in germ line-specific protein Oct-4. Nature 1990, 344, 435–439. [Google Scholar] [CrossRef]
- Tong, Z.B.; Gold, L.; Pfeifer, K.E.; Dorward, H.; Lee, E.; Bondy, C.A.; Dean, J.; Nelson, L.M. Mater, a maternal effect gene required for early embryonic development in mice. Nat. Genet. 2000, 26, 267–268. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.H.; Yuan, H.Y.; Cao, C.Y.; Gao, Z.P.; Zhu, B.Y.; Huang, H.L.; Liao, D.F. Heat shock protein 90 acts as a molecular chaperone in late-phase activation of extracellular signal-regulated kinase 1/2 stimulated by oxidative stress in vascular smooth muscle cells. Acta Pharmacol. Sin. 2007, 28, 1907–1913. [Google Scholar] [CrossRef]
- Wang, X.H.; Kang, L. Differences in egg thermotolerance between tropical and temperate populations of the migratory locust Locusta migratoria (Orthoptera: Acridiidae). J. Insect Physiol. 2005, 51, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Haase, M.; Fitze, G. HSP90AB1: Helping the good and the bad. Gene 2016, 575, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.S.; Narasimhan, P.; Kim, G.S.; Jung, J.E.; Park, E.H.; Chan, P.H. The role of Akt signaling in oxidative stress mediates NF-kappaB activation inmild transient focal cerebral ischemia. J. Cereb. Blood Flow Metab. 2008, 28, 1917–1926. [Google Scholar] [CrossRef] [PubMed]
| Gene | Symbol | Sequence | Accession Number | Annealing Temp. | Size (bps) |
|---|---|---|---|---|---|
| Actin β | ACTB | F: 5′ ttcctgggtatggatcctg 3′ R: 5′ ggtgatctccttctgcatcc 3′ | NM_001009784 | 60 °C | 162 |
| BCL2 associated X Protein | BAX | F: 5′ ctccccgagaggtctttttc R: 5′ tcgaaggaagtccaatgtcc | XM_004015363 | 58 °C | 176 |
| Cold inducible RNA binding protein | CIRBP | F: 5′ gagggctgagttttgacacc 3′ R: 5′ atgggaagtctgtggatggg | XM_004008776 | 60 °C | 190 |
| Follicle stimulating hormone receptor | FSHR | F: 5′ agtcttcctctgccaggaca 3′ R: 5′ cttctgggatgactcgaagc 3′ | NM_001009289 | 60 °C | 107 |
| Heat shock protein 90 b | HSP90B | F: 5′ tggagatcaaccctgacca R: 5′ gggatcctcaagcgagaag | XM_004018854 | 58 °C | 143 |
| NLR family pyrin domain containing 5 | NLRP5 | F: 5′ cagcctccaggagttctttg 3′ R:5′ gacagcctaggagggtttcc 3′ | XM_027978862 | 59 °C | 212 |
| POU domain, class 5, transcription factor 1 | POU5F1 (OCT4) | F: 5′ gaggagtcccaggacatcaa 3′ R:5′ ccgcagcttacacatgttct 3′ | XM_012101009 | 56 °C | 204 |
| Ribosomal protein L9 | RLP19 | F: 5′caactcccgccagcagat 3′ R:5′ ccgggaatggacagtcaca 3′ | XM_004012836 | 56 °C | 127 |
| Steroidogenic acute regulatory protein | STAR | 5′ cccatggagaggctttatga 3′ 5′ cagccaactcgtgagtgatg 3′ | NM_001009243 | 60 °C | 130 |
| Succinate Dehydrogenase | SDHA | F: 5′catccactacatgacggagca 3′ R:5′ atcttgccatcttcagttctgcta 3′ | XM_012125144 | 60 °C | 90 |
| Superoxide dismutase 1 | SOD1 | F: 5′ ggcaatgtgaaggctgacaa 3′ R:5′ aagaccagatgacttgggca 3′ | NM_001145185 | 58 °C | 130 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bebbere, D.; Arav, A.; Nieddu, S.M.; Burrai, G.P.; Succu, S.; Patrizio, P.; Ledda, S. Molecular and Histological Evaluation of Sheep Ovarian Tissue Subjected to Lyophilization. Animals 2021, 11, 3407. https://doi.org/10.3390/ani11123407
Bebbere D, Arav A, Nieddu SM, Burrai GP, Succu S, Patrizio P, Ledda S. Molecular and Histological Evaluation of Sheep Ovarian Tissue Subjected to Lyophilization. Animals. 2021; 11(12):3407. https://doi.org/10.3390/ani11123407
Chicago/Turabian StyleBebbere, Daniela, Amir Arav, Stefano Mario Nieddu, Giovanni Pietro Burrai, Sara Succu, Pasquale Patrizio, and Sergio Ledda. 2021. "Molecular and Histological Evaluation of Sheep Ovarian Tissue Subjected to Lyophilization" Animals 11, no. 12: 3407. https://doi.org/10.3390/ani11123407
APA StyleBebbere, D., Arav, A., Nieddu, S. M., Burrai, G. P., Succu, S., Patrizio, P., & Ledda, S. (2021). Molecular and Histological Evaluation of Sheep Ovarian Tissue Subjected to Lyophilization. Animals, 11(12), 3407. https://doi.org/10.3390/ani11123407






